The Phylogeny of Class B Flavoprotein Monooxygenases and the Origin of the YUCCA Protein Family
Abstract
:1. Introduction
2. Results
2.1. Analysis of the Proteins of Class B Flavoprotein Monooxygenases
2.2. Comparative Analysis of the Functional Sites and Domains of Class B Flavoprotein Monooxygenases
2.3. Abundance of the Sequences Homologous to Type IIb FMOs in the Main Taxa
2.4. Analysis of the Plant Class B Flavoprotein Monooxygenases Represented in Transcriptome Projects
3. Discussion
3.1. Type IIb FMOs Is a Novel Family of Class B Flavoprotein Monooxygenases
3.2. Different Functions of Type IIb FMO and YUCCA Proteins
3.3. The Origin of the Main Auxin Biosynthesis Pathway in Higher Plants
4. Materials and Methods
4.1. Sampling of Protein and Transcriptome Sequences and Their Alignment
4.2. Phylogenetic Analysis
4.3. Analysis of Conserved Sites, Protein Domains, and Taxonomic Representation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BVMO | Baeyer–Villiger monooxygenases |
| FAD | Flavin adenine dinucleotide |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NADH | Nicotinamide adenine dinucleotide |
| CDD | Conserved Domains Database |
| 1KP | 1000 plants |
| FMO | Flavin-containing monooxygenase |
| MRCA | Most recent common ancestor |
| IAA | Indole acetic acid |
| IPA | Indole-3-pyruvate |
| NMO | N-hydroxylating monooxygenases |
| YUCCA | YUCCA flavin-containing monooxygenase |
| TAA | Tryptophan aminotransferase enzyme |
| HGT | Horizontal gene transfer |
References
- Weijers, D.; Nemhauser, J.; Yang, Z. Auxin: Small molecule, big impact. J. Exp. Bot. 2018, 69, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Doležal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mironova, V.; Teale, W.; Shahriari, M.; Dawson, J.; Palme, K. The systems biology of auxin in developing embryos. Trends Plant Sci. 2017, 22, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A. Auxin: A regulator of cold stress response. Physiol. Plant. 2013, 147, 28–35. [Google Scholar] [CrossRef]
- Blakeslee, J.J.; Rossi, T.S.; Kriechbaumer, V. Auxin biosynthesis: Spatial regulation and adaptation to stress. J. Exp. Bot. 2019, 70, 5041–5049. [Google Scholar] [CrossRef]
- Kunkel, B.N.; Harper, C.P. The roles of auxin during interactions between bacterial plant pathogens and their hosts. J. Exp. Bot. 2018, 69, 245–254. [Google Scholar] [CrossRef]
- Kazan, K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 2013, 112, 1655–1665. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.H.; Savina, M.; Du, J.; Devendran, A.; Ramakanth, K.K.; Tian, X.; Sim, W.S.; Victoria, V.; Mironova, V.V.; Xu, J. A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress. Cell 2017, 170, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Rozov, S.M.; Zagorskaya, A.A.; Deineko, E.V.; Shumny, V.K. Auxins: Biosynthesis, metabolism, and transport. Biol. Bull. Rev. 2013, 3, 286–295. [Google Scholar] [CrossRef]
- Kasahara, H. Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 2015, 80, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Tivendale, N.D.; Ross, J.J.; Cohen, J.D. The shifting paradigms of auxin biosynthesis. Trends Plant Sci. 2014, 19, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Brumos, J.; Alonso, J.M.; Stepanova, A.N. Genetic aspects of auxin biosynthesis and its regulation. Physiol. Plant. 2014, 151, 3–12. [Google Scholar] [CrossRef]
- Matthes, M.S.; Best, N.B.; Robil, J.M.; Malcomber, S.; Gallavotti, A.; McSteen, P. Auxin EvoDevo: Conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol. Plant 2019, 12, 298–320. [Google Scholar] [CrossRef] [Green Version]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef] [Green Version]
- Won, C.; Shen, X.; Mashiguchi, K.; Zheng, Z.; Dai, X.; Cheng, Y.; Kasahara, H.; Kamiya, Y.; Chory, J.; Zhao, Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18518–18523. [Google Scholar] [CrossRef] [Green Version]
- Stepanova, A.N.; Yun, J.; Robles, L.M.; Novak, O.; He, W.; Guo, H.; Ljung, K.; Alonso, J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 2011, 23, 3961–3973. [Google Scholar] [CrossRef] [Green Version]
- Tivendale, N.D.; Davidson, S.E.; Davies, N.W.; Smith, J.A.; Dalmais, M.; Bendahmane, A.I.; Quittenden, L.J.; Sutton, L.; Bala, R.K.; Le Signor, C.; et al. Biosynthesis of the Halogenated Auxin, 4-Chloroindole-3-Acetic Acid. Plant Physiol. 2012, 159, 1055–1063. [Google Scholar] [CrossRef] [Green Version]
- Di, D.-W.; Zhang, C.; Luo, P.; An, C.-W.; Guang-Qin Guo, G.-Q. The biosynthesis of auxin: How many paths truly lead to IAA? Plant Growth Regul. 2016, 78, 275–285. [Google Scholar] [CrossRef]
- Poulet, A.; Kriechbaumer, V. Bioinformatics Analysis of Phylogeny and Transcription of TAA/YUC Auxin Biosynthetic Genes. Int. J. Mol. Sci. 2017, 18, 1791. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Yang, H.; Shang, C.; Ma, S.; Liu, L.; Cheng, J. The Roles of Auxin Biosynthesis YUCCA Gene Family in Plants. Int. J. Mol. Sci. 2019, 20, 6343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, J.; Hu, X.; Huang, J. Origin of plant auxin biosynthesis. Trends Plant Sci. 2014, 19, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Klebsormidium Nitens NIES-2285 Genome Project. Available online: http://www.plantmorphogenesis.bio.titech.ac.jp/~algae_genome_project/klebsormidium/ (accessed on 24 July 2020).
- Wang, C.; Liu, Y.; Li, S.-H.; Guan-Zhu Han, G.-Z. Origin of plant auxin biosynthesis in charophyte algae. Trends Plant Sci. 2014, 19, 741–743. [Google Scholar] [CrossRef]
- McCourt, R.M.; Delwiche, C.F.; Karol, K.G. Charophyte alga and land plant origins. Trends Ecol. Evol. 2004, 19, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Turnaev, I.I.; Gunbin, K.V.; Afonnikov, D.A. Plant auxin biosynthesis did not originate in charophytes. Trends Plant Sci. 2015, 20, 463–465. [Google Scholar] [CrossRef]
- Ke, M.; Zheng, Y.; Zhu, Z. Rethinking the origin of auxin biosynthesis in plants. Front. Plant Sci. 2015, 6, 1093. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Li, S.-S.; Han, G.-Z. Commentary: Plant auxin biosynthesis did not originate in charophytes. Front. Plant Sci. 2016, 7, 158. [Google Scholar] [CrossRef] [Green Version]
- Romani, F. Origin of TAA Genes in Charophytes: New Insights into the Controversy over the Origin of Auxin Biosynthesis. Front. Plant Sci. 2017, 8, 1616. [Google Scholar] [CrossRef]
- Thodberg, S.; Neilson, E.H.J. The “Green” FMOs: Diversity, functionality and application of plant flavoproteins. Catalysts 2020, 10, 329. [Google Scholar] [CrossRef] [Green Version]
- Huijbers, M.M.E.; Montersino, S.; Westphal, A.H.; Tischler, D.; van Berkel, W.J.H. Flavin dependent monooxygenases. Arch. Biochem. Biophys. 2014, 544, 2–17. [Google Scholar] [CrossRef] [PubMed]
- Ozols, J. Covalent structure of liver microsomal flavin-containing monooxygenase form 1. J. Biol. Chem. 1990, 265, 10289–10299. [Google Scholar] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Mascotti, M.L.; Lapadula, W.J.; Juri, A.M. The Origin and evolution of baeyer-villiger monooxygenases (BVMOs): An ancestral family of flavin monooxygenases. PLoS ONE 2015, 10, e0132689. [Google Scholar] [CrossRef] [Green Version]
- van Berkel, W.J.H.; Kamerbeek, N.M.; Fraaije, M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 2006, 124, 670–689. [Google Scholar] [CrossRef] [Green Version]
- Fraaije, M.W.; Kamerbeek, N.M.; van Berkel, W.J.; Janssen, D.B. Identification of a Baeyer-Villiger monooxygenase sequence motif. FEBS Lett. 2002, 518, 43–47. [Google Scholar] [CrossRef] [Green Version]
- Schlaich, N.L. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007, 12, 412–418. [Google Scholar] [CrossRef]
- Riebel, A.; Dudek, H.M.; de Gonzalo, G.; Stepniak, P.; Rychlewski, L.; Fraaije, M.W. Expanding the set of rhodococcal Baeyer–Villiger monooxygenases by high-throughput cloning, expression and substrate screening. Appl. Microbiol. Biotechnol. 2012, 95, 1479–1489. [Google Scholar] [CrossRef] [Green Version]
- Riebel, A.; de Gonzalo, G.; Fraaije, M.W. Expanding the biocatalytic toolbox of flavoprotein monooxygenases from Rhodococcus jostii RHA1. J. Mol. Catal. B Enzym. 2013, 88, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Stehr, M.; Diekmann, H.; Smau, L.; Seth, O.; Ghisla, S.; Singh, M.; Macheroux, P. A hydrophobic sequence motif common to N-hydroxylating enzymes. Trends Biochem. Sci. 1998, 23, 56–57. [Google Scholar] [CrossRef] [Green Version]
- Riebel, A.; Fink, M.J.; Mihovilovic, M.D.; Fraaije, M.W. Type II flavin-containing monooxygenases: A new class of biocatalysts that harbors baeyer–villiger monooxygenases with a relaxed coenzyme specificity. ChemCatChem 2014, 6, 1112–1117. [Google Scholar] [CrossRef]
- Krueger, S.K.; Williams, D.E. Mammalian flavin-containing monooxygenases: Structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol. Ther. 2005, 106, 357–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziegler, D.M. Flavin-containing monooxygenases: Enzymes adapted for multisubstrate specificity. Trends Pharm. Sci. 1990, 11, 321–324. [Google Scholar] [CrossRef]
- Dolphin, C.T.; Janmohamed, A.; Smith, R.L.; Shephard, E.A.; Phillips, I.R. Missense mutation in flavin-containing mono-oxygenase 3 gene, FMO3, underlies fish-odour syndrome. Nat. Genet. 1997, 17, 491–494. [Google Scholar] [CrossRef]
- Rioz-Martínez, A.; Kopacz, M.; de Gonzalo, G.; Torres Pazmiño, D.E.; Gotor, V.; Fraaije, M.W. Exploring the biocatalytic scope of a bacterial flavin-containing monooxygenase. Org. Biomol. Chem. 2011, 9, 1337–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The 1000 Plants. Available online: https://sites.google.com/a/ualberta.ca/onekp/ (accessed on 24 July 2020).
- One Thousand Plant Transcriptomes Initiative. One Thousand Plant Transcriptomes and the Phylogenomics of Green Plants. Nature 2019, 574, 679–685. [Google Scholar] [CrossRef] [Green Version]
- Cooper, E.D.; Delwiche, C.F. Green Algal Transcriptomes for Phylogenetics and Comparative Genomics. 2016. Available online: https://figshare.com/articles/Green_algal_transcriptomes_for_phylogenetics_and_comparative_genomics/1604778 (accessed on 24 July 2020).
- Lewis, L.A.; McCourt, R.M. Green algae and the origin of land plants. Am. J. Bot. 2004, 91, 1535–1556. [Google Scholar] [CrossRef] [Green Version]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; De Clerck, O. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef] [Green Version]
- Timme, R.E.; Bachvaroff, T.R.; Delwiche, C.F. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 2012, 7, e29696. [Google Scholar] [CrossRef] [Green Version]
- Kolaczkowski, B.; Thornton, J.W. Long-branch attraction bias and inconsistency in bayesian phylogenetics. PLoS ONE 2009, 4, e7891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Grimwood, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into Land Plant Evolution Garnered from the Marchantia polymorpha Genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, R.Y.; Chang, Y.; Bateman, A.; Murzin, A.G.; Axelrod, H.L.; Hwang, W.C.; Aravind, L. Filling out the structural map of the NTF2-like superfamily. BMC Bioinform. 2013, 14, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, C.; Sørensen, I.; Sun, X.; Sun, H.; Behar, H.; Alseekh, S.; Philippe, G.; Lopez, K.P.; Sun, L.; Reed, R.; et al. The genome of the charophyte alga penium margaritaceum bears footprints of the evolutionary origins of land plants. J. Cell 2019. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, T.; Sakayama, H.; De Vries, J.; Buschmann, H.; Saint-Marcoux, D.; Ullrich, K.K.; Haas, F.B.; Vanderstraeten, L.; Becker, D.; Lang, D.; et al. The Chara Genome: Secondary Complexity and Implications for Plant Terrestrialization. Cell 2018, 174, 448–464. [Google Scholar] [CrossRef] [Green Version]
- Ai, Y.; Zhang, Z.-H.; Zheng, Y.-Y.; Zhong, B.-J.; Zhu, Z.-Q. Preliminary study on the function of TAA1, a key enzyme in auxin biosynthesis. Kleb. Flaccidum Zhiwu Shengli Xuebao/Plant Physiol. J. 2018, 54, 1451–1458. [Google Scholar] [CrossRef]
- Basu, S.; Sun, H.; Brian, L.; Quatrano, R.L.; Muday, G.K. Early embryo development in Fucus distichus is auxin sensitive. Plant Physiol. 2002, 130, 292–302. [Google Scholar] [CrossRef] [Green Version]
- Le Bail, A.; Billoud, B.; Kowalczyk, N.; Kowalczyk, M.; Gicquel, M.; Le Panse, S.; Stewart, S.; Scornet, D.; Cock, J.M.; Ljung, K.; et al. Auxin metabolism and function in the multicellular brown alga Ectocarpus siliculosus. Plant Physiol. 2010, 153, 128–144. [Google Scholar] [CrossRef] [Green Version]
- Mikami, K.; Mori, I.C.; Matsuura, T.; Ikeda, Y.; Kojima, M.; Sakakibara, H.; Hirayama, T. Comprehensive quantification and genome survey reveal the presence of novel phytohormone action modes in red seaweeds. J. Appl. Phycol. 2016, 28, 2539–2548. [Google Scholar] [CrossRef]
- Ohtaka, K.; Hori, K.; Kanno, Y.; Seo, M.; Ohta, H. Primitive auxin response without TIR1 and Aux/IAA in the charophyte alga klebsormidium nitens. Plant Physiol. 2017, 174, 1621–1632. [Google Scholar] [CrossRef] [Green Version]
- Labeeuw, L.; Khey, J.; Bramucci, A.R.; Atwal, H.; de la Mata, A.P.; Harynuk, J.; Case, R.J. Indole-3-acetic acid is produced by emiliania huxleyi coccolith-bearing cells and triggers a physiological response in bald cells. Front Microbiol. 2016, 7, 828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smet, I.; Voß, U.; Lau, S.; Wilson, M.; Shao, N.; Timme, R.E.; Swarup, R.; Kerr, I.; Hodgman, C.; Bock, R.; et al. Unraveling the evolution of auxin signaling. Plant Physiol. 2011, 155, 209–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PLAZA 2.5 Database. Available online: https://bioinformatics.psb.ugent.be/plaza/versions/plaza_v2_5/blast/index (accessed on 24 July 2020).
- Congenie.org. Available online: http://congenie.org (accessed on 24 July 2020).
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.-C.; Scofield, D.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Congenie.org/blast. Available online: http://congenie.org/blast (accessed on 24 July 2020).
- Mascotti, M.L.; Ayub, M.J.; Furnham, N.; Thornton, J.M.; Laskowski, R.A. Chopping and changing: The evolution of the flavin-dependent monooxygenases. J. Mol. Biol. 2016, 428, 3131–3146. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [Green Version]
- SAMEM v. 0.83—Computer System for Analysis of Molecular Evolution Modes. Available online: http://pixie.bionet.nsc.ru/cgi-bin/pipeline/index.pl?nodes_file=/apache/www/cgidata/xmldata/samem/nodes_prot.xml&programs_file=/apache/www/cgidata/xmldata/samem/programs_prot.xml#Mafft (accessed on 24 July 2020).
- Gunbin, K.V.; Suslov, V.V.; Genaev, M.A.; Afonnikov, D.A. Computer System for Analysis of Molecular Evolution Modes (SAMEM): Analysis of molecular evolution modes at deep inner branches of the phylogenetic tree. Silico Biol. 2011, 11, 109–123. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Pei, J.; Grishin, N.V. PROMALS: Towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 2007, 23, 802–808. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Meier, M.; Mirdita, M.; Vöhringer, H.; Haunsberger, S.J.; Söding, J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019, 473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D220–D230. [Google Scholar] [CrossRef] [Green Version]

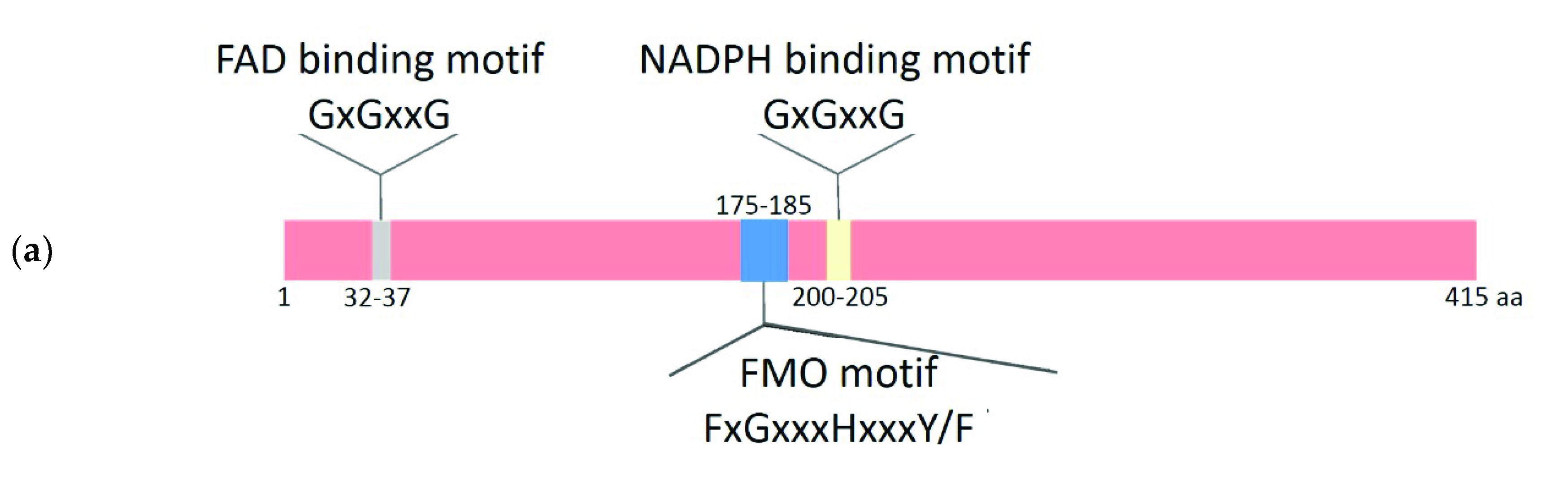
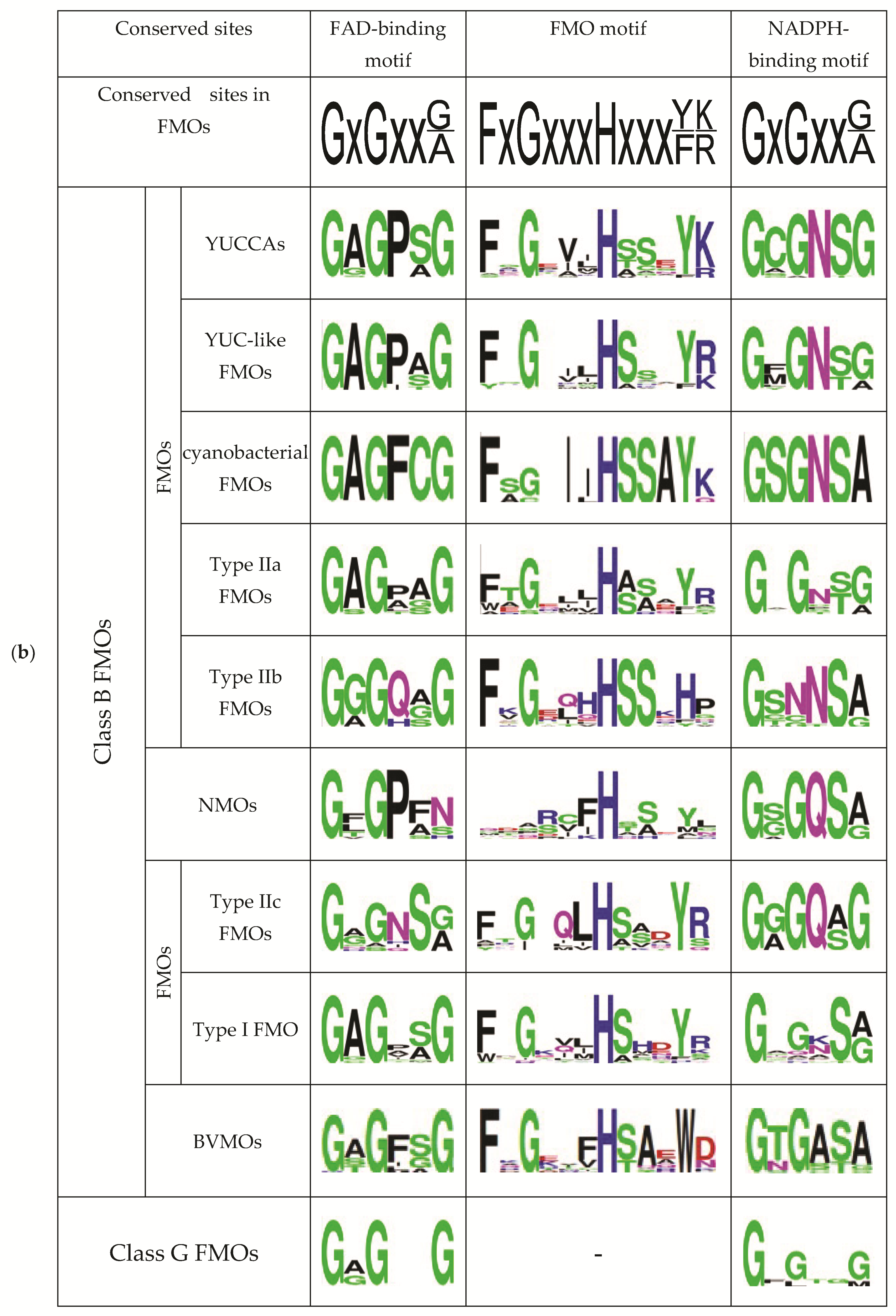
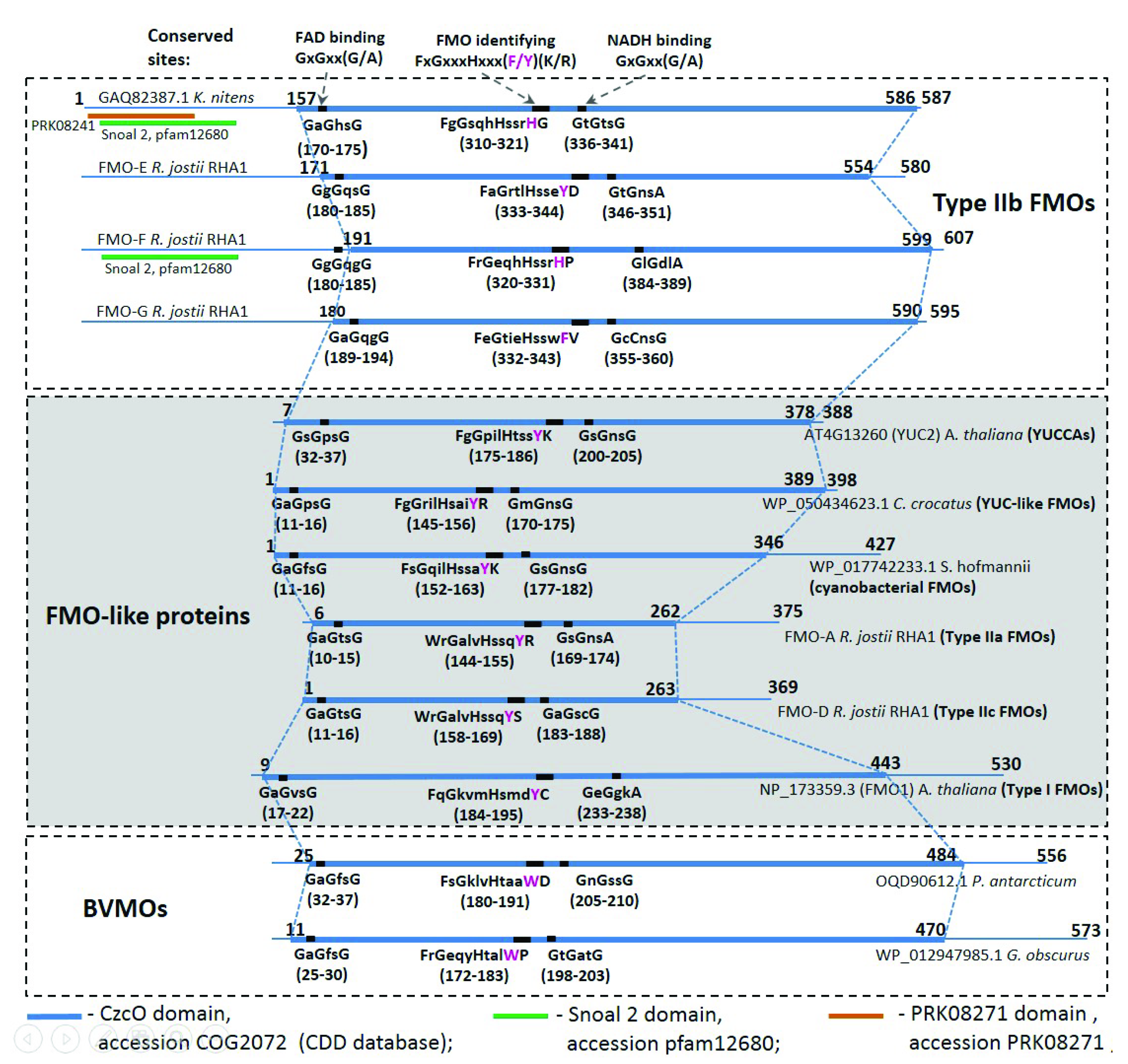
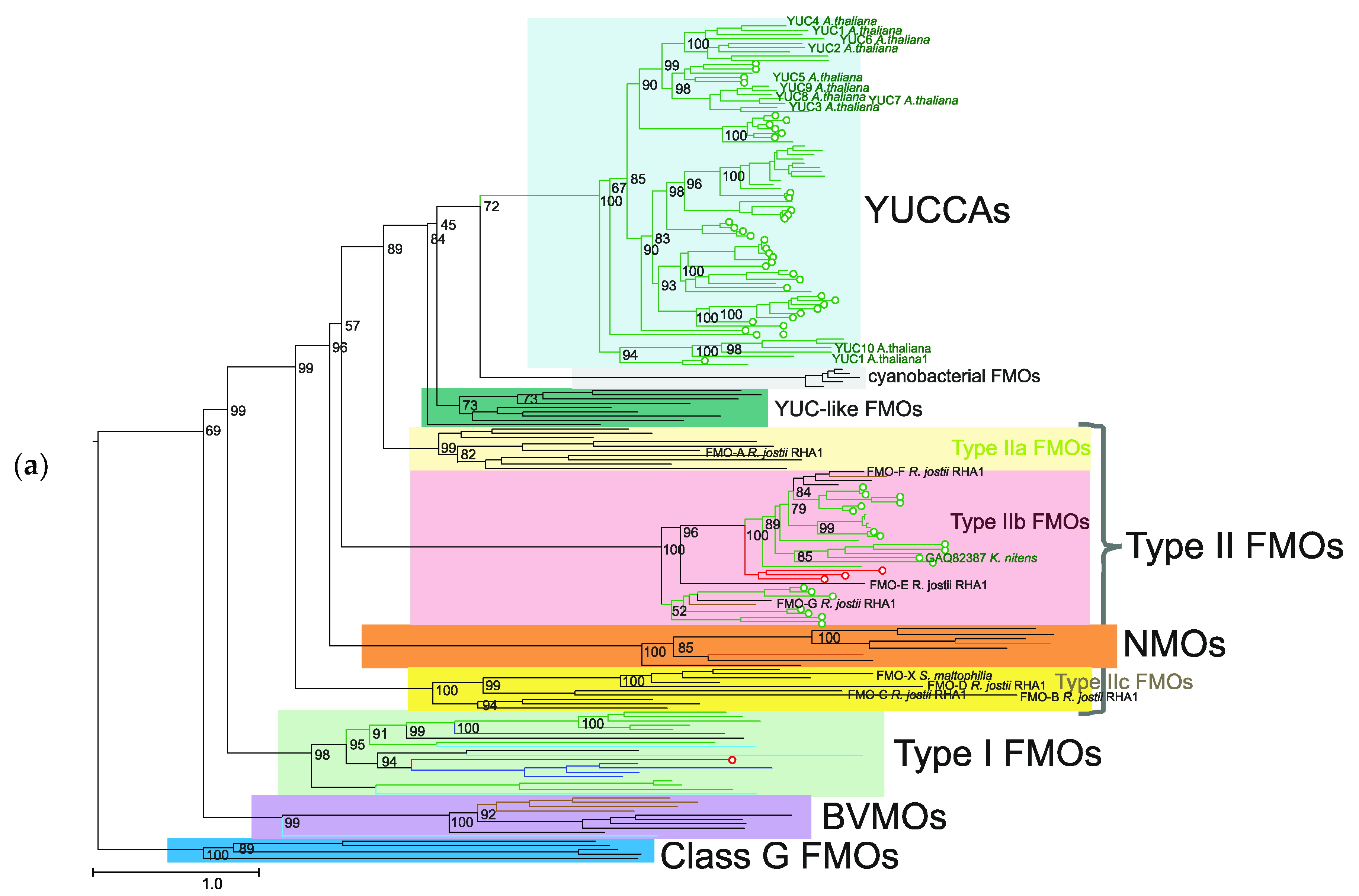

| Protein Groups | Animal | Fungi | Plants | Bacteria | Archaea |
|---|---|---|---|---|---|
| Type IIb FMOs * | |||||
| 0 to 10−70 ** | 0 | 419 | 4 | 1387 | 1 |
| 10−70 to 10−40 | 0 | 80 | 0 | 2 | 0 |
| 10−40 to 10−5 | 0 | 8 | 3 | 19 | 0 |
| All homologs | 0 | 507(285) *** | 7(4) | 1408(1041) | 1(1) |
| BVMOs | |||||
| 0 to 10−70 | 8 | 918 | 1 | 3155 | 1 |
| 10−70 to 10−40 | 4 | 3475 | 2 | 1090 | 9 |
| 10−40 to 10−5 | 4 | 1198 | 1 | 1605 | 1 |
| All homologs | 16(6) | 5591(549) | 4(2) | 5850(1656) | 11(9) |
| FMO-like plant | |||||
| 0 to 10−70 | 0 | 0 | 512 | 0 | 0 |
| 10−70 to 10−40 | 0 | 0 | 16 | 4 | 0 |
| 10−40 to 10−5 | 2486 | 417 | 302 | 2052 | 0 |
| All homologs | 2486(409) | 417(244) | 831(106) | 2056(1306) | 0 |
| FMO-like bacteria | |||||
| 0 to 10−70 | 0 | 0 | 0 | 329 | 0 |
| 10−70 to 10−40 | 2 | 0 | 612 | 537 | 0 |
| 10−40 to 10−05 | 2917 | 1305 | 993 | 8660 | 0 |
| All homologs | 2919(187) | 1305(433) | 1605(114) | 9526(837) | 0 |
| Taxa | GAQ82387.1 Homologs in the 1KP Database | GAQ82387.1 Homologs in the NCBI Database | YUCCA Homologs in the 1KP Database | YUCCA Homologs in the NCBI Database |
|---|---|---|---|---|
| Eudicots (596) | 4(3) | - | 434(333) | 1680(114) |
| Monocots (104) | 1(1) | - | 47(35) | 444(26) |
| Conifers (73) | 1(1) | - | 4(4) | - |
| Cycadales (4) | - | - | 2(2) | - |
| Leptosporangiate monilophytes (65) | 90(33) | - | 25(19) | - |
| Eusporangiate monilophytes (12) | 1(1) | - | - | - |
| Lycophytes (22) | 5(4) | 6(1) | - | 10(1) |
| Hornworts (9) | 9(5) | - | 3(3) | - |
| Liverworts (28) | 17(9) | 2(1) | 10(10) | 6(2) |
| Bryophyta (41) | 1(1) | - | 24(18) | 6(1) |
| Zygnemophyceae (5) | - | - | - | - |
| Coleochaetophyceae (3) | - | - | - | - |
| Charophyceae (1) | - | - | - | - |
| Mesostigmatophyceae (1) | - | - | - | - |
| Chlorokybophyceae (1) | - | - | - | - |
| Kebsormidiophyceae (2) | 2(2) | 1(1) | - | - |
| Green algae (152) | 5(4) | 1(1) | - | - |
| Red algae (28) | 3(3) | - | - | - |
| Species Identifier in the 1000 Plants (1KP) Database | Taxa | Number of GAQ82387.1 Homologs | Number of YUCCA Homologs |
|---|---|---|---|
| TVSH_201823_Bituminaria_bituminosa | Core Eudicots/Fabaceae | 1 | 2 |
| WWQZ_211706_Medinilla_magnifica | Core Eudicots/Myrtiflorae | 1 | 1 |
| OCWZ_200432_Dioscorea_villosa | Monocots/Dioscoreaceae | 1 | 2 |
| AFPO_201018_Blechnum_spicant | Leptosporangiate monilophytes | 4 | 1 |
| BMJR_200209_Adiantum_tenerum | Leptosporangiate monilophytes | 2 | 2 |
| DCDT_207190_Cheilanthes_arizonica | Leptosporangiate monilophytes | 3 | 1 |
| FLTD_200266_Pteris_ensigormis | Leptosporangiate monilophytes | 1 | 2 |
| GANB_201380_Cyathea_spinulosa | Leptosporangiate monilophytes | 4 | 1 |
| KIIX_201108_Pilularia_globulifera | Leptosporangiate monilophytes | 2 | 1 |
| KJZG_200972_Asplenium_platyneuron | Leptosporangiate monilophytes | 5 | 1 |
| NDUV_201591_Vittaria_appalachiana | Leptosporangiate monilophytes | 1 | 2 |
| NOKI_201577_Lindsaea_linearis | Leptosporangiate monilophytes | 5 | 1 |
| PNZO_215202_Culcita_macrocarpa | Leptosporangiate monilophytes | 1 | 1 |
| RICC_200988_Cystopteris_reevesiana | Leptosporangiate monilophytes | 3 | 1 |
| UFJN_208949_Diplazium_wichurae | Leptosporangiate monilophytes | 3 | 1 |
| UOMY_200602_Osmunda_sp. | Leptosporangiate monilophytes | 1 | 1 |
| WQML_200900_Cryptogramma_ acrostichoides | Leptosporangiate monilophytes | 1 | 2 |
| YLJA_207326_Polypodium_amorphum | Leptosporangiate monilophytes | 2 | 1 |
| RXRQ_201835_Phaeoceros_carolinianus | Hornworts | 3 | 1 |
| TCBC_200001_Megaceros_vincentianus | Hornworts | 1 | 1 |
| HMHL_201008_Marchantia_paleacea | Liverworts | 2 | 1 |
| ILBQ_200700_Conocephalum_conicum | Liverworts | 2 | 1 |
| TXVB_207470_Lunularia_cruciata | Liverworts | 3 | 1 |
| RCBT_Sphagnum palustre | Mosses | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turnaev, I.I.; Gunbin, K.V.; Suslov, V.V.; Akberdin, I.R.; Kolchanov, N.A.; Afonnikov, D.A. The Phylogeny of Class B Flavoprotein Monooxygenases and the Origin of the YUCCA Protein Family. Plants 2020, 9, 1092. https://doi.org/10.3390/plants9091092
Turnaev II, Gunbin KV, Suslov VV, Akberdin IR, Kolchanov NA, Afonnikov DA. The Phylogeny of Class B Flavoprotein Monooxygenases and the Origin of the YUCCA Protein Family. Plants. 2020; 9(9):1092. https://doi.org/10.3390/plants9091092
Chicago/Turabian StyleTurnaev, Igor I., Konstantin V. Gunbin, Valentin V. Suslov, Ilya R. Akberdin, Nikolay A. Kolchanov, and Dmitry A. Afonnikov. 2020. "The Phylogeny of Class B Flavoprotein Monooxygenases and the Origin of the YUCCA Protein Family" Plants 9, no. 9: 1092. https://doi.org/10.3390/plants9091092
APA StyleTurnaev, I. I., Gunbin, K. V., Suslov, V. V., Akberdin, I. R., Kolchanov, N. A., & Afonnikov, D. A. (2020). The Phylogeny of Class B Flavoprotein Monooxygenases and the Origin of the YUCCA Protein Family. Plants, 9(9), 1092. https://doi.org/10.3390/plants9091092






