Transcriptome Profiling Provides Molecular Insights into Auxin-Induced Adventitious Root Formation in Sugarcane (Saccharum spp. Interspecific Hybrids) Microshoots
Abstract
:1. Introduction
2. Results
2.1. A Combination of IBA and NAA Increases the Number and Length of Adventitious Roots
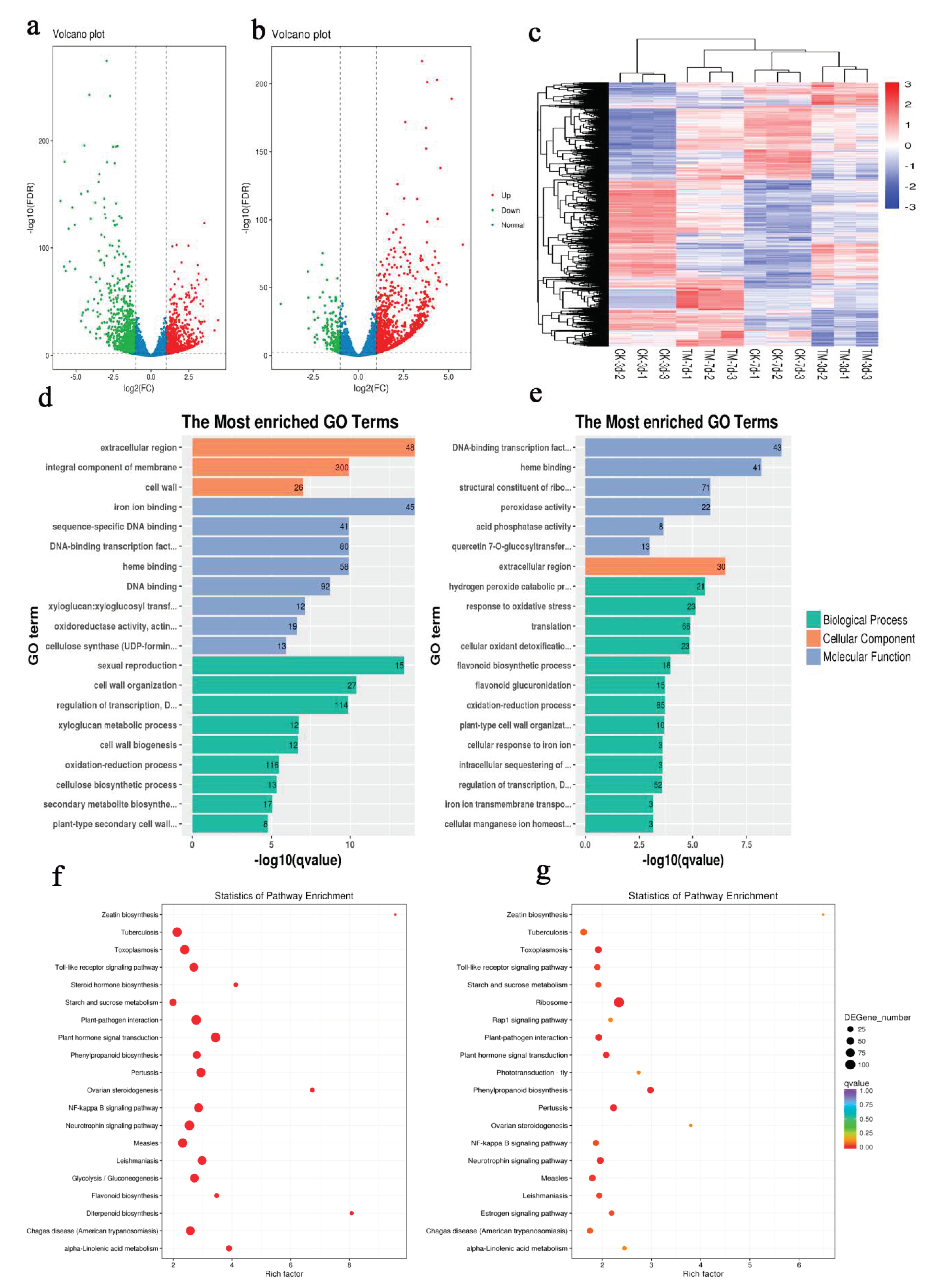
2.2. Transcriptome Sequencing and Assembly
2.3. Transcription Annotation
2.4. Differential Expression and Gene Functional Classification
2.5. Sugarcane Genes Responding to Auxin Treatment
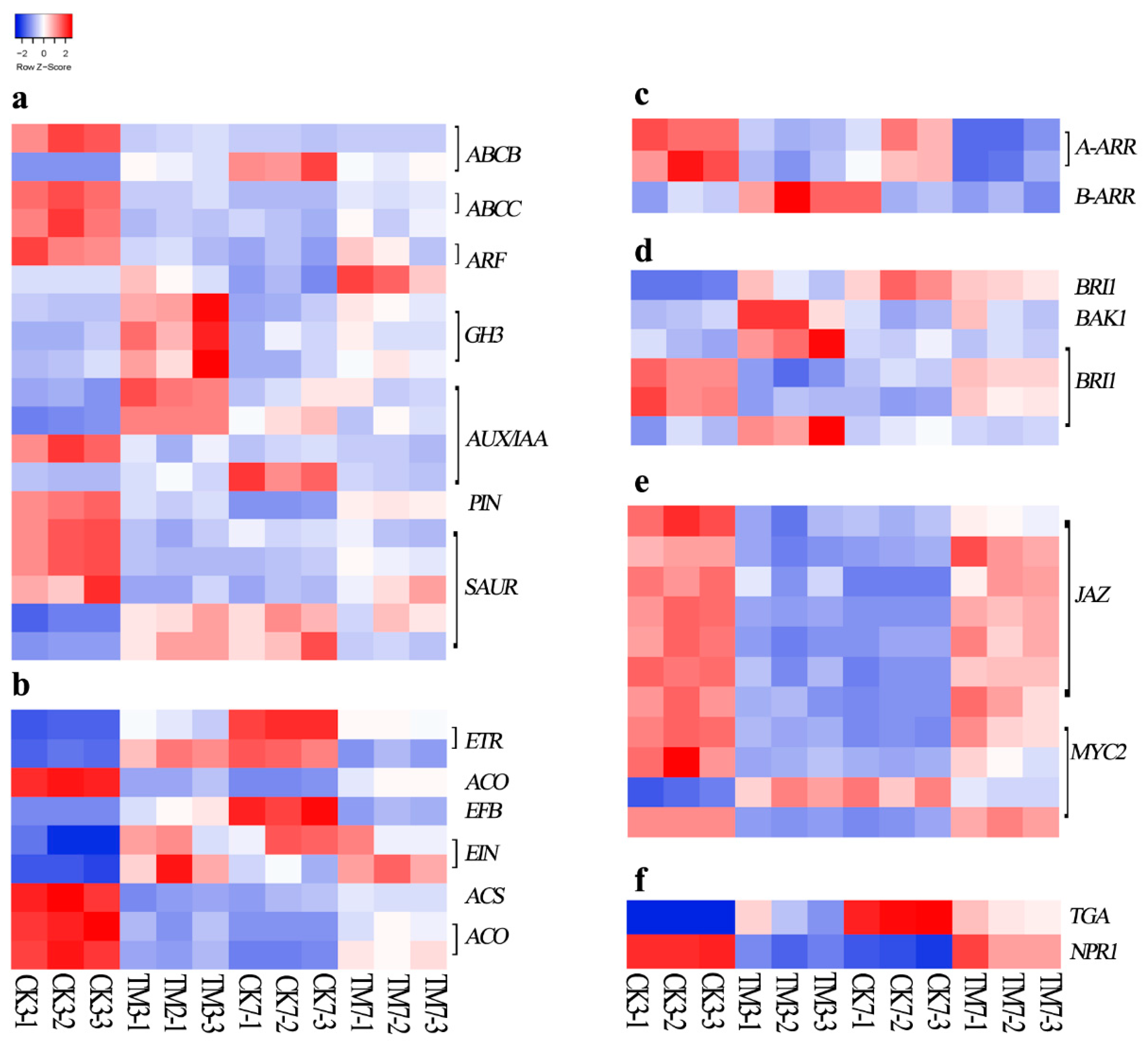
2.5.1. Plant Hormone-Related Genes
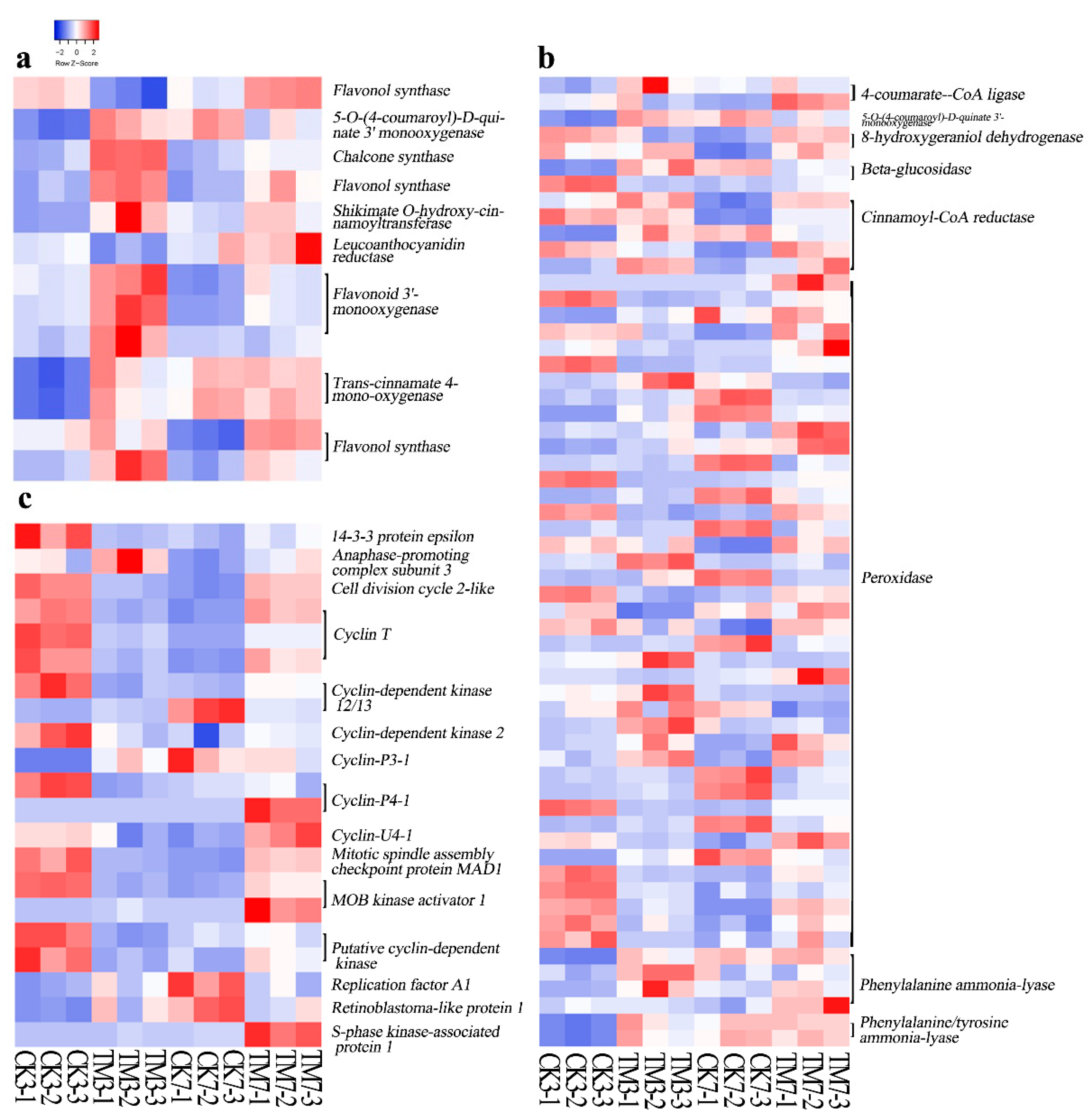
2.5.2. Flavonoid and Phenylpropanoid Biosynthesis-Related Genes
2.5.3. Cell Cycle-Related Genes
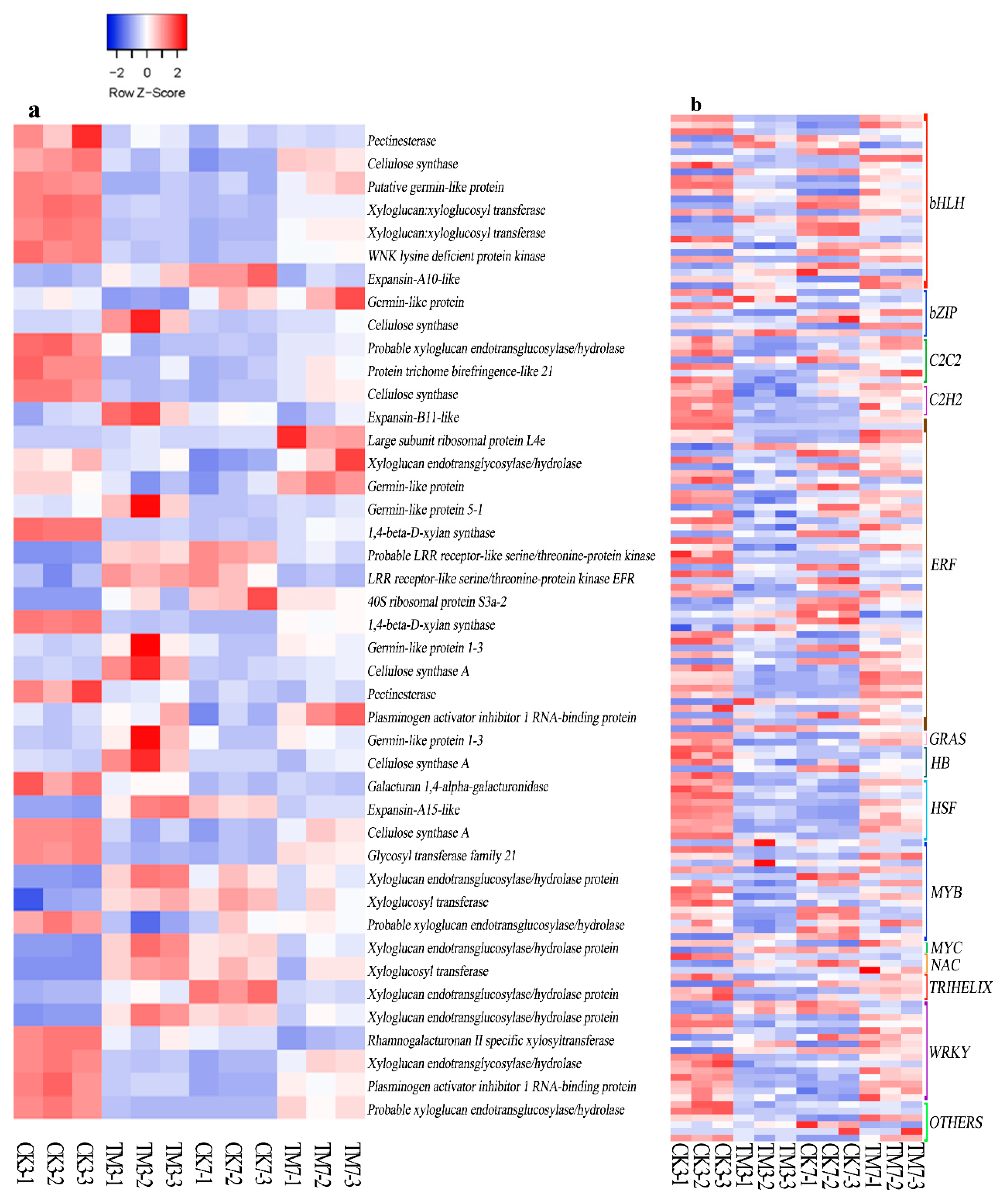
2.5.4. Cell Wall Modification-Related Genes
2.5.5. Transcription Factors
2.6. Verification of DEGs Using Quantitative Reverse Transcription–Polymerase Chain Reaction Analysis
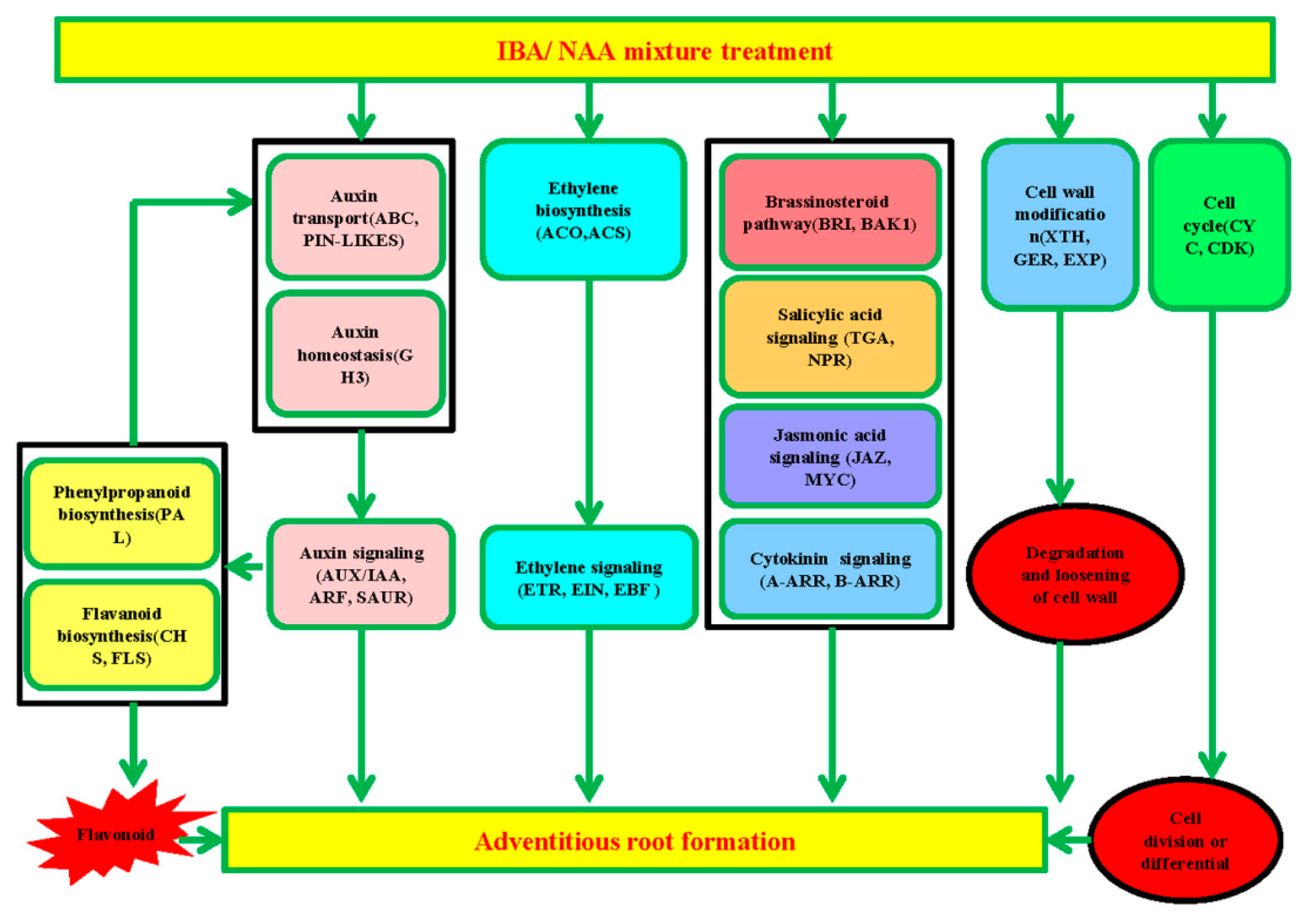
3. Discussion
3.1. Genes Related to Plant Hormone Signaling
3.2. Genes Related to Flavonoid and Phenylpropanoid Biosyntheses
3.3. Cell Cycle
3.4. Cell Wall Modification
3.5. Transcription Factors
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. RNA Extraction, Library Preparation, Sequencing, and Transcriptome Assembly
4.3. Gene Functional Annotation
4.4. Identification and Annotation of Differentially Expressed Genes
4.5. Validation of RNA-Seq Data by qRT–PCR
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ali, A.; Khan, M.; Sharif, R.; Mujtaba, M.; Gao, S.-J. Sugarcane Omics: An Update on the Current Status of Research and Crop Improvement. Plants 2019, 8, 344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C.; et al. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Pierre, J.S.; Perroux, J.; Whan, A.; Rae, A.L.; Bonnett, G.D. Poor fertility, short longevity, and low abundance in the soil seed bank limit volunteer sugarcane from seed. Front. Bioeng. Biotechnol. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, T.S.G. Micropropagation of sugarcane (Saccharum spp.). Plant. Cell. Tissue Organ. Cult. 1987, 10, 47–55. [Google Scholar] [CrossRef]
- Cardoso, J.C.; Sheng Gerald, L.T.; Teixeira da Silva, J.A. Micropropagation in the Twenty-First Century. Methods Mol. Biol. 2018, 1815, 17–46. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious Roots and Lateral Roots: Similarities and Differences. Annu. Rev. Plant. Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Lakehal, A.; Bellini, C. Control of adventitious root formation: Insights into synergistic and antagonistic hormonal interactions. Physiol. Plant. 2019, 165, 90–100. [Google Scholar] [CrossRef] [Green Version]
- Zaman, M.; Kurepin, L.V.; Catto, W.; Pharis, R.P. Evaluating the use of plant hormones and biostimulators in forage pastures to enhance shoot dry biomass production by perennial ryegrass (Lolium perenne L.). J. Sci. Food Agric. 2016, 96, 715–726. [Google Scholar] [CrossRef]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plant. 2014, 151, 83–96. [Google Scholar] [CrossRef]
- Guan, L.; Murphy, A.S.; Peer, W.A.; Gan, L.; Li, Y.; Cheng, Z.M. (Max) Physiological and Molecular Regulation of Adventitious Root Formation. CRC. Crit. Rev. Plant. Sci. 2015, 34, 506–521. [Google Scholar] [CrossRef]
- Gonin, M.; Bergougnoux, V.; Nguyen, T.D.; Gantet, P.; Champion, A. What makes adventitious roots? Plants 2019, 8, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutierrez, L.; Bussell, J.D.; Pǎcurar, D.I.; Schwambach, J.; Pǎcurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [Green Version]
- Lakehal, A.; Chaabouni, S.; Cavel, E.; Le Hir, R.; Ranjan, A.; Raneshan, Z.; Novák, O.; Păcurar, D.I.; Perrone, I.; Jobert, F.; et al. A Molecular Framework for the Control of Adventitious Rooting by TIR1/AFB2-Aux/IAA-Dependent Auxin Signaling in Arabidopsis. Mol. Plant 2019, 12, 1499–1514. [Google Scholar] [CrossRef]
- Lakehal, A.; Dob, A.; Rahneshan, Z.; Novák, O.; Escamez, S.; Alallaq, S.; Strnad, M.; Tuominen, H.; Bellini, C. Ethylene response factor 115 integrates jasmonate and cytokinin signaling machineries to repress adventitious rooting in Arabidopsis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Z.; Hua, J.; Yin, Y.; Gu, C.; Yu, C.; Shi, Q.; Guo, J.; Xuan, L.; Yu, F. An integrated transcriptome and proteome analysis reveals putative regulators of adventitious root formation in taxodium ‘zhongshanshan’. Int. J. Mol. Sci. 2019, 20, 1225. [Google Scholar] [CrossRef] [Green Version]
- Li, S.W.; Xue, L.; Xu, S.; Feng, H.; An, L. IBA-induced changes in antioxidant enzymes during adventitious rooting in mung bean seedlings: The role of H2O2. Environ. Exp. Bot. 2009, 66, 442–450. [Google Scholar] [CrossRef]
- Fattorini, L.; Veloccia, A.; Della Rovere, F.; D’Angeli, S.; Falasca, G.; Altamura, M.M. Indole-3-butyric acid promotes adventitious rooting in Arabidopsis thaliana thin cell layers by conversion into indole-3-acetic acid and stimulation of anthranilate synthase activity. BMC Plant Biol. 2017, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Ruan, L.; Wang, L.; Cheng, H. Auxin-Induced Adventitious Root Formation in Nodal Cuttings of Camellia sinensis. Int. J. Mol. Sci. 2019, 20, 4817. [Google Scholar] [CrossRef] [Green Version]
- Kubota, C.; Afreen, F.; Zobayed, S.M.A. Plant species successfully micropropagated photoautotrophically. In Photoautotrophic (Sugar-Free Mediu. Micropropag. as a New Micropropagation and Transplant Production System; Springer: Dordrecht, The Netherlands, 2005; pp. 243–266. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Philip, D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Macmanes, M.D.; et al. De novo Transcript Sequence Recostruction from RNA-Seq: Reference Generation and Analysis with Trinity. Nat. Protocol. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.Y.; Wu, L.Y.; Zhang, C.C.; Li, H.L.; Tan, L.Q.; Cao, H.L.; Cheng, H. Transcriptome analysis of indole-3-butyric acid-induced adventitious root formation in nodal cuttings of Camellia sinensis (L.). PLoS ONE 2014, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Li, S.W.; Shi, R.F.; Leng, Y.; Zhou, Y. Transcriptomic analysis reveals the gene expression profile that specifically responds to IBA during adventitious rooting in mung bean seedlings. BMC Genom. 2016, 17, 1–23. [Google Scholar] [CrossRef] [Green Version]
- De Klerk, G.J.; Van Der Krieken, W.; De Jong, J.C. The formation of adventitious roots: New concepts, new possibilities. Vitr. Cell. Dev. Biol. Plant 1999, 35, 189–199. [Google Scholar] [CrossRef]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Xue, L.; Xu, S.; Feng, H.; An, L. Hydrogen peroxide involvement in formation and development of adventitious roots in cucumber. Plant Growth Regul. 2007, 52, 173–180. [Google Scholar] [CrossRef]
- Kasirajan, L.; Hoang, N.V.; Furtado, A.; Botha, F.C.; Henry, R.J. Transcriptome analysis highlights key differentially expressed genes involved in cellulose and lignin biosynthesis of sugarcane genotypes varying in fiber content. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Cunha, C.P.; Roberto, G.G.; Vicentini, R.; Lembke, C.G.; Souza, G.M.; Ribeiro, R.V.; Machado, E.C.; Lagôa, A.M.M.A.; Menossi, M. Ethylene-induced transcriptional and hormonal responses at the onset of sugarcane ripening. Sci. Rep. 2017, 7, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liang, Z.; Zeng, Y.; Jing, Y.; Wu, K.; Liang, J.; He, S.; Wang, G.; Mo, Z.; Tan, F.; et al. De novo analysis of transcriptome reveals genes associated with leaf abscission in sugarcane (Saccharum officinarum L.). BMC Genom. 2016, 17, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional Genomic Analysis of the AUXIN RESPONSE FACTOR Gene Family Members in Arabidopsis thaliana. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [Green Version]
- Ori, N. Dissecting the biological functions of ARF and aux/IAA genes. Plant Cell 2019, 31, 1210–1211. [Google Scholar] [CrossRef] [Green Version]
- Brunoni, F.; Collani, S.; Casanova-Saéz, R.; Šimura, J.; Karady, M.; Schmid, M.; Ljung, K.; Bellini, C. Conifers exhibit a characteristic inactivation of auxin to maintain tissue homeostasis. bioRxiv 2019, 789420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludwig-Müller, J.; Jülke, S.; Bierfreund, N.M.; Decker, E.L.; Reski, R. Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol. 2009, 181, 323–338. [Google Scholar] [CrossRef]
- Barbez, E.; Kubeš, M.; Rolčík, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef] [Green Version]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Hu, H.; Luo, X.; Zhang, C.; Li, X.; Li, P.; Li, W.; Khawar, A.; Sun, X.; Ren, Z.; et al. Dynamic changes of phytohormone signaling in the base of Taxus media stem cuttings during adventitious root formation. Sci. Hortic. 2019, 246, 338–346. [Google Scholar] [CrossRef]
- Zimmerman, P.W.; Wilcoxon, F. Several Chemical Growth Substances which Cause Initiation of Roots and Other Responses in Plants. Contrib. Boyce Thompson Inst. 1935, 7, 209–229. [Google Scholar]
- Negi, S.; Sukumar, P.; Liu, X.; Cohen, J.D.; Muday, G.K. Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 2010, 61, 3–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.L.-C.; Li, H.; Ecker, J.R. Ethylene biosynthesis and signaling networks. Plant Cell 2002, 14, 131–151. [Google Scholar] [CrossRef] [Green Version]
- Wilmowicz, E.; Frankowski, K.; Kesy, J.; Glazińska, P.; Wojciechowski, W.; Kućko, A.; Kopcewicz, J. The role of PnACO1 in light- and IAA-regulated flower inhibition in Pharbitis nil. Acta Physiol. Plant. 2013, 35, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; An, F.; Feng, Y.; Li, P.; Xue, L.; Mu, A.; Jiang, Z.; Kim, J.M.; To, T.K.; Li, W.; et al. Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 12539–12544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naseem, M.; Kaltdorf, M.; Dandekar, T. The nexus between growth and defence signalling: Auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 2015, 66, 4885–4896. [Google Scholar] [CrossRef] [Green Version]
- Ramirez-Carvajal, G.A.; Morse, A.M.; Dervinis, C.; Davis, J.M. The cytokinin type-B response regulator PtRR13 is a negative regulator of adventitious root development in populus 1[C][W][OA]. Plant Physiol. 2009, 150, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Meng, W.J.; Cheng, Z.J.; Sang, Y.L.; Zhang, M.M.; Rong, X.F.; Wang, Z.W.; Tang, Y.Y.; Zhang, X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 2017, 29, 1357–1372. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Hacham, Y.; Holland, N.; Butterfield, C.; Ubeda-Tomas, S.; Bennett, M.J.; Chory, J.; Savaldi-Goldstein, S. Brassinosteroid perception in the epidermis controls root meristem size. Development 2011, 138, 839–848. [Google Scholar] [CrossRef] [Green Version]
- Lischweski, S.; Muchow, A.; Guthörl, D.; Hause, B. Jasmonates act positively in adventitious root formation in petunia cuttings. BMC Plant Biol. 2015, 15, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.-T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U.; et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009, 181, 613–625. [Google Scholar] [CrossRef]
- Hoo, S.C.; Koo, A.J.K.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef] [Green Version]
- Curir, P.; VanSumere, C.F.; Termini, A.; Barthe, P.; Marchesini, A.; Dolci, M. Flavonoid accumulation is correlated with adventitious roots formation in Eucalyptus gunnii hook micropropagated through axillary bud stimulation. Plant Physiol. 1990, 92, 1148–1153. [Google Scholar] [CrossRef] [Green Version]
- Lewis, D.R.; Ramirez, M.V.; Miller, N.D.; Vallabhaneni, P.; Keith Ray, W.; Helm, R.F.; Winkel, B.S.J.; Muday, G.K. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 2011, 156, 144–164. [Google Scholar] [CrossRef] [Green Version]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Brinker, M.; Van Zyl, L.; Liu, W.; Craig, D.; Sederoff, R.R.; Clapham, D.H.; Von Arnold, S. Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol. 2004, 135, 1526–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurepa, J.; Shull, T.E.; Karunadasa, S.S.; Smalle, J.A. Modulation of auxin and cytokinin responses by early steps of the phenylpropanoid pathway. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef]
- Komaki, S.; Sugimoto, K. Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol. 2012, 53, 953–964. [Google Scholar] [CrossRef] [Green Version]
- Wildwater, M.; Campilho, A.; Perez-Perez, J.M.; Heidstra, R.; Blilou, I.; Korthout, H.; Chatterjee, J.; Mariconti, L.; Gruissem, W.; Scheres, B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005, 123, 1337–1349. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Ramírez, A.; Díaz-Triviño, S.; Wachsman, G.; Du, Y.; Arteága-Vázquez, M.; Zhang, H.; Benjamins, R.; Blilou, I.; Neef, A.B.; Chandler, V.; et al. A SCARECROW-RETINOBLASTOMA Protein Network Controls Protective Quiescence in the Arabidopsis Root Stem Cell Organizer. PLoS Biol. 2013, 11. [Google Scholar] [CrossRef] [Green Version]
- Swarup, K.; Benková, E.; Swarup, R.; Casimiro, I.; Péret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef]
- Majda, M.; Robert, S. The role of auxin in cell wall expansion. Int. J. Mol. Sci. 2018, 19, 951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar] [CrossRef] [Green Version]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Hutchison, K.W.; Singer, P.B.; McInnis, S.; Diaz-Sala, C.; Greenwood, M.S. Expansins are conserved in conifers and expressed in hypocotyls in response to exogenous auxin. Plant Physiol. 1999, 120, 827–831. [Google Scholar] [CrossRef] [Green Version]
- Mathieu, M.; Lelu-Walter, M.A.; Blervacq, A.S.; David, H.; Hawkins, S.; Neutelings, G. Germin-like genes are expressed during somatic embryogenesis and early development of conifers. Plant Mol. Biol. 2006, 61, 615–627. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Verslues, P.E.; Zheng, X.; Lee, B.H.; Zhan, X.; Manabe, Y.; Sokolchik, I.; Zhu, Y.; Dong, C.H.; Zhu, J.K.; et al. Erratum: HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proc. Natl. Acad. Sci. USA 2010, 107, 13972. [Google Scholar] [CrossRef] [Green Version]
- Rigal, A.; Yordanov, Y.S.; Perrone, I.; Karlberg, A.; Tisserant, E.; Bellini, C.; Busov, V.B.; Martin, F.; Kohler, A.; Bhalerao, R.; et al. The AINTEGUMENTA LIKE1 homeotic transcription factor PtAIL1 controls the formation of adventitious root primordia in poplar. Plant Physiol. 2012, 160, 1996–2006. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, J.; Mao, Y.; Zhang, M.; Wang, R.; Hu, Y.; Mao, Z.; Shen, X. Transcriptional regulation of MdPIN3 and MdPIN10 by MdFLP during apple self-rooted stock adventitious root gravitropism. BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Bao, M.; Bian, H.; Zha, Y.; Li, F.; Sun, Y.; Bai, B.; Chen, Z.; Wang, J.; Zhu, M.; Han, N. MiR396a-mediated basic helix-loop-helix transcription factor bHLH74 repression acts as a regulator for root growth in arabidopsis seedlings. Plant Cell Physiol. 2014, 55, 1343–1353. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Hu, Y.; Vannozzi, A.; Wu, K.; Cai, H.; Qin, Y.; Mullis, A.; Lin, Z.; Zhang, L. The WRKY Transcription Factor Family in Model Plants and Crops. CRC. Crit. Rev. Plant Sci. 2017, 36, 311–335. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Peng, Y.; Guo, Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008, 18, 508–521. [Google Scholar] [CrossRef] [Green Version]
- Sabrina, S.; Renze, H.; Marjolein, W.; Ben, S. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 2003, 17, 1–6. [Google Scholar] [CrossRef]
- Zhou, W.; Lozano-Torres, J.L.; Blilou, I.; Zhang, X.; Zhai, Q.; Smant, G.; Li, C.; Scheres, B. A Jasmonate Signaling Network Activates Root Stem Cells and Promotes Regeneration. Cell 2019, 177, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Ramírez, A.; Díaz-Triviño, S.; Blilou, I.; Grieneisen, V.A.; Sozzani, R.; Zamioudis, C.; Miskolczi, P.; Nieuwland, J.; Benjamins, R.; Dhonukshe, P.; et al. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 2012, 150, 1002–1015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabherr, M.G.; Haas, B.J.; Moran, Y.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Sabeh, M.; Duceppe, M.-O.; Mimee, B.; Lafond-Lapalme, J.; Moffett, P.; Palomares-Rius, J.E.; Blok, V. Analysis of survival and hatching transcriptomes from potato cyst nematodes, Globodera rostochiensis and G. pallida. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11. [Google Scholar] [CrossRef] [Green Version]
- Ling, H.; Wu, Q.; Guo, J.; Xu, L.; Que, Y. Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, A.; Lakshmanan, P.; He, W.; Tan, H.; Liu, L.; Liu, H.; Liu, J.; Huang, D.; Chen, Z. Transcriptome Profiling Provides Molecular Insights into Auxin-Induced Adventitious Root Formation in Sugarcane (Saccharum spp. Interspecific Hybrids) Microshoots. Plants 2020, 9, 931. https://doi.org/10.3390/plants9080931
Li A, Lakshmanan P, He W, Tan H, Liu L, Liu H, Liu J, Huang D, Chen Z. Transcriptome Profiling Provides Molecular Insights into Auxin-Induced Adventitious Root Formation in Sugarcane (Saccharum spp. Interspecific Hybrids) Microshoots. Plants. 2020; 9(8):931. https://doi.org/10.3390/plants9080931
Chicago/Turabian StyleLi, Aomei, Prakash Lakshmanan, Weizhong He, Hongwei Tan, Limin Liu, Hongjian Liu, Junxian Liu, Dongliang Huang, and Zhongliang Chen. 2020. "Transcriptome Profiling Provides Molecular Insights into Auxin-Induced Adventitious Root Formation in Sugarcane (Saccharum spp. Interspecific Hybrids) Microshoots" Plants 9, no. 8: 931. https://doi.org/10.3390/plants9080931
APA StyleLi, A., Lakshmanan, P., He, W., Tan, H., Liu, L., Liu, H., Liu, J., Huang, D., & Chen, Z. (2020). Transcriptome Profiling Provides Molecular Insights into Auxin-Induced Adventitious Root Formation in Sugarcane (Saccharum spp. Interspecific Hybrids) Microshoots. Plants, 9(8), 931. https://doi.org/10.3390/plants9080931




