A Block Coordinate Descent-Based Projected Gradient Algorithm for Orthogonal Non-Negative Matrix Factorization
Abstract
:1. Introduction
1.1. Motivation
1.2. Problem Formulation
1.3. Related Work
1.4. Our Contribution
- We develop an algorithm for (ONMF) and (bi-ONMF), which is essentially a BCD algorithm, in the literature also known as alternating minimization, coordinate relaxation, the Gauss-Seidel method, subspace correction, domain decomposition, etc., see e.g., [35,36]. For each block optimization, we use a PG method and Armijo rule to find a suitable step-size.
- We construct synthetic data sets of instances for (ONMF) and (bi-ONMF), for which we know the optimum value by construction.
- The implemented algorithms are compared on the constructed synthetic data-sets in terms of: (i) the accuracy of the reconstruction, and (ii) the deviation of the factors from the orthonormality. This deviation is a measure the feasibility of the obtained solution and has not been analyzed in the work Of Ding [11] and Mirzal [28]. Accuracy is measured by the so-called root-square error (RSE), defined asPlease note that we added 1 to the denominator in the formula above to prevent numerical difficulties when the data matrix R has a very small Frobenius norm.Deviations from the orthonormality are computed using Formulas (17) and (18) from Section 4. Our numerical results show that our algorithm is very competitive and almost always outperforms the MU based algorithms.
1.5. Notations
1.6. Structure of the Paper
2. Existing Methods to Solve (NMF)
2.1. The Method of Ding
| Algorithm 1: Ding’s MU algorithm for (bi-ONMF). |
| INPUT: , |
| 1. Initialize: generate as an random matrix and as a random matrix. |
| 2. Repeat |
| 3. Until convergence or a maximum number of iterations or maximum time is reached. |
| OUTPUT:. |
2.2. The Method of Mirzal
| Algorithm 2: Mirzal’s algorithm for bi-ONMF [40] |
| INPUT: inner dimension p, maximum number of iterations: maxit; small positive , small positive to increase . |
| 1. Compute initial and . |
| 2. For |
| ; |
| Repeat |
| ; |
| ; |
| Until |
| ; |
| Repeat |
| ; |
| ; |
| Until |
| ; |
| OUTPUT: . |
3. PG Method for (ONMF) and (bi-ONMF)
3.1. Main Steps of PG Method
| Algorithm 3: BCD method for (pen-ONMF) |
| INPUT: inner dimension p, initial matrices . |
| 1. Set . |
| 2. Repeat |
| Fix and compute new G as follows: |
| Fix and compute new H as follows: |
| 3. Until some stopping criteria is satisfied |
| OUTPUT: . |
| Algorithm 4: PG method using Armijo rule to solve sub-problem (9) |
| INPUT: , and initial . |
| 1. Set |
| 2. Repeat |
| Find a (using updating factor ) such that for |
| we have |
| Set |
| Set ; |
| 3. Until some stopping criteria is satisfied. |
| OUTPUT: . |
3.2. Stopping Criteria for Algorithms 3 and 4
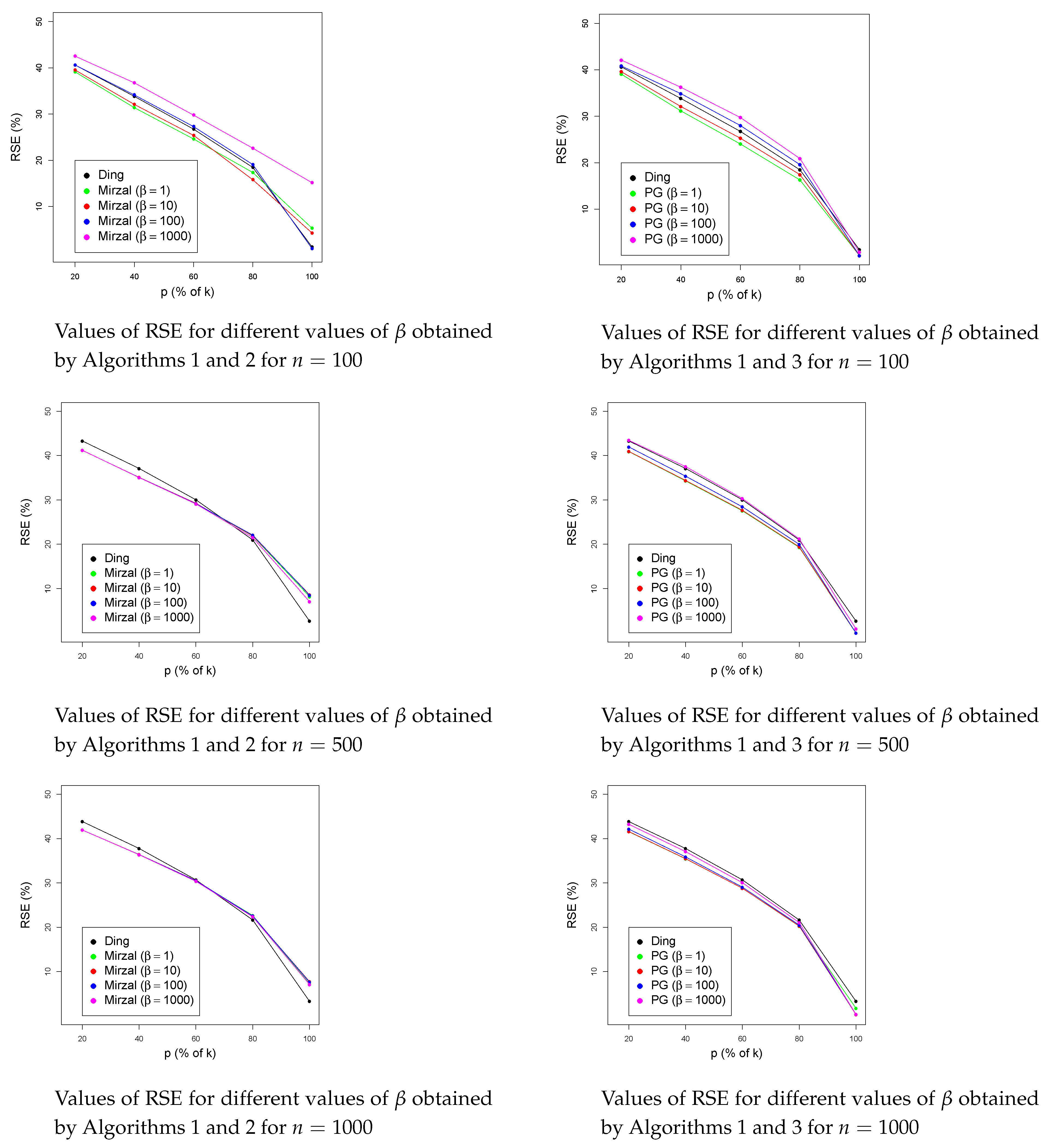
4. Numerical Results
4.1. Artificial Data
4.2. Numerical Results for UNION
4.3. Numerical Results for Bi-Orthonormal Data (BION)
4.4. Numerical Results on the Noisy BION Dataset
4.5. Time Complexity of All Algorithms
5. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berry, M.W.; Browne, M.; Langville, A.N.; Pauca, V.P.; Plemmons, R.J. Algorithms and applications for approximate nonnegative matrix factorization. Comput. Stat. Data Anal. 2007, 52, 155–173. [Google Scholar] [CrossRef] [Green Version]
- Pauca, V.P.; Shahnaz, F.; Berry, M.W.; Plemmons, R.J. Text mining using non-negative matrix factorizations. In Proceedings of the 2004 SIAM International Conference on Data Mining, Lake Buena Vista, FL, USA, 22–24 April 2004; pp. 452–456. [Google Scholar]
- Shahnaz, F.; Berry, M.W.; Pauca, V.P.; Plemmons, R.J. Document clustering using nonnegative matrix factorization. Inf. Process. Manag. 2006, 42, 373–386. [Google Scholar] [CrossRef]
- Berry, M.W.; Gillis, N.; Glineur, F. Document classification using nonnegative matrix factorization and underapproximation. In Proceedings of the 2009 IEEE International Symposium on Circuits and Systems, Taipei, Taiwan, 24–27 May 2009; pp. 2782–2785. [Google Scholar]
- Li, T.; Ding, C. The relationships among various nonnegative matrix factorization methods for clustering. In Proceedings of the IEEE Sixth International Conference on Data Mining (ICDM’06), Hong Kong, China, 18–22 December 2006; pp. 362–371. [Google Scholar]
- Xu, W.; Liu, X.; Gong, Y. Document clustering based on non-negative matrix factorization. In Proceedings of the 26th Annual International ACM SIGIR Conference on Research and Development in Informaion Retrieval, Toronto, ON, Canada, 28 July–1 August 2003; pp. 267–273. [Google Scholar]
- Kaarna, A. Non-negative matrix factorization features from spectral signatures of AVIRIS images. In Proceedings of the 2006 IEEE International Symposium on Geoscience and Remote Sensing, Denver, CO, USA, 31 July–4 August 2006; pp. 549–552. [Google Scholar]
- Zafeiriou, S.; Tefas, A.; Buciu, I.; Pitas, I. Exploiting discriminant information in nonnegative matrix factorization with application to frontal face verification. IEEE Trans. Neural Netw. 2006, 17, 683–695. [Google Scholar] [CrossRef] [Green Version]
- Golub, G.H.; Reinsch, C. Singular value decomposition and least squares solutions. In Linear Algebra; Springer: Berlin, Germany, 1971; pp. 134–151. [Google Scholar]
- Jolliffe, I. Principal Component Analysis; Wiley Online Library: Hoboken, NJ, USA, 2005. [Google Scholar]
- Ding, C.; Li, T.; Peng, W.; Park, H. Orthogonal nonnegative matrix t-factorizations for clustering. In Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Philadelphia, PA, USA, 20–23 August 2006; pp. 126–135. [Google Scholar]
- Gillis, N. Nonnegative Matrix Factorization; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2020. [Google Scholar]
- Paatero, P.; Tapper, U. Positive matrix factorization: A non-negative factor model with optimal utilization of error estimates of data values. Environmetrics 1994, 5, 111–126. [Google Scholar] [CrossRef]
- Anttila, P.; Paatero, P.; Tapper, U.; Järvinen, O. Source identification of bulk wet deposition in Finland by positive matrix factorization. Atmos. Environ. 1995, 29, 1705–1718. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.D.; Seung, H.S. Algorithms for non-negative matrix factorization. In Proceedings of the Advances in Neural Information Processing Systems, Denver, CO, USA, 27 November–2 December 2000; pp. 556–562. [Google Scholar]
- Chu, M.; Diele, F.; Plemmons, R.; Ragni, S. Optimality, computation, and interpretation of nonnegative matrix factorizations. SIAM J. Matrix Anal. 2004, 4, 8030. [Google Scholar]
- Kim, H.; Park, H. Nonnegative matrix factorization based on alternating nonnegativity constrained least squares and active set method. SIAM J. Matrix Anal. Appl. 2008, 30, 713–730. [Google Scholar] [CrossRef]
- Lin, C. Projected gradient methods for nonnegative matrix factorization. Neural Comput. 2007, 19, 2756–2779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cichocki, A.; Zdunek, R.; Amari, S.i. Hierarchical ALS algorithms for nonnegative matrix and 3D tensor factorization. In International Conference on Independent Component Analysis and Signal Separation; Springer: Berlin, Germany, 2007; pp. 169–176. [Google Scholar]
- Halko, N.; Martinsson, P.G.; Tropp, J.A. Finding structure with randomness: Probabilistic algorithms for constructing approximate matrix decompositions. SIAM Rev. 2011, 53, 217–288. [Google Scholar] [CrossRef]
- Yoo, J.; Choi, S. Orthogonal nonnegative matrix factorization: Multiplicative updates on Stiefel manifolds. In International Conference on Intelligent Data Engineering and Automated Learning; Springer: Berlin, Germany, 2008; pp. 140–147. [Google Scholar]
- Yoo, J.; Choi, S. Orthogonal nonnegative matrix tri-factorization for co-clustering: Multiplicative updates on stiefel manifolds. Inf. Process. Manag. 2010, 46, 559–570. [Google Scholar] [CrossRef]
- Choi, S. Algorithms for orthogonal nonnegative matrix factorization. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 1828–1832. [Google Scholar]
- Kim, D.; Sra, S.; Dhillon, I.S. Fast Projection-Based Methods for the Least Squares Nonnegative Matrix Approximation Problem. Stat. Anal. Data Mining 2008, 1, 38–51. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Sra, S.; Dhillon, I.S. Fast Newton-type methods for the least squares nonnegative matrix approximation problem. In Proceedings of the 2007 SIAM International Conference on Data Mining, Minneapolis, MN, USA, 26–28 April 2007; pp. 343–354. [Google Scholar]
- Kim, J.; Park, H. Toward faster nonnegative matrix factorization: A new algorithm and comparisons. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15–19 December 2008; pp. 353–362. [Google Scholar]
- Mirzal, A. A convergent algorithm for orthogonal nonnegative matrix factorization. J. Comput. Appl. Math. 2014, 260, 149–166. [Google Scholar] [CrossRef]
- Lin, C.J. On the convergence of multiplicative update algorithms for nonnegative matrix factorization. IEEE Trans. Neural Netw. 2007, 18, 1589–1596. [Google Scholar]
- Esposito, F.; Boccarelli, A.; Del Buono, N. An NMF-Based methodology for selecting biomarkers in the landscape of genes of heterogeneous cancer-associated fibroblast Populations. Bioinform. Biol. Insights 2020, 14, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Ser, W.; Chen, B.; Lin, Z. Robust orthogonal nonnegative matrix tri-factorization for data representation. Knowl.-Based Syst. 2020, 201, 106054. [Google Scholar] [CrossRef]
- Leplat, N.V.; Gillis, A.A. Blind audio source separation with minimum-volume beta-divergence NMF. IEEE Trans. Signal Process. 2020, 68, 3400–3410. [Google Scholar] [CrossRef]
- Casalino, G.; Coluccia, M.; Pati, M.L.; Pannunzio, A.; Vacca, A.; Scilimati, A.; Perrone, M.G. Intelligent microarray data analysis through non-negative matrix factorization to study human multiple myeloma cell lines. Appl. Sci. 2019, 9, 5552. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.; Luo, L.; Li, H. Orthogonal incremental non-negative matrix factorization algorithm and its application in image classification. Comput. Appl. Math. 2020, 39, 1–16. [Google Scholar] [CrossRef]
- Bertsekas, D. Nonlinear Programming; Athena Scientific optimization and Computation Series; Athena Scientific: Nashua, NH, USA, 2016. [Google Scholar]
- Richtárik, P.; Takác, M. Iteration complexity of randomized block-coordinate descent methods for minimizing a composite function. Math. Program. 2014, 144, 1–38. [Google Scholar] [CrossRef] [Green Version]
- The MathWorks. MATLAB Version R2019a; The MathWorks: Natick, MA, USA, 2019. [Google Scholar]
- Cichocki, A.; Zdunek, R.; Phan, A.H.; Amari, S.i. Nonnegative Matrix and Tensor Factorizations: Applications to Exploratory Multi-Way Data Analysis and Blind Source Separation; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Piper, J.; Pauca, V.P.; Plemmons, R.J.; Giffin, M. Object Characterization from Spectral Data Using Nonnegative Factorization and Information theory. In Proceedings of the AMOS Technical Conference; 2004. Available online: http://users.wfu.edu/plemmons/papers/Amos2004_2.pdf (accessed on 8 November 2020).
- Mirzal, A. A Convergent Algorithm for Bi-orthogonal Nonnegative Matrix Tri-Factorization. arXiv 2017, arXiv:1710.11478. [Google Scholar]
- Armijo, L. Minimization of functions having Lipschitz continuous first partial derivatives. Pac. J. Math. 1966, 16, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.J.; Moré, J.J. Newton’s method for large bound-constrained optimization problems. SIAM J. Optim. 1999, 9, 1100–1127. [Google Scholar] [CrossRef] [Green Version]





| n | 50 | 100 | 200 | 500 | 1000 |
|---|---|---|---|---|---|
| k | 10 | 20 | 40 | 100 | 200 |
| k | 20 | 40 | 80 | 200 | 400 |
| n | p | RSE of | RSE of Algorithm 2 | RSE of Algorithm 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (% of k) | Algorithm 1 | |||||||||
| 50 | 40 | 0.3143 | 0.2965 | 0.3070 | 0.3329 | 0.3898 | 0.2963 | 0.3081 | 0.3425 | 0.3508 |
| 50 | 60 | 0.2348 | 0.2227 | 0.2356 | 0.2676 | 0.3459 | 0.2201 | 0.2382 | 0.2733 | 0.2765 |
| 50 | 80 | 0.1738 | 0.1492 | 0.1634 | 0.1894 | 0.3277 | 0.1468 | 0.1620 | 0.1953 | 0.2053 |
| 50 | 100 | 0.0002 | 0.0133 | 0.0004 | 0.0932 | 0.2973 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| 100 | 20 | 0.4063 | 0.3914 | 0.3955 | 0.4063 | 0.4254 | 0.3906 | 0.3959 | 0.4083 | 0.4210 |
| 100 | 40 | 0.3384 | 0.3139 | 0.3210 | 0.3415 | 0.3677 | 0.3116 | 0.3210 | 0.3488 | 0.3625 |
| 100 | 60 | 0.2674 | 0.2462 | 0.2541 | 0.2730 | 0.2978 | 0.2403 | 0.2528 | 0.2801 | 0.2974 |
| 100 | 80 | 0.1847 | 0.1737 | 0.1581 | 0.1909 | 0.2263 | 0.1629 | 0.1744 | 0.1959 | 0.2090 |
| 100 | 100 | 0.0126 | 0.0532 | 0.0427 | 0.0089 | 0.1515 | 0.0000 | 0.0000 | 0.0000 | 0.0075 |
| 200 | 20 | 0.4213 | 0.4024 | 0.4077 | 0.4080 | 0.4257 | 0.4005 | 0.4032 | 0.4162 | 0.4337 |
| 200 | 40 | 0.3562 | 0.3315 | 0.3398 | 0.3401 | 0.3647 | 0.3270 | 0.3313 | 0.3497 | 0.3738 |
| 200 | 60 | 0.2845 | 0.2675 | 0.2746 | 0.2748 | 0.2955 | 0.2573 | 0.2617 | 0.2812 | 0.3061 |
| 200 | 80 | 0.1959 | 0.1958 | 0.2013 | 0.1996 | 0.2085 | 0.1773 | 0.1819 | 0.1960 | 0.2133 |
| 200 | 100 | 0.0191 | 0.0753 | 0.0632 | 0.0622 | 0.0415 | 0.0000 | 0.0000 | 0.0069 | 0.0181 |
| 500 | 20 | 0.4332 | 0.4120 | 0.4119 | 0.4120 | 0.4121 | 0.4092 | 0.4096 | 0.4197 | 0.4346 |
| 500 | 40 | 0.3711 | 0.3506 | 0.3509 | 0.3507 | 0.3505 | 0.3430 | 0.3440 | 0.3537 | 0.3753 |
| 500 | 60 | 0.3003 | 0.2919 | 0.2923 | 0.2916 | 0.2909 | 0.2756 | 0.2766 | 0.2845 | 0.3031 |
| 500 | 80 | 0.2098 | 0.2186 | 0.2192 | 0.2207 | 0.2151 | 0.1931 | 0.1941 | 0.1999 | 0.2122 |
| 500 | 100 | 0.0273 | 0.0822 | 0.0864 | 0.0853 | 0.0713 | 0.0002 | 0.0003 | 0.0002 | 0.0097 |
| 1000 | 20 | 0.4386 | 0.4195 | 0.4194 | 0.4193 | 0.4195 | 0.4156 | 0.4160 | 0.4216 | 0.4324 |
| 1000 | 40 | 0.3777 | 0.3641 | 0.3640 | 0.3638 | 0.3637 | 0.3545 | 0.3548 | 0.3588 | 0.3707 |
| 1000 | 60 | 0.3070 | 0.3047 | 0.3055 | 0.3051 | 0.3036 | 0.2881 | 0.2880 | 0.2906 | 0.3006 |
| 1000 | 80 | 0.2164 | 0.2265 | 0.2248 | 0.2254 | 0.2236 | 0.2024 | 0.2029 | 0.2050 | 0.2106 |
| 1000 | 100 | 0.0329 | 0.0725 | 0.0772 | 0.0761 | 0.0709 | 0.0173 | 0.0030 | 0.0035 | 0.0035 |
| n | p | Infeas. of | Infeas. of Algorithm 2 | Infeas. of Algorithm 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (% of k) | Algorithm 1 | |||||||||
| 50 | 20 | 0.0964 | 0.2490 | 0.0924 | 0.0155 | 0.0038 | 0.2298 | 0.0909 | 0.0154 | 0.0022 |
| 50 | 40 | 0.0740 | 0.1886 | 0.0676 | 0.0131 | 0.0040 | 0.1845 | 0.0670 | 0.0135 | 0.0023 |
| 50 | 60 | 0.0553 | 0.1324 | 0.0465 | 0.0068 | 0.0040 | 0.1245 | 0.0440 | 0.0091 | 0.0015 |
| 50 | 80 | 0.0324 | 0.0964 | 0.0241 | 0.0053 | 0.0034 | 0.0789 | 0.0250 | 0.0069 | 0.0020 |
| 50 | 100 | 0.0023 | 0.0257 | 0.0022 | 0.0023 | 0.0039 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| 100 | 20 | 0.0774 | 0.2624 | 0.1441 | 0.0258 | 0.0064 | 0.2588 | 0.1308 | 0.0258 | 0.0036 |
| 100 | 40 | 0.0539 | 0.1754 | 0.0928 | 0.0168 | 0.0036 | 0.1654 | 0.0819 | 0.0182 | 0.0035 |
| 100 | 60 | 0.0400 | 0.1205 | 0.0545 | 0.0102 | 0.0024 | 0.1109 | 0.0487 | 0.0138 | 0.0033 |
| 100 | 80 | 0.0239 | 0.0890 | 0.0324 | 0.0062 | 0.0022 | 0.0623 | 0.0258 | 0.0083 | 0.0018 |
| 100 | 100 | 0.0062 | 0.0452 | 0.0153 | 0.0009 | 0.0016 | 0.0002 | 0.0000 | 0.0000 | 0.0000 |
| 200 | 20 | 0.0584 | 0.2157 | 0.1437 | 0.1433 | 0.0054 | 0.2087 | 0.1512 | 0.0348 | 0.0074 |
| 200 | 40 | 0.0356 | 0.1379 | 0.1004 | 0.1000 | 0.0036 | 0.1240 | 0.0806 | 0.0207 | 0.0053 |
| 200 | 60 | 0.0260 | 0.0955 | 0.0791 | 0.0793 | 0.0031 | 0.0754 | 0.0434 | 0.0143 | 0.0047 |
| 200 | 80 | 0.0154 | 0.0657 | 0.0634 | 0.0629 | 0.0017 | 0.0416 | 0.0218 | 0.0080 | 0.0026 |
| 200 | 100 | 0.0059 | 0.0412 | 0.0517 | 0.0512 | 0.0016 | 0.0002 | 0.0001 | 0.0002 | 0.0001 |
| 500 | 20 | 0.0332 | 0.1587 | 0.1894 | 0.1908 | 0.1908 | 0.1475 | 0.1268 | 0.0436 | 0.0087 |
| 500 | 40 | 0.0189 | 0.1155 | 0.1343 | 0.1349 | 0.1347 | 0.0770 | 0.0621 | 0.0227 | 0.0069 |
| 500 | 60 | 0.0134 | 0.0889 | 0.1095 | 0.1102 | 0.1055 | 0.0412 | 0.0312 | 0.0123 | 0.0038 |
| 500 | 80 | 0.0084 | 0.0656 | 0.0946 | 0.0954 | 0.0826 | 0.0300 | 0.0154 | 0.0061 | 0.0021 |
| 500 | 100 | 0.0050 | 0.0499 | 0.0847 | 0.0853 | 0.0693 | 0.0249 | 0.0003 | 0.0001 | 0.0001 |
| 1000 | 20 | 0.0211 | 0.1200 | 0.1344 | 0.1349 | 0.1350 | 0.1043 | 0.0970 | 0.0471 | 0.0097 |
| 1000 | 40 | 0.0122 | 0.0863 | 0.0951 | 0.0954 | 0.0954 | 0.0542 | 0.0422 | 0.0199 | 0.0059 |
| 1000 | 60 | 0.0073 | 0.0662 | 0.0776 | 0.0779 | 0.0779 | 0.0414 | 0.0205 | 0.0098 | 0.0037 |
| 1000 | 80 | 0.0045 | 0.0539 | 0.0671 | 0.0675 | 0.0675 | 0.0336 | 0.0103 | 0.0047 | 0.0018 |
| 1000 | 100 | 0.0040 | 0.0475 | 0.0600 | 0.0603 | 0.0604 | 0.0296 | 0.0066 | 0.0005 | 0.0003 |
| n | p | RSE of | RSE of Algorithm 2 | RSE of Algorithm 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (% of k) | Algorithm 1 | |||||||||
| 50 | 20 | 0.7053 | 0.7053 | 0.7053 | 0.7053 | 0.8283 | 0.7053 | 0.7053 | 0.7055 | 0.8259 |
| 50 | 40 | 0.6108 | 0.6108 | 0.6108 | 0.6108 | 0.9066 | 0.6108 | 0.6108 | 0.6108 | 0.6631 |
| 50 | 60 | 0.4987 | 0.4987 | 0.4987 | 0.5442 | 0.9665 | 0.4987 | 0.4987 | 0.4987 | 0.5000 |
| 50 | 80 | 0.3526 | 0.3671 | 0.3742 | 0.4497 | 1.0282 | 0.3526 | 0.3796 | 0.3527 | 0.4374 |
| 50 | 100 | 0.0607 | 0.1712 | 0.2786 | 0.5198 | 1.0781 | 0.1145 | 0.1820 | 0.2604 | 0.3689 |
| 100 | 20 | 0.7516 | 0.7516 | 0.7516 | 0.7517 | 0.9070 | 0.7516 | 0.7516 | 0.7517 | 0.8224 |
| 100 | 40 | 0.6509 | 0.6509 | 0.6509 | 0.7174 | 0.9779 | 0.6509 | 0.6509 | 0.6509 | 0.6514 |
| 100 | 60 | 0.5315 | 0.5315 | 0.5315 | 0.5504 | 1.0401 | 0.5315 | 0.5315 | 0.5315 | 0.5352 |
| 100 | 80 | 0.3758 | 0.3787 | 0.4106 | 0.4542 | 1.1082 | 0.3801 | 0.3888 | 0.3917 | 0.3898 |
| 100 | 100 | 0.1377 | 0.1993 | 0.3311 | 0.4898 | 1.1734 | 0.0457 | 0.1016 | 0.2758 | 0.3757 |
| 200 | 20 | 0.7884 | 0.7884 | 0.7884 | 0.7884 | 0.9499 | 0.7884 | 0.7884 | 0.7884 | 0.7888 |
| 200 | 40 | 0.6828 | 0.6828 | 0.6828 | 0.6828 | 1.0325 | 0.6828 | 0.6828 | 0.6828 | 0.6828 |
| 200 | 60 | 0.5575 | 0.5575 | 0.5575 | 0.5647 | 1.0938 | 0.5575 | 0.5575 | 0.5575 | 0.5610 |
| 200 | 80 | 0.3942 | 0.3942 | 0.3965 | 0.5019 | 1.1618 | 0.3942 | 0.3942 | 0.3942 | 0.4373 |
| 200 | 100 | 0.1447 | 0.1851 | 0.3014 | 0.5400 | 1.2297 | 0.0202 | 0.1429 | 0.2964 | 0.3315 |
| 500 | 20 | 0.8242 | 0.8242 | 0.8242 | 0.8242 | 0.9956 | 0.8242 | 0.8242 | 0.8242 | 0.8243 |
| 500 | 40 | 0.7138 | 0.7138 | 0.7138 | 0.7138 | 1.0679 | 0.7138 | 0.7138 | 0.7138 | 0.7138 |
| 500 | 60 | 0.5828 | 0.5828 | 0.5828 | 0.6045 | 1.1534 | 0.5828 | 0.5828 | 0.5828 | 0.5828 |
| 500 | 80 | 0.4121 | 0.4121 | 0.4203 | 0.5285 | 1.2160 | 0.4121 | 0.4121 | 0.4121 | 0.4334 |
| 500 | 100 | 0.1405 | 0.1814 | 0.3401 | 0.5854 | 1.2822 | 0.0067 | 0.1059 | 0.2044 | 0.3378 |
| 1000 | 20 | 0.8436 | 0.8436 | 0.8436 | 0.8436 | 1.0261 | 0.8436 | 0.8436 | 0.8436 | 0.8436 |
| 1000 | 40 | 0.7306 | 0.7306 | 0.7306 | 0.7309 | 1.0916 | 0.7306 | 0.7306 | 0.7306 | 0.7306 |
| 1000 | 60 | 0.5965 | 0.5965 | 0.5965 | 0.6121 | 1.1669 | 0.5965 | 0.5965 | 0.5965 | 0.5968 |
| 1000 | 80 | 0.4218 | 0.4218 | 0.4256 | 0.5338 | 1.2389 | 0.4218 | 0.4218 | 0.4218 | 0.4397 |
| 1000 | 100 | 0.1346 | 0.1635 | 0.3324 | 0.5755 | 1.3080 | 0.0096 | 0.0697 | 0.1661 | 0.2188 |
| n | p | Infeas. of | Infeas. of Algorithm 2 | Infeas. of Algorithm 3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (% of k) | Algorithm 1 | |||||||||
| 50 | 20 | 0.0001 | 0.0070 | 0.0036 | 0.0010 | 0.0068 | 0.0017 | 0.0021 | 0.0021 | 0.0026 |
| 50 | 40 | 0.0000 | 0.0041 | 0.0021 | 0.0004 | 0.0056 | 0.0008 | 0.0012 | 0.0012 | 0.0014 |
| 50 | 60 | 0.0000 | 0.0030 | 0.0009 | 0.0032 | 0.0038 | 0.0005 | 0.0008 | 0.0009 | 0.0009 |
| 50 | 80 | 0.0000 | 0.0183 | 0.0030 | 0.0021 | 0.0028 | 0.0004 | 0.0202 | 0.0006 | 0.0013 |
| 50 | 100 | 0.0355 | 0.0533 | 0.0127 | 0.0045 | 0.0027 | 0.0418 | 0.0478 | 0.0123 | 0.0021 |
| 100 | 20 | 0.0001 | 0.0051 | 0.0024 | 0.0006 | 0.0063 | 0.0010 | 0.0012 | 0.0013 | 0.0016 |
| 100 | 40 | 0.0000 | 0.0029 | 0.0017 | 0.0066 | 0.0040 | 0.0004 | 0.0006 | 0.0007 | 0.0007 |
| 100 | 60 | 0.0000 | 0.0019 | 0.0008 | 0.0009 | 0.0027 | 0.0003 | 0.0004 | 0.0005 | 0.0005 |
| 100 | 80 | 0.0000 | 0.0039 | 0.0048 | 0.0015 | 0.0021 | 0.0062 | 0.0149 | 0.0037 | 0.0006 |
| 100 | 100 | 0.0606 | 0.0454 | 0.0105 | 0.0022 | 0.0018 | 0.0106 | 0.0228 | 0.0173 | 0.0028 |
| 200 | 20 | 0.0002 | 0.0033 | 0.0019 | 0.0005 | 0.0043 | 0.0005 | 0.0007 | 0.0007 | 0.0007 |
| 200 | 40 | 0.0001 | 0.0017 | 0.0010 | 0.0002 | 0.0027 | 0.0002 | 0.0003 | 0.0004 | 0.0003 |
| 200 | 60 | 0.0001 | 0.0010 | 0.0005 | 0.0004 | 0.0019 | 0.0001 | 0.0002 | 0.0002 | 0.0004 |
| 200 | 80 | 0.0000 | 0.0006 | 0.0006 | 0.0015 | 0.0014 | 0.0001 | 0.0001 | 0.0002 | 0.0013 |
| 200 | 100 | 0.0425 | 0.0280 | 0.0064 | 0.0019 | 0.0015 | 0.0046 | 0.0224 | 0.0240 | 0.0034 |
| 500 | 20 | 0.0001 | 0.0017 | 0.0011 | 0.0003 | 0.0025 | 0.0002 | 0.0003 | 0.0003 | 0.0003 |
| 500 | 40 | 0.0001 | 0.0008 | 0.0005 | 0.0001 | 0.0016 | 0.0001 | 0.0001 | 0.0002 | 0.0002 |
| 500 | 60 | 0.0000 | 0.0005 | 0.0003 | 0.0006 | 0.0013 | 0.0001 | 0.0001 | 0.0001 | 0.0002 |
| 500 | 80 | 0.0000 | 0.0003 | 0.0009 | 0.0009 | 0.0008 | 0.0000 | 0.0001 | 0.0001 | 0.0016 |
| 500 | 100 | 0.0258 | 0.0184 | 0.0045 | 0.0013 | 0.0007 | 0.0017 | 0.0101 | 0.0175 | 0.0053 |
| 1000 | 20 | 0.0001 | 0.0010 | 0.0006 | 0.0002 | 0.0024 | 0.0001 | 0.0002 | 0.0002 | 0.0002 |
| 1000 | 40 | 0.0000 | 0.0005 | 0.0003 | 0.0001 | 0.0009 | 0.0001 | 0.0002 | 0.0003 | 0.0002 |
| 1000 | 60 | 0.0000 | 0.0003 | 0.0002 | 0.0004 | 0.0009 | 0.0003 | 0.0002 | 0.0003 | 0.0003 |
| 1000 | 80 | 0.0000 | 0.0002 | 0.0005 | 0.0007 | 0.0006 | 0.0040 | 0.0001 | 0.0002 | 0.0020 |
| 1000 | 100 | 0.0173 | 0.0117 | 0.0031 | 0.0009 | 0.0005 | 0.0043 | 0.0050 | 0.0121 | 0.0060 |
| Algorithm 1 | Algorithm 2 | Algorithm 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RSE | RSE | RSE | |||||||||
| 20 | 200 | 0.7877 | 0.0019 | 0.0019 | 0.7877 | 0.0041 | 0.0041 | 0.7877 | 0.0042 | 0.0042 | |
| 40 | 200 | 0.6821 | 0.0014 | 0.0014 | 0.6823 | 0.0024 | 0.0024 | 0.6821 | 0.0027 | 0.0028 | |
| 60 | 200 | 0.5570 | 0.0010 | 0.0010 | 0.5571 | 0.0015 | 0.0015 | 0.5569 | 0.0020 | 0.0020 | |
| 80 | 200 | 0.3939 | 0.0007 | 0.0007 | 0.3939 | 0.0010 | 0.0010 | 0.3938 | 0.0015 | 0.0015 | |
| 100 | 200 | 0.0072 | 0.0003 | 0.0003 | 0.1327 | 0.0097 | 0.0098 | 0.0039 | 0.0010 | 0.0010 | |
| 120 | 200 | 0.0278 | 0.0483 | 0.0483 | 0.1325 | 0.0283 | 0.0326 | 0.0036 | 0.0465 | 0.0141 | |
| 140 | 200 | 0.0344 | 0.0589 | 0.0590 | 0.1909 | 0.0366 | 0.0363 | 0.0034 | 0.0565 | 0.0188 | |
| 20 | 200 | 0.7884 | 0.0002 | 0.0002 | 0.7884 | 0.0018 | 0.0018 | 0.7884 | 0.0003 | 0.0003 | |
| 40 | 200 | 0.6828 | 0.0001 | 0.0001 | 0.6828 | 0.0009 | 0.0009 | 0.6828 | 0.0001 | 0.0001 | |
| 60 | 200 | 0.5575 | 0.0001 | 0.0001 | 0.5575 | 0.0006 | 0.0006 | 0.5575 | 0.0001 | 0.0001 | |
| 80 | 200 | 0.3942 | 0.0000 | 0.0000 | 0.3942 | 0.0004 | 0.0003 | 0.3942 | 0.0001 | 0.0001 | |
| 100 | 200 | 0.0575 | 0.0086 | 0.0086 | 0.1717 | 0.0089 | 0.0192 | 0.0001 | 0.0004 | 0.0004 | |
| 120 | 200 | 0.0043 | 0.0490 | 0.0489 | 0.1407 | 0.0275 | 0.0321 | 0.0003 | 0.0468 | 0.0159 | |
| 140 | 200 | 0.0049 | 0.0596 | 0.0596 | 0.1743 | 0.0363 | 0.0390 | 0.0003 | 0.0558 | 0.0221 | |
| 20 | 200 | 0.7884 | 0.0001 | 0.0001 | 0.7884 | 0.0017 | 0.0017 | 0.7884 | 0.0002 | 0.0002 | |
| 40 | 200 | 0.6828 | 0.0000 | 0.0000 | 0.6828 | 0.0009 | 0.0008 | 0.6828 | 0.0001 | 0.0001 | |
| 60 | 200 | 0.5575 | 0.0000 | 0.0000 | 0.5575 | 0.0005 | 0.0005 | 0.5575 | 0.0001 | 0.0001 | |
| 80 | 200 | 0.3942 | 0.0000 | 0.0000 | 0.3942 | 0.0003 | 0.0003 | 0.3942 | 0.0001 | 0.0001 | |
| 100 | 200 | 0.0624 | 0.0092 | 0.0091 | 0.1966 | 0.0159 | 0.0167 | 0.0137 | 0.0029 | 0.0006 | |
| 120 | 200 | 0.0031 | 0.0490 | 0.0492 | 0.1250 | 0.0301 | 0.0309 | 0.0003 | 0.0478 | 0.0179 | |
| 140 | 200 | 0.0051 | 0.0595 | 0.0597 | 0.1692 | 0.0367 | 0.0381 | 0.0003 | 0.0562 | 0.0268 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asadi, S.; Povh, J. A Block Coordinate Descent-Based Projected Gradient Algorithm for Orthogonal Non-Negative Matrix Factorization. Mathematics 2021, 9, 540. https://doi.org/10.3390/math9050540
Asadi S, Povh J. A Block Coordinate Descent-Based Projected Gradient Algorithm for Orthogonal Non-Negative Matrix Factorization. Mathematics. 2021; 9(5):540. https://doi.org/10.3390/math9050540
Chicago/Turabian StyleAsadi, Soodabeh, and Janez Povh. 2021. "A Block Coordinate Descent-Based Projected Gradient Algorithm for Orthogonal Non-Negative Matrix Factorization" Mathematics 9, no. 5: 540. https://doi.org/10.3390/math9050540
APA StyleAsadi, S., & Povh, J. (2021). A Block Coordinate Descent-Based Projected Gradient Algorithm for Orthogonal Non-Negative Matrix Factorization. Mathematics, 9(5), 540. https://doi.org/10.3390/math9050540







