Leaf Trichome Distribution Pattern in Arabidopsis Reveals Gene Expression Variation Associated with Environmental Adaptation
Abstract
1. Introduction
2. Results
2.1. Computational Simulation of the Effect of Variation in Gene Expression on Trichome Patterning
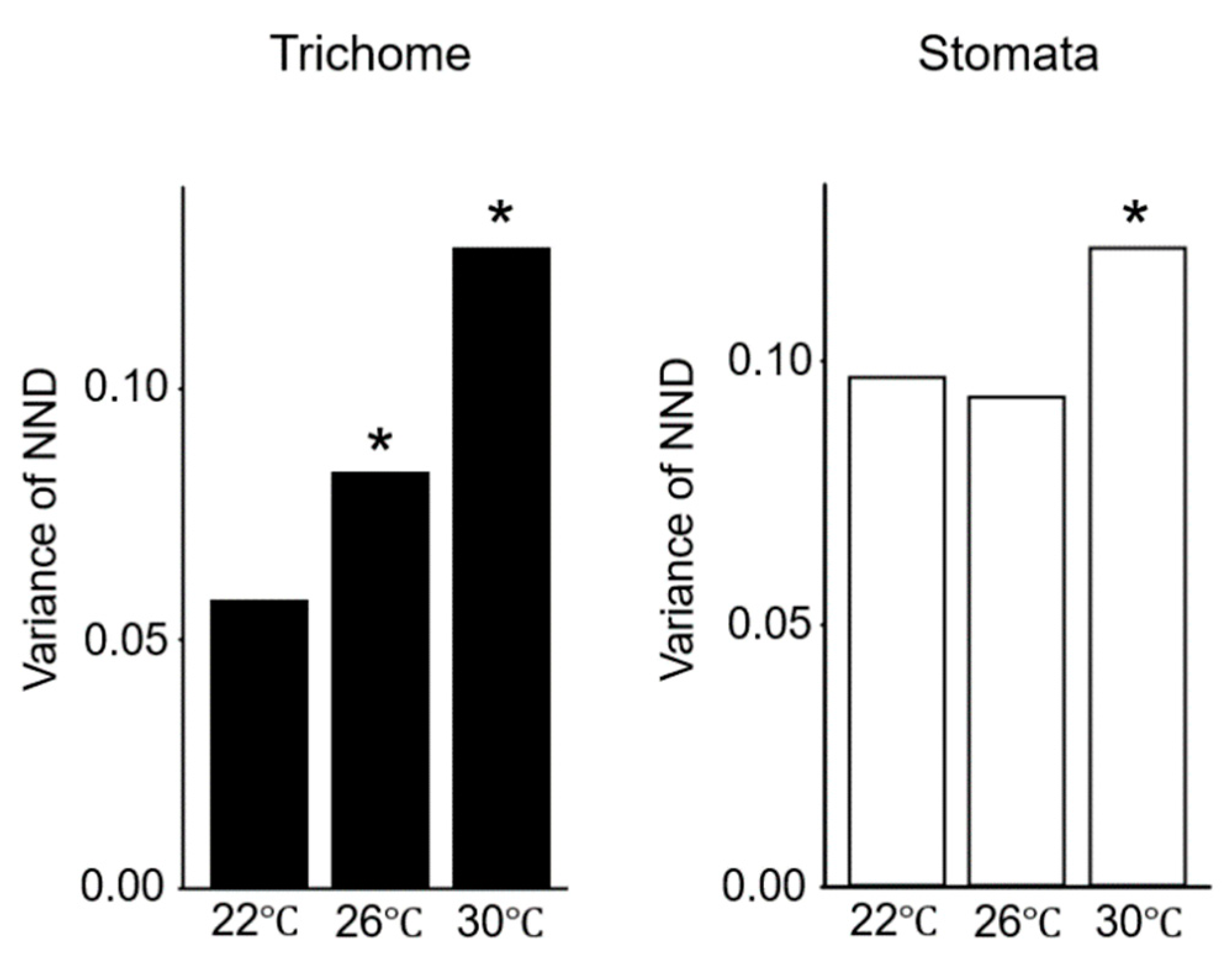
2.2. Gene Expression Variations Between Cells Increase at Elevated Temperatures
2.3. Trichome Distribution Patterns Are Variable in Accessions and Climate
2.4. Trichome Distribution Patterns Are Altered by Histone-Modifying Agents
2.5. Variations in the Trichome Distribution Patterns and H2A.Z
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Coefficient of Variation (CV) of Gene Expression Variations
4.3. Quantification of the Trichome Distribution Pattern
4.4. Estimation of the Range of Biologically Appropriate Stochastic Variations
4.5. Mathematical Modeling and Simulations
4.6. Comparison of Climate Data
4.7. MNase Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elowitz, M.B.; Levine, A.J.; Siggia, E.D.; Swain, P.S. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar] [CrossRef]
- Chalancon, G.; Ravarani, C.N.J.; Balaji, S.; Martinez-Arias, A.; Aravind, L.; Babu, M.M. Interplay between gene expressionnoise and regulatory networkarchitecture. Trends Genet. 2012, 28, 221–232. [Google Scholar] [CrossRef]
- Eldar, A.; Elowitz, M.B. Functional roles for noise in genetic circuits. Nature 2010, 467, 167–173. [Google Scholar] [CrossRef]
- Shalek, A.K.; Satija, R.; Adiconis, X.; Gertner, R.S.; Gaublomme, J.T.; Raychowdhury, R.; Schwartz, S.; Yosef, N.; Malboeuf, C.; Lu, D.; et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [Google Scholar] [CrossRef]
- Guo, G.; Pinello, L.; Han, X.; Lai, S.; Shen, L.; Lin, T.-W.; Zou, K.; Yuan, G.-C.; Orkin, S.H. Serum-based culture conditions provoke gene expression variability in mouse embryonic stem cells as revealed by single-cell analysis. Cell Rep. 2016, 14, 956–965. [Google Scholar] [CrossRef]
- Meyer, H.M.; Teles, J.; Formosa-Jordan, P.; Refahi, Y.; San-Bento, R.; Ingram, G.; Jönsson, H.; Locke, J.C.W.; Roeder, A.H.K. Fluctuations of the transcription factor ATML1 generate the pattern of giant cells in the Arabidopsis sepal. eLife 2017, 6, e19131. [Google Scholar] [CrossRef]
- Besnard, F.; Refahi, Y.; Morin, V.; Marteaux, B.; Brunoud, G.; Chambrier, P.; Rozier, F.; Mirabet, V.; Legrand, J.; Lainé, S.; et al. Cytokinin signalling inhibitory fields provide robustness to phyllotaxis. Nature 2014, 505, 417–421. [Google Scholar] [CrossRef]
- Meyer, H.M.; Roeder, A.H.K. Stochasticity in plant cellular growth and patterning. Front. Plant Sci. 2014, 5, 420. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Gomez, J.M.; Corwin, J.A.; Joseph, B.; Maloof, J.N.; Kliebenstein, D.J. Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genet 2011, 7, e1002295-17. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.O.; Roeder, A.H. Themes and variations in cell type patterning in the plant epidermis. Curr. Opin. Genet. Dev. 2015, 32, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Araújo, I.S.; Pietsch, J.M.; Keizer, E.M.; Greese, B.; Balkunde, R.; Fleck, C.; Hülskamp, M. Stochastic gene expression in Arabidopsis thaliana. Nat. Commun. 2017, 8, 2132. [Google Scholar] [CrossRef] [PubMed]
- Hülskamp, M. Plant trichomes: A model for cell differentiation. Nature 2004, 5, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Digiuni, S.; Schellmann, S.; Geier, F.; Greese, B.; Pesch, M.; Wester, K.; Dartan, B.; Mach, V.; Srinivas, B.P.; Timmer, J.; et al. A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 2008, 4, 217. [Google Scholar] [CrossRef] [PubMed]
- Morohashi, K.; Grotewold, E. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 2009, 5, e1000396. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Kurata, T.; Okada, K.; Wada, T. A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 2008, 59, 365–386. [Google Scholar] [CrossRef]
- Pattanaik, S.; Patra, B.; Singh, S.K.; Yuan, L. An overview of the gene regulatory network controlling trichome development in the model plant, Arabidopsis. Front. Plant Sci. 2014, 5, 259. [Google Scholar] [CrossRef]
- Kondo, S.; Miura, T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 2010, 329, 1616–1620. [Google Scholar] [CrossRef]
- Torii, K.U. Two-dimensional spatial patterning in developmental systems. Trends Cell Biol. 2012, 22, 438–446. [Google Scholar] [CrossRef]
- Greese, B.; Hülskamp, M.; Fleck, C. Quantification of variability in trichome patterns. Front. Plant Sci. 2014, 5, 596. [Google Scholar] [CrossRef]
- Treutlein, B.; Brownfield, D.G.; Wu, A.R.; Neff, N.F.; Mantalas, G.L.; Espinoza, F.H.; Desai, T.J.; Krasnow, M.A.; Quake, S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 2014, 509, 371–375. [Google Scholar] [CrossRef]
- Pomeranz, M.; Campbell, J.; Siegal-Gaskins, D.; Engelmeier, J.; Wilson, T.; Fernandez, V.; Brkljacic, J. High-resolution computational imaging of leaf hair patterning using polarized light microscopy. Plant J. 2013, 73, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Simmons, A.R.; Bergmann, D.C. Transcriptional control of cell fate in the stomatal lineage. Curr. Opin. Plant Biol. 2016, 29, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Staff, L.; Hurd, P.; Reale, L.; Seoighe, C.; Rockwood, A.; Gehring, C. The hidden geometries of the Arabidopsis thaliana epidermis. PLoS ONE 2012, 7, e43546. [Google Scholar] [CrossRef] [PubMed]
- Majda, M.; Grones, P.; Sintorn, I.-M.; Vain, T.; Milani, P.; Krupinski, P.; Zagórska-Marek, B.; Viotti, C.; Jönsson, H.; Mellerowicz, E.J.; et al. Mechanochemical polarization of contiguous cell walls shapes plant pavement cells. Dev. Cell. 2017, 43, 290–304.e4. [Google Scholar] [CrossRef] [PubMed]
- Ringli, C.; Bigler, L.; Kuhn, B.M.; Leiber, R.-M.; Diet, A.; Santelia, D.; Frey, B.; Pollmann, S.; Klein, M. The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 2008, 20, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Kaiserli, E.; Perrella, G.; Davidson, M.L. Light and temperature shape nuclear architecture and gene expression. Curr. Opin. Plant Biol. 2018, 45, 103–111. [Google Scholar] [CrossRef]
- Morohashi, K.; Zhao, M.; Yang, M.; Read, B.; Lloyd, A.; Lamb, R. Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 2007, 145, 736–746. [Google Scholar] [CrossRef]
- Schnittger, A.; Folkers, U.; Schwab, B.; Jürgens, G.; Hülskamp, M. Generation of a spacing pattern: The role of triptychon in trichome patterning in Arabidopsis. Plant Cell 1999, 11, 1105–1116. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Korte, A.; Cooper, M.D.; Nordborg, M.; Schmitt, J.; Wilczek, A.M. A map of local adaptation in Arabidopsis thaliana. Science 2011, 334, 86–89. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Huang, S.-S.C.; Jupe, F.; Sasaki, E.; Schmitz, R.J.; Urich, M.A.; Castanon, R.; Nery, J.R.; Barragan, C.; He, Y.; et al. Epigenomic diversity in a global collection of arabidopsis thaliana accessions. Cell 2016, 166, 492–505. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- Worldclim. Available online: http://www.worldclim.org/current (accessed on 17 July 2020).
- Dey, S.S.; Foley, J.E.; Limsirichai, P.; Schaffer, D.V.; Arkin, A.P. Orthogonal control of expression mean and variance by epigenetic features at different genomic loci. Mol. Syst. Biol. 2015, 11, 806. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Pikaard, C.S. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997, 11, 2124–2136. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A.Z-containing nucleosomes mediate the thermosensory response in arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef]
- Quint, M.; Delker, C.; Franklin, K.A.; Wigge, P.A.; Halliday, K.J.; van Zanten, M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2016, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kärkkäinen, K.; Løe, G.; Agren, J. Population structure in Arabidopsis lyrata: Evidence for divergent selection on trichome production. Evolution 2004, 58, 2831–2836. [Google Scholar] [CrossRef] [PubMed]
- Rédei, G.P. Methods in Arabidopsis Research; Koncz, C., Chua, N.-H., Schell, J., Eds.; World Scientific: Singapore, 1992; pp. 1–15. [Google Scholar]
- Dai, X.; Bai, Y.; Zhao, L.; Dou, X.; Liu, Y.; Wang, L.; Li, Y.; Li, W.; Hui, Y.; Huang, X.; et al. H2A.Z represses gene expression by modulating promoter nucleosome structure and enhancer histone modifications in arabidopsis. Mol. Plant 2017, 10, 1274–1292. [Google Scholar] [CrossRef] [PubMed]
- Armengot, L.; Moreno-Romero, J. Micrococcal Nuclease (MNase) Assay of Arabidopsis thaliana nuclei. Bio Protoc. 2013, 3, e455. [Google Scholar] [CrossRef]
- Franklin, K.A. Plant chromatin feels the heat. Cell 2010, 140, 26–28. [Google Scholar] [CrossRef][Green Version]









| Parameter | Function | Value |
|---|---|---|
| σ1 | Basal GL1 transcription rate | 8.2707 |
| α1 | AC-regulated GL1 transcription rate | 3.4869 |
| ρ1 | GL1 degradation rate | 1 |
| β1 | GL1-GL3 interaction rate | 1 |
| σ2 | Basal GL3 transcription rate | 15.0952 |
| α2 | AC-regulated GL3 transcription rate | 1.3488 |
| ρ2 | GL3 degradation rate | 0.4503 |
| β2 | GL3-R3MYB interaction rate | 7.9509 |
| α3 | AC-regulated R3MYB transcription rate | 0.4117 |
| ρ3 | R3MYB degradation rate | 0.9565 |
| γ1 | R3MYB transport rate | 9.565 |
| ρ4 | AC degradation rate | 0.2703 |
| BioClim Index | Description |
|---|---|
| BIO1 | Annual Mean Temperature |
| BIO2 | Mean Diurnal Range (Mean of monthly (max temp − min temp)) |
| BIO3 | Isothermality (BIO2/BIO7) (×100) |
| BIO4 | Temperature Seasonality (standard deviation × 100) |
| BIO5 | Max Temperature of Warmest Month |
| BIO6 | Min Temperature of Coldest Month |
| BIO7 | Temperature Annual Range (BIO5-BIO6) |
| BIO8 | Mean Temperature of Wettest Quarter |
| BIO9 | Mean Temperature of Driest Quarter |
| BIO10 | Mean Temperature of Warmest Quarter |
| BIO11 | Mean Temperature of Coldest Quarter |
| BIO12 | Annual Precipitation |
| BIO13 | Precipitation of Wettest Month |
| BIO14 | Precipitation of Driest Month |
| BIO15 | Precipitation Seasonality (Coefficient of Variation) |
| BIO16 | Precipitation of Wettest Quarter |
| BIO17 | Precipitation of Driest Quarter |
| BIO18 | Precipitation of Warmest Quarter |
| BIO19 | Precipitation of Coldest Quarter |
| SO_MNase_At4g07700_Fwd | ACTGGTTGCTAGCTGGGAGA |
| SO_MNase_At4g07700_Rev | CCAGTGTTGGTTCTCCTTGG |
| SO_MNase_GL3_1_Fwd | AAAAGTTCAGCCTTGACGTG |
| SO_MNase_GL3_1_Rev | CCATTGTTGTGGTCTTGTCTTC |
| SO_MNase_GL3_2_Fwd | TCGAGTTCAAGCTCAAACAAC |
| SO_MNase_GL3_2_Rev | TTTCGCCGGAACAAACAG |
| SO_MNase_GL3_3_Fwd | ATGGCTACCGGACAAAACAG |
| SO_MNase_GL3_3_Rev | TTACCCAGACTGAGAAGCAGAG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okamoto, S.; Negishi, K.; Toyama, Y.; Ushijima, T.; Morohashi, K. Leaf Trichome Distribution Pattern in Arabidopsis Reveals Gene Expression Variation Associated with Environmental Adaptation. Plants 2020, 9, 909. https://doi.org/10.3390/plants9070909
Okamoto S, Negishi K, Toyama Y, Ushijima T, Morohashi K. Leaf Trichome Distribution Pattern in Arabidopsis Reveals Gene Expression Variation Associated with Environmental Adaptation. Plants. 2020; 9(7):909. https://doi.org/10.3390/plants9070909
Chicago/Turabian StyleOkamoto, Shotaro, Kohei Negishi, Yuko Toyama, Takeo Ushijima, and Kengo Morohashi. 2020. "Leaf Trichome Distribution Pattern in Arabidopsis Reveals Gene Expression Variation Associated with Environmental Adaptation" Plants 9, no. 7: 909. https://doi.org/10.3390/plants9070909
APA StyleOkamoto, S., Negishi, K., Toyama, Y., Ushijima, T., & Morohashi, K. (2020). Leaf Trichome Distribution Pattern in Arabidopsis Reveals Gene Expression Variation Associated with Environmental Adaptation. Plants, 9(7), 909. https://doi.org/10.3390/plants9070909






