Exploiting the Molecular Basis of Oesophageal Cancer for Targeted Therapies and Biomarkers for Drug Response: Guiding Clinical Decision-Making
Abstract
:1. Introduction
2. Altered Gene Expression in OSCC
2.1. Gene Mutations
2.2. Expression Changes
2.3. Genetic Drivers of OSCC
3. Tumor Mutational Burden
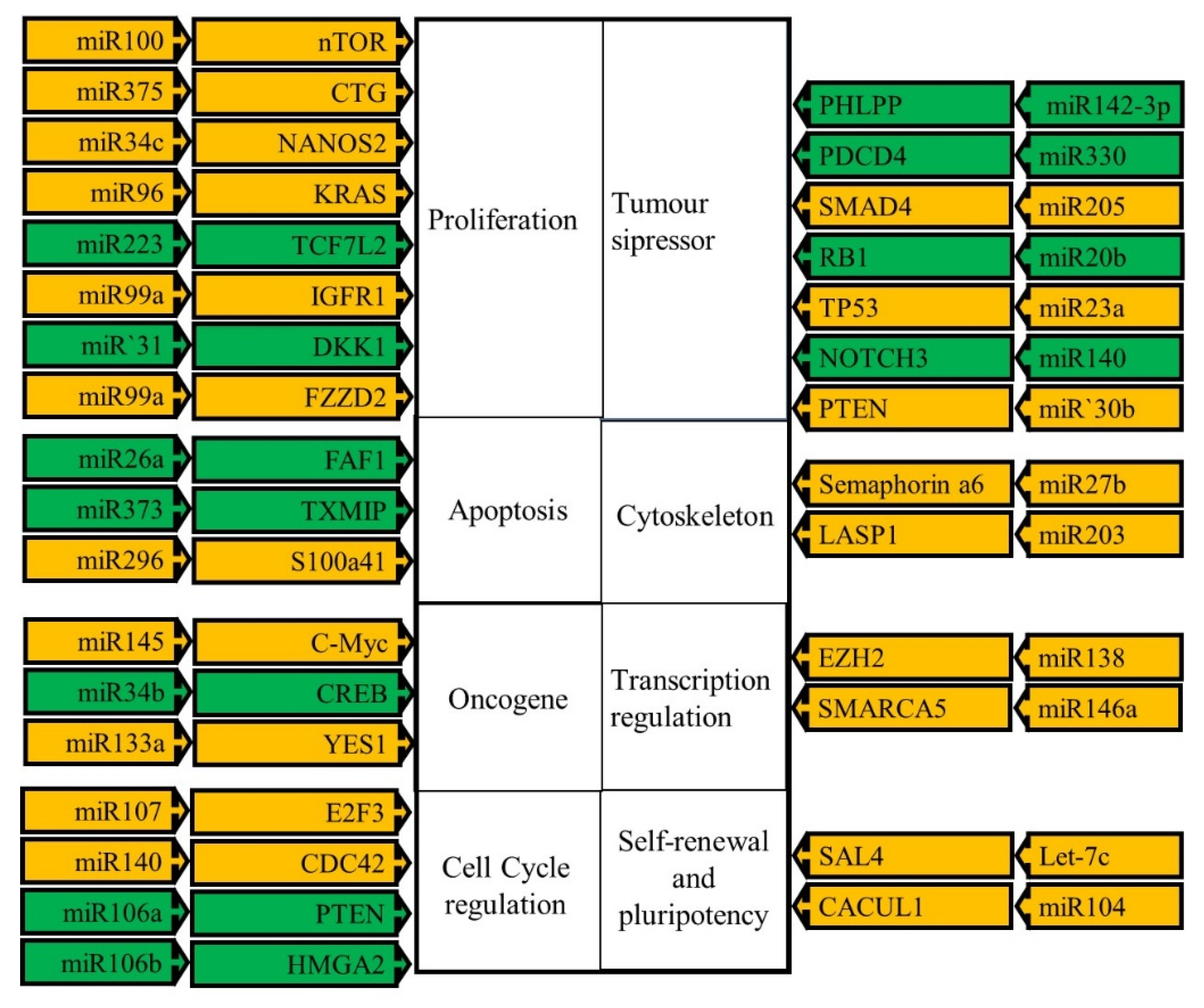
4. MicroRNAs
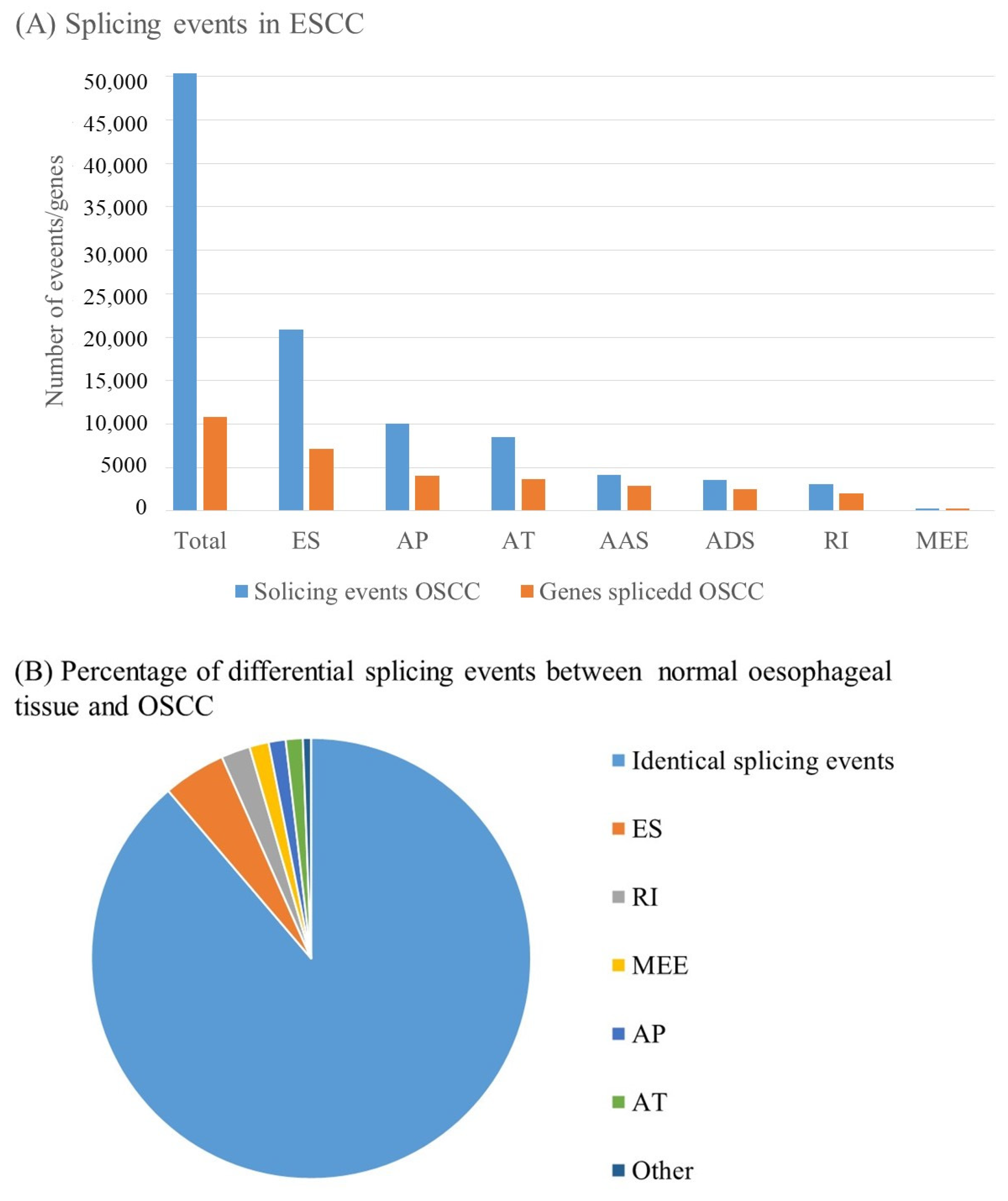
5. Alternative Splicing
6. Viral Oncogenesis
7. Treatment of OSCC and the Clinical Application of Molecular Profile Data
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smyth, E.C.; Lagergren, J.; Fitzgerald, R.C.; Lordick, F.; Shah, M.A.; Lagergren, P.; Cunningham, D. Oesophageal cancer. Nat. Rev. Dis. Prim. 2017, 3, 17048. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.; Mbele, M.; Makhafola, T.; Hicks, C.; Wang, S.M.; Reis, R.M.; Mehrotra, R.; Mkhize-Kwitshana, Z.; Hussain, S.; Kibiki, G.; et al. A multinational review: Oesophageal cancer in low to middle-income countries. Oncol. Lett. 2020, 20, 42. [Google Scholar] [CrossRef] [PubMed]
- Kaz, A.M.; Grady, W.M. Epigenetic biomarkers in esophageal cancer. Cancer Lett 2014, 342, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Kunzmann, A.T.; McMenamin, Ú.C.; Spence, A.D.; Gray, R.T.; Murray, L.J.; Turkington, R.C.; Coleman, H.G. Blood biomarkers for early diagnosis of oesophageal cancer: A systematic review. Eur. J. Gastroenterol. Hepatol. 2018, 30, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Mardis, E.R. The emerging clinical relevance of genomics in cancer medicine. Nat. Rev. Clin. Oncol. 2018, 15, 353–365. [Google Scholar] [CrossRef] [PubMed]
- El-Deiry, W.S.; Goldberg, R.M.; Lenz, H.-J.; Shields, A.F.; Gibney, G.T.; Tan, A.R.; Brown, J.; Eisenberg, B.; Heath, E.I.; Phuphanich, S.; et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J. Clin. 2019, 69, 305–343. [Google Scholar] [CrossRef]
- Chen, X.X.; Zhong, Q.; Liu, Y.; Yan, S.M.; Chen, Z.H.; Jin, S.Z.; Xia, T.L.; Li, R.Y.; Zhou, A.J.; Su, Z.; et al. Genomic comparison of esophageal squamous cell carcinoma and its precursor lesions by multi-region whole-exome sequencing. Nat. Commun. 2017, 8, 524. [Google Scholar] [CrossRef]
- Walker, R.C.; Underwood, T.J. Molecular pathways in the development and treatment of oesophageal cancer. Best Pract. Res. Clin. Gastroenterol. 2018, 36–37, 9–15. [Google Scholar] [CrossRef]
- Walline, H.M.; Carey, T.E.; Goudsmit, C.M.; Bellile, E.L.; D’Souza, G.; Peterson, L.A.; McHugh, J.B.; Pai, S.I.; Lee, J.J.; Shin, D.M.; et al. High-Risk HPV, Biomarkers, and Outcome in Matched Cohorts of Head and Neck Cancer Patients Positive and Negative for HIV. Mol. Cancer Res. 2017, 15, 179–188. [Google Scholar] [CrossRef]
- Gen, Y.; Yasui, K.; Nishikawa, T.; Yoshikawa, T. SOX2 promotes tumor growth of esophageal squamous cell carcinoma through the AKT/mammalian target of rapamycin complex 1 signaling pathway. Cancer Sci. 2013, 104, 810–816. [Google Scholar] [CrossRef]
- Di Resta, C.; Galbiati, S.; Carrera, P.; Ferrari, M. Next-generation sequencing approach for the diagnosis of human diseases: Open challenges and new opportunities. EJIFCC 2018, 29, 4–14. [Google Scholar] [PubMed]
- Takahashi, M.; Hosomichi, K.; Nakaoka, H.; Sakata, H.; Uesato, N.; Murakami, K.; Kano, M.; Toyozumi, T.; Matsumoto, Y.; Isozaki, T.; et al. Biased expression of mutant alleles in cancer-related genes in esophageal squamous cell carcinoma. Esophagus 2022, 19, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahzadeh, D.; Roshandel, G.; Delhomme, T.M.; Avogbe, P.H.; Foll, M.; Saidi, F.; Poustchi, H.; Sotoudeh, M.; Malekzadeh, R.; Brennan, P.; et al. TP53 Targeted Deep Sequencing of Cell-Free DNA in Esophageal Squamous Cell Carcinoma Using Low-Quality Serum: Concordance with Tumor Mutation. Int. J. Mol. Sci. 2021, 22, 5627. [Google Scholar] [CrossRef] [PubMed]
- Munari, F.F.; Dos Santos, W.; Evangelista, A.F.; Carvalho, A.C.; Pastrez, P.A.; Bugatti, D.; Wohnrath, D.R.; Scapulatempo-Neto, C.; Guimarães, D.P.; Longatto-Filho, A.; et al. Profile of esophageal squamous cell carcinoma mutations in Brazilian patients. Sci. Rep. 2021, 11, 20596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Shi, J.; Shi, X.; Chen, W.; Liu, J. Mutational Characterization and Potential Prognostic Biomarkers of Chinese Patients with Esophageal Squamous Cell Carcinoma. OncoTargets Ther. 2020, 13, 12797–12809. [Google Scholar] [CrossRef]
- Moody, S.; Senkin, S.; Islam, S.M.A.; Wang, J.; Nasrollahzadeh, D.; Penha, R.C.C.; Fitzgerald, S.; Bergstrom, E.N.; Atkins, J.; He, Y.; et al. Mutational signatures in esophageal squamous cell carcinoma from eight countries with varying incidence. Nat. Genet. 2021, 53, 1553–1563. [Google Scholar] [CrossRef]
- Salem, M.E.; Puccini, A.; Xiu, J.; Raghavan, D.; Lenz, H.J.; Korn, W.M.; Shields, A.F.; Philip, P.A.; Marshall, J.L.; Goldberg, R.M. Comparative Molecular Analyses of Esophageal Squamous Cell Carcinoma, Esophageal Adenocarcinoma, and Gastric Adenocarcinoma. Oncolologist 2018, 23, 1319–1327. [Google Scholar] [CrossRef]
- Agrawal, N.; Jiao, Y.; Bettegowda, C.; Hutfless, S.M.; Wang, Y.; David, S.; Cheng, Y.; Twaddell, W.S.; Latt, N.L.; Shin, E.J.; et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012, 2, 899–905. [Google Scholar] [CrossRef]
- Lin, D.-C.; Wang, M.-R.; Koeffler, H.P. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology 2018, 154, 374–389. [Google Scholar] [CrossRef]
- Gao, D.; Zhang, Z.; Yang, Q.; Li, B. Multiregion sequencing and subclonal analysis reveal intratumoral heterogeneity in esophageal squamous cell carcinoma. J. Cancer Res. Ther. 2021, 17, 756–763. [Google Scholar]
- Yang, J.W.; Choi, Y.L. Genomic profiling of esophageal squamous cell carcinoma (ESCC)-Basis for precision medicine. Pathol. Res. Pract. 2017, 213, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Snell, J.M.; Jeck, W.R.; Hoadley, K.A.; Wilkerson, M.D.; Parker, J.S.; Patel, N.; Mlombe, Y.B.; Mulima, G.; Liomba, N.G.; et al. Subtyping sub-Saharan esophageal squamous cell carcinoma by comprehensive molecular analysis. JCI Insight 2016, 1, e88755. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Wei, Y.; Hu, Y.; Chen, X.; Zheng, Y.; Liu, M.; Wang, Y.; Zhou, Y. Identification of prognostic biomarkers associated with tumor microenvironment in ceRNA network for esophageal squamous cell carcinoma: A bioinformatics study based on TCGA database. Discov. Oncol. 2021, 12, 46. [Google Scholar] [CrossRef]
- Kashyap, M.K.; Marimuthu, A.; Kishore, C.J.; Peri, S.; Keerthikumar, S.; Prasad, T.S.; Mahmood, R.; Rao, S.; Ranganathan, P.; Sanjeeviah, R.C.; et al. Genomewide mRNA profiling of esophageal squamous cell carcinoma for identification of cancer biomarkers. Cancer Biol. Ther. 2009, 8, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Lopes, N.; Correia, M.P.; Henrique, R.; Jerónimo, C. Epigenetic Alterations in Oesophageal Cancer: Expression and Role of the Involved Enzymes. Int. J. Mol. Sci. 2020, 21, 3522. [Google Scholar] [CrossRef]
- Huang, F.L.; Yu, S.J. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J. Surg. 2018, 41, 210–215. [Google Scholar] [CrossRef]
- Spitzwieser, M.; Entfellner, E.; Werner, B.; Pulverer, W.; Pfeiler, G.; Hacker, S.; Cichna-Markl, M. Hypermethylation of CDKN2A exon 2 in tumor, tumor-adjacent and tumor-distant tissues from breast cancer patients. BMC Cancer 2017, 17, 260. [Google Scholar] [CrossRef]
- Song, Y.; Li, L.; Ou, Y.; Gao, Z.; Li, E.; Li, X.; Zhang, W.; Wang, J.; Xu, L.; Zhou, Y.; et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014, 509, 91–95. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, Y.; Li, C.; Xu, S.; Li, M.; Liu, W.; Ma, Y.; Wang, H. The Expression and Prognostic Value of FGF2, FGFR3, and FGFBP1 in Esophageal Squamous Cell Carcinoma. Anal. Cell. Pathol. 2020, 2020, 2872479. [Google Scholar] [CrossRef]
- Fan, Z.L.; Yang, L.Y.; Zhang, N.; Feng, D.; Guo, J.; Chang, C.; Yuan, Q.; Cai, Y.; Zhang, Y.; Wei, W.Q.; et al. NEFL promotes invasion and migration of esophageal squamous carcinoma cells via the EGFR/AKT/S6 pathway. Yi Chuan Hered. 2022, 44, 322–334. [Google Scholar]
- Zhang, X.; Chen, Y.; Li, Z.; Han, X.; Liang, Y. TGFBR3 is an independent unfavourable prognostic marker in oesophageal squamous cell cancer and is positively correlated with Ki-67. Int. J. Exp. Pathol. 2020, 101, 223–229. [Google Scholar] [CrossRef]
- Saito, T.; Mitomi, H.; Imamhasan, A.; Hayashi, T.; Mitani, K.; Takahashi, M.; Kajiyama, Y.; Yao, T. Downregulation of sFRP-2 by epigenetic silencing activates the β-catenin/Wnt signaling pathway in esophageal basaloid squamous cell carcinoma. Virchows Arch. 2014, 464, 135–143. [Google Scholar] [CrossRef]
- Fukukawa, C.; Hanaoka, H.; Nagayama, S.; Tsunoda, T.; Toguchida, J.; Endo, K.; Nakamura, Y.; Katagiri, T. Radioimmunotherapy of human synovial sarcoma using a monoclonal antibody against FZD10. Cancer Sci. 2008, 99, 432–440. [Google Scholar] [CrossRef]
- Businello, G.; Parente, P.; Mastracci, L.; Pennelli, G.; Traverso, G.; Milione, M.; Bellan, E.; Michelotto, M.; Kotsafti, A.; Grillo, F.; et al. The Pathologic and Molecular Landscape of Esophageal Squamous Cell Carcinogenesis. Cancers 2020, 12, 2160. [Google Scholar] [CrossRef]
- Zsákai, L.; Sipos, A.; Dobos, J.; Erős, D.; Szántai-Kis, C.; Bánhegyi, P.; Pató, J.; Őrfi, L.; Matula, Z.; Mikala, G.; et al. Targeted drug combination therapy design based on driver genes. Oncotarget 2019, 10, 5255–5266. [Google Scholar] [CrossRef]
- Kadian, L.K.; Arora, M.; Prasad, C.P.; Pramanik, R.; Chauhan, S.S. Signaling pathways and their potential therapeutic utility in esophageal squamous cell carcinoma. Clin. Transl. Oncol. 2022, 24, 1014–1032. [Google Scholar] [CrossRef]
- Yuan, C.; Xiang, L.; Cao, K.; Zhang, J.; Luo, Y.; Sun, W.; Zhang, N.; Ren, J.; Zhang, J.; Gong, Y.; et al. The prognostic value of tumor mutational burden and immune cell infiltration in esophageal cancer patients with or without radiotherapy. Aging 2020, 12, 4603–4616. [Google Scholar] [CrossRef]
- Sha, D.; Jin, Z.; Budczies, J.; Kluck, K.; Stenzinger, A.; Sinicrope, F.A. Tumor Mutational Burden as a Predictive Biomarker in Solid Tumors. Cancer Discov. 2020, 10, 1808–1825. [Google Scholar] [CrossRef]
- Pestinger, V.; Smith, M.; Sillo, T.; Findlay, J.M.; Laes, J.-F.; Martin, G.; Middleton, G.; Taniere, P.; Beggs, A.D. Use of an Integrated Pan-Cancer Oncology Enrichment Next-Generation Sequencing Assay to Measure Tumour Mutational Burden and Detect Clinically Actionable Variants. Mol. Diagn. Ther. 2020, 24, 339–349. [Google Scholar] [CrossRef]
- Sawada, G.; Niida, A.; Uchi, R.; Hirata, H.; Shimamura, T.; Suzuki, Y.; Shiraishi, Y.; Chiba, K.; Imoto, S.; Takahashi, Y.; et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology 2016, 150, 1171–1182. [Google Scholar] [CrossRef]
- Zhang, R.; Hu, Y.; Zhou, T.; Han, W.; Liu, Y.; Qian, J.; Chen, Y.; Liu, X.; Liu, S.; Yu, Y.; et al. The mutation profiles of cell-free DNA in patients with oesophageal squamous cell carcinoma who were responsive and non-responsive to neoadjuvant chemotherapy. J. Thorac. Dis. 2020, 12, 4274–4283. [Google Scholar] [CrossRef]
- Kumar, N. Checkmate 274 trial: Is Nivolumab the new standard in adjuvant setting for high-risk muscle invasive urothelial carcinoma? Indian J. Urol. 2021, 37, 369–371. [Google Scholar] [CrossRef]
- Shao, C.; Li, G.; Huang, L.; Pruitt, S.; Castellanos, E.; Frampton, G.; Carson, K.R.; Snow, T.; Singal, G.; Fabrizio, D.; et al. Prevalence of High Tumor Mutational Burden and Association with Survival in Patients with Less Common Solid Tumors. JAMA Netw. Open 2020, 3, e2025109. [Google Scholar] [CrossRef]
- Sakai, N.S.; Samia-Aly, E.; Barbera, M.; Fitzgerald, R.C. A review of the current understanding and clinical utility of miRNAs in esophageal cancer. Semin. Cancer Biol. 2013, 23 Pt B, 512–521. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Y.; Wu, X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr. Pharm. Des. 2013, 19, 1292–1300. [Google Scholar]
- Bai, X.; Wang, Q.; Rui, X.; Li, X.; Wang, X. Upregulation of miR-1269 Contributes to the Progression of Esophageal Squamous Cell Cancer Cells and Is Associated with Poor Prognosis. Technol. Cancer Res. Treat. 2021, 20, 1533033820985858. [Google Scholar] [CrossRef]
- Zhou, S.-L.; Wang, L.-D. Circulating microRNAs: Novel biomarkers for esophageal cancer. World J. Gastroenterol. 2010, 16, 2348–2354. [Google Scholar] [CrossRef]
- Feber, A.; Xi, L.; Luketich, J.D.; Pennathur, A.; Landreneau, R.J.; Wu, M.; Swanson, S.J.; Godfrey, T.E.; Litle, V.R. MicroRNA expression profiles of esophageal cancer. J. Thorac. Cardiovasc. Surg. 2008, 135, 255–260. [Google Scholar] [CrossRef]
- Lin, R.J.; Xiao, D.W.; Liao, L.D.; Chen, T.; Xie, Z.F.; Huang, W.Z.; Wang, W.S.; Jiang, T.F.; Wu, B.L.; Li, E.M.; et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J. Surg. Oncol. 2012, 105, 175–182. [Google Scholar] [CrossRef]
- Wu, C.; Li, M.; Hu, C.; Duan, H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol. Biol. Rep. 2014, 41, 1257–1266. [Google Scholar] [CrossRef]
- Li, J.; Shan, F.; Xiong, G.; Wang, J.M.; Wang, W.L.; Xu, X.; Bai, Y. Transcriptional regulation of miR-146b by C/EBPβ LAP2 in esophageal cancer cells. Biochem. Biophys. Res. Commun. 2014, 446, 267–271. [Google Scholar] [CrossRef]
- Yu, J.; Chen, S.; Niu, Y.; Liu, M.; Zhang, J.; Yang, Z.; Gao, P.; Wang, W.; Han, X.; Sun, G. Functional significance and therapeutic potential of miRNA-20b-5p in esophageal squamous cell carcinoma. Mol. Ther. Nucleic Acids 2020, 21, 315–331. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, C.; Liu, Z.; Guo, H.; Lu, W.; Hu, W.; Lin, Z. PABPC1-induced stabilization of IFI27 mRNA promotes angiogenesis and malignant progression in esophageal squamous cell carcinoma through exosomal miRNA-21-5p. J. Exp. Clin. Cancer Res. 2022, 41, 111. [Google Scholar] [CrossRef]
- Isayeva, T.; Brandwein-Gensler, M.; Somarathna, M.; Moore-Smith, L.D.; Lee, T. Micro-RNA profiling as a predictor of clinical outcomes for head and neck cancer patients. Curr. Pharm. Des. 2017, 23, 4729–4744. [Google Scholar] [CrossRef]
- Guo, B.; Tian, Z. Mir-25 Promotes Metastasis of Esophageal Cancer by Targeting BTG2. Appl. Biochem. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Harata, K.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitsui, A.; Ogawa, R.; Katada, T.; Tanaka, T.; Shiozaki, M.; Fujii, Y. MicroRNA-34b has an oncogenic role in esophageal squamous cell carcinoma. Oncol. Lett. 2010, 1, 685–689. [Google Scholar] [CrossRef]
- Xia, H.; Chen, S.; Chen, K.; Huang, H.; Ma, H. Pharmacotherapy, MiR-96 promotes proliferation and chemo-or radioresistance by down-regulating RECK in esophageal cancer. Biomed. Pharmacother. 2014, 68, 951–958. [Google Scholar] [CrossRef]
- Jin, J.; Guo, T.; Guo, Y.; Liu, J.; Qu, F.; He, Y. Methylation-associated silencing of miR-128 promotes the development of esophageal cancer by targeting COX-2 in areas with a high incidence of esophageal cancer. Int. J. Oncol. 2019, 54, 644–654. [Google Scholar] [CrossRef]
- Ogawa, R.; Ishiguro, H.; Kuwabara, Y.; Kimura, M.; Mitsui, A.; Katada, T.; Harata, K.; Tanaka, T.; Fujii, Y. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med. Mol. Morphol. 2009, 42, 102–109. [Google Scholar] [CrossRef]
- Yu, T.; Cao, R.; Li, S.; Fu, M.; Ren, L.; Chen, W.; Zhu, H.; Zhan, Q.; Shi, R. MiR-130b plays an oncogenic role by repressing PTEN expression in esophageal squamous cell carcinoma cells. BMC Cancer 2015, 15, 29. [Google Scholar] [CrossRef]
- Gao, S.; Zhao, Z.Y.; Zhang, Z.Y.; Zhang, Y.; Wu, R. Prognostic Value of MicroRNAs in Esophageal Carcinoma: A Meta-Analysis. Clin. Transl. Gastroenterol. 2018, 9, 203. [Google Scholar] [CrossRef]
- Liu, R.; Yang, M.; Meng, Y.; Liao, J.; Sheng, J.; Pu, Y.; Yin, L.; Kim, S.J. Tumor-suppressive function of miR-139-5p in esophageal squamous cell carcinoma. PLoS ONE 2013, 8, e77068. [Google Scholar] [CrossRef]
- Meng, H.; Wang, K.; Chen, X.; Guan, X.; Hu, L.; Xiong, G.; Li, J.; Bai, Y. MicroRNA-330-3p functions as an oncogene in human esophageal cancer by targeting programmed cell death 4. Am. J. Cancer Res. 2015, 5, 1062–1075. [Google Scholar]
- Liu, W.; Li, M.; Chen, X.; Zhang, D.; Wei, L.; Zhang, Z.; Wang, S.; Meng, L.; Zhu, S.; Li, B. MicroRNA-373 promotes migration and invasion in human esophageal squamous cell carcinoma by inhibiting TIMP3 expression. Am. J. Cancer Res. 2016, 6, 1–14. [Google Scholar]
- Zhang, L.; Chen, J.; Cheng, T.; Yang, H.; Pan, C.; Li, H. Identification of Differentially Expressed Genes and miRNAs Associated with Esophageal Squamous Cell Carcinoma by Integrated Analysis of Microarray Data. BioMed Res. Int. 2020, 2020, 1980921. [Google Scholar] [CrossRef]
- Lu, Q.; Wang, X.; Zhu, J.; Fei, X.; Chen, H.; Li, C. Hypoxic Tumor-Derived Exosomal Circ0048117 Facilitates M2 Macrophage Polarization Acting as miR-140 Sponge in Esophageal Squamous Cell Carcinoma. OncoTargets Ther. 2020, 13, 11883–11897. [Google Scholar] [CrossRef]
- Wu, B.L.; Xu, L.Y.; Du, Z.P.; Liao, L.D.; Zhang, H.F.; Huang, Q.; Fang, G.Q.; Li, E.M. MiRNA profile in esophageal squamous cell carcinoma: Downregulation of miR-143 and miR-145. World J. Gastroenterol. 2011, 17, 79–88. [Google Scholar] [CrossRef]
- Jia, Z.; Yang, Y.; Dengyan, Z.; Chunyang, Z.; Donglei, L.; Kai, W.; Song, Z. RAP1B, a DVL2 binding protein, activates Wnt/beta-catenin signaling in esophageal squamous cell carcinoma. Gene 2017, 611, 15–20. [Google Scholar] [CrossRef]
- Yang, C.; Zheng, S.; Liu, T.; Liu, Q.; Dai, F.; Zhou, J.; Chen, Y.; Sheyhidin, I.; Lu, X. Down-regulated miR-26a promotes proliferation, migration, and invasion via negative regulation of MTDH in esophageal squamous cell carcinoma. FASEB J. 2017, 31, 2114–2122. [Google Scholar] [CrossRef]
- Ding, D.P.; Chen, Z.L.; Zhao, X.H.; Wang, J.W.; Sun, J.; Wang, Z.; Tan, F.W.; Tan, X.G.; Li, B.Z.; Zhou, F.; et al. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis 2011, 32, 1025–1032. [Google Scholar] [CrossRef]
- Qi, B.; Wang, Y.; Chen, Z.J.; Li, X.N.; Qi, Y.; Yang, Y.; Cui, G.H.; Guo, H.Z.; Li, W.H.; Zhao, S. Down-regulation of miR-30a-3p/5p promotes esophageal squamous cell carcinoma cell proliferation by activating the Wnt signaling pathway. World J. Gastroenterol. 2017, 23, 7965–7977. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yi, J.; Zhang, K.; Bai, F.; Feng, B.; Wang, R.; Chu, X.; Chen, L.; Song, H. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 161. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Cen, K. MiR-92a inhibits proliferation and promotes apoptosis of OSCC cells through Wnt/β-catenin signaling pathway. J. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4803–4809. [Google Scholar]
- Mei, L.-L.; Qiu, Y.-T.; Huang, M.-B.; Wang, W.-J.; Bai, J.; Shi, Z.-Z. MiR-99a suppresses proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inhibiting the IGF1R signaling pathway. Cancer Biomarks 2017, 20, 527–537. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Z.; Tan, X.; Zhou, F.; Tan, F.; Gao, Y.; Sun, N.; Xu, X.; Shao, K.; He, J. MicroRNA-99a/100 promotes apoptosis by targeting mTOR in human esophageal squamous cell carcinoma. Med. Oncol. 2013, 30, 411. [Google Scholar] [CrossRef]
- Zhang, N.; Fu, H.; Song, L.; Ding, Y.; Wang, X.; Zhao, C.; Zhao, Y.; Jiao, F.; Zhao, Y. MicroRNA-100 promotes migration and invasion through mammalian target of rapamycin in esophageal squamous cell carcinoma. Oncol. Rep. 2014, 32, 1409–1418. [Google Scholar] [CrossRef]
- Sharma, P.; Saini, N.; Sharma, R. miR-107 functions as a tumor suppressor in human esophageal squamous cell carcinoma and targets Cdc42. Oncol. Rep. 2017, 37, 3116–3127. [Google Scholar] [CrossRef]
- Kano, M.; Seki, N.; Kikkawa, N.; Fujimura, L.; Hoshino, I.; Akutsu, Y.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; Matsubara, H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 2010, 127, 2804–2814. [Google Scholar] [CrossRef]
- Zhang, B.X.; Yu, T.; Yu, Z.; Yang, X.G. MicroRNA-148a regulates the MAPK/ERK signaling pathway and suppresses the development of esophagus squamous cell carcinoma via targeting MAP3K9. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 6497–6504. [Google Scholar]
- Yokobori, T.; Suzuki, S.; Tanaka, N.; Inose, T.; Sohda, M.; Sano, A.; Sakai, M.; Nakajima, M.; Miyazaki, T.; Kato, H.; et al. MiR-150 is associated with poor prognosis in esophageal squamous cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci. 2013, 104, 48–54. [Google Scholar] [CrossRef]
- Yu, X.; Jiang, X.; Li, H.; Guo, L.; Jiang, W.; Lu, S.H. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev. 2014, 23, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Z.; Luo, Y.R.; Du, J.; Yu, Y.; Yang, X.Z.; Cui, Y.J.; Jin, X.F. MiR-296-5p inhibits cell invasion and migration of esophageal squamous cell carcinoma by downregulating STAT3 signaling. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5206–5214. [Google Scholar] [PubMed]
- Yan, S.; Jiang, H.; Fang, S.; Yin, F.; Wang, Z.; Jia, Y.; Sun, X.; Wu, S.; Jiang, T.; Mao, A. MicroRNA-340 Inhibits Esophageal Cancer Cell Growth and Invasion by Targeting Phosphoserine Aminotransferase 1. Cell. Physiol. Biochem. 2015, 37, 375–386. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, Y.; Li, X.; Wang, X.; Yuan, Y. miR-375 Inhibits the Proliferation, Migration and Invasion of Esophageal Squamous Cell Carcinoma by Targeting XPR1. Curr. Gene Ther. 2021, 21, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Jia, E.; Ren, N.; Xin, H. Identification of prognostic miRNA biomarkers for esophageal cancer based on The Cancer Genome Atlas and Gene Expression Omnibus. Medicine 2021, 100, e24832. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, D.; Liu, X.; Wang, X.; Li, B. MicroRNA-27a downregulates the expression of Hsp90 and enhances the radiosensitivity in esophageal squamous cell carcinoma. OncoTargets Ther. 2019, 12, 5967–5977. [Google Scholar] [CrossRef] [PubMed]
- El Marabti, E.; Younis, I. The Cancer Spliceome: Reprograming of Alternative Splicing in Cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Qian, J.; Gu, C.; Yang, Y. Alternative splicing and cancer: A systematic review. Signal. Transduct. Target. Ther. 2021, 6, 78. [Google Scholar] [CrossRef]
- Sun, J.-R.; Kong, C.-F.; Lou, Y.-N.; Yu, R.; Qu, X.-K.; Jia, L.-Q. Genome-Wide Profiling of Alternative Splicing Signature Reveals Prognostic Predictor for Esophageal Carcinoma. Front. Genet. 2020, 11, 796. [Google Scholar] [CrossRef]
- Chen, J.; Weiss, W.A. Alternative splicing in cancer: Implications for biology and therapy. Oncogene 2015, 34, 1–14. [Google Scholar] [CrossRef]
- Climente-González, H.; Porta-Pardo, E.; Godzik, A.; Eyras, E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017, 20, 2215–2226. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Pham, M.H.C.; Ko, K.S.; Rhee, B.D.; Han, J. Alternative splicing isoforms in health and disease. Pflügers Arch. Eur. J. Physiol. 2018, 470, 995–1016. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Li, C.; Cheng, Y.; Du, Z.; Wang, Q.; Tang, Z.; Song, C.; Xia, Q.; Bai, W.; Lin, L.; et al. Alterations of RNA splicing patterns in esophagus squamous cell carcinoma. Cell Biosci. 2021, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.-C.; Wu, H.-Y.; Ma, F.-C.; Dang, Y.-W.; Peng, Z.-G.; Zhou, H.-F.; Chen, G. Prognostic alternative splicing signatures and underlying regulatory network in esophageal carcinoma. Am. J. Transl. Res. 2019, 11, 4010–4028. [Google Scholar]
- Dlamini, Z.; Hull, R.; Mbatha, S.Z.; Alaouna, M.; Qiao, Y.-L.; Yu, H.; Chatziioannou, A. Prognostic Alternative Splicing Signatures in Esophageal Carcinoma. Cancer Manag. Res. 2021, 13, 4509–4527. [Google Scholar] [CrossRef]
- Clower, C.V.; Chatterjee, D.; Wang, Z.; Cantley, L.C.; Vander Heiden, M.G.; Krainer, A.R. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc. Natl. Acad. Sci. USA 2010, 107, 1894–1899. [Google Scholar] [CrossRef]
- Nowak, D.G.; Amin, E.M.; Rennel, E.S.; Hoareau-Aveilla, C.; Gammons, M.; Damodoran, G.; Hagiwara, M.; Harper, S.J.; Woolard, J.; Ladomery, M.R.; et al. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: A novel therapeutic strategy for angiogenesis. J. Biol. Chem. 2010, 285, 5532–5540. [Google Scholar] [CrossRef]
- Dou, X.Q.; Chen, X.J.; Zhou, Q.; Wen, M.X.; Zhang, S.Z.; Zhang, S.Q. miR-335 modulates Numb alternative splicing via targeting RBM10 in endometrial cancer. Kaohsiung J. Med. Sci. 2020, 36, 171–177. [Google Scholar] [CrossRef] [Green Version]
- Lu, F.; Gladden, A.B.; Diehl, J.A. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer Res. 2003, 63, 7056–7061. [Google Scholar]
- Ogura, Y.; Hoshino, T.; Tanaka, N.; Ailiken, G.; Kobayashi, S.; Kitamura, K.; Rahmutulla, B.; Kano, M.; Murakami, K.; Akutsu, Y.; et al. Disturbed alternative splicing of FIR (PUF60) directed cyclin E overexpression in esophageal cancers. Oncotarget 2018, 9, 22929. [Google Scholar] [CrossRef]
- Bessman, M.J.; Frick, D.N.; O’Handley, S.F. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 1996, 271, 25059–25062. [Google Scholar] [CrossRef] [PubMed]
- Rekasi, Z.; Varga, J.L.; Schally, A.V.; Halmos, G.; Armatis, P.; Groot, K.; Czompoly, T. Antagonists of growth hormone-releasing hormone and vasoactive intestinal peptide inhibit tumor proliferation by different mechanisms: Evidence from in vitro studies on human prostatic and pancreatic cancers. Endocrinology 2000, 141, 2120–2128. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.-K.; Xu, L.-Y.; Lu, X.-F.; Liao, L.-D.; Cai, W.-J.; Shen, Z.-Y.; Li, E.-M. A novel alternative spliced variant of neutrophil gelatinase-associated lipocalin receptor in oesophageal carcinoma cells. Biochem. J. 2007, 403, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.-Y.; Lv, G.-Q.; Dai, L.-H.; Zhan, X.-H.; Jiao, J.-W.; Liao, L.-D.; Zhou, T.-M.; Li, C.-Q.; Wu, B.-L.; Xu, L.-Y.; et al. A truncated splice variant of human lysyl oxidase-like 2 promotes migration and invasion in esophageal squamous cell carcinoma. Int. J. Biochem. Cell Biol. 2016, 75, 85–98. [Google Scholar] [CrossRef]
- Lin, J.; Lin, L.; Thomas, D.G.; Greenson, J.K.; Giordano, T.J.; Robinson, G.S.; Barve, R.A.; Weishaar, F.A.; Taylor, J.M.; Orringer, M.B.; et al. Melanoma-associated antigens in esophageal adenocarcinoma: Identification of novel MAGE-A10 splice variants. J. Am. Coll. Surg. 2004, 10, 5708–5716. [Google Scholar] [CrossRef]
- Song, Z.-B.; Gao, S.-S.; Yi, X.-N.; Li, Y.-J.; Wang, Q.-M.; Zhuang, Z.-H.; Wang, L.-D. Expression of MUC1 in esophageal squamous-cell carcinoma and its relationship with prognosis of patients from Linzhou city, a high incidence area of northern China. World J. Gastroenterol. 2003, 9, 404–407. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Zhu, J.; Thompson, C.B. The hallmarks of cancer metabolism: Still emerging. Cell Metab. 2022, 34, 355–377. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Hull, R.; Mbita, Z.; Dlamini, Z. Long non-coding RNAs (LncRNAs), viral oncogenomics, and aberrant splicing events: Therapeutics implications. Am. J. Cancer Res. 2021, 11, 866–883. [Google Scholar]
- National Health Laboratory Services Cancer in South Africa. 2019. Available online: https://www.nicd.ac.za/wp-content/uploads/2021/12/NCR_Path_2019_Full_Report_8dec2021.pdf (accessed on 12 April 2022).
- Thrift, A.P.; Kramer, J.R.; Hartman, C.M.; Royse, K.; Richardson, P.; Dong, Y.; Raychaudhury, S.; Desiderio, R.; Sanchez, D.; Anandasabapathy, S.; et al. Risk and Predictors of Esophageal and Stomach Cancers in HIV-Infected Veterans: A Matched Cohort Study. J. Acquir. Immune Defic. Syndr. 2019, 81, e65–e72. [Google Scholar] [CrossRef]
- Poljak, M.; Šterbenc, A.; Lunar, M.M. Prevention of human papillomavirus (HPV)-related tumors in people living with human immunodeficiency virus (HIV). Expert Rev. Anti-Infect. Ther. 2017, 15, 987–999. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, X.; Liu, F.; Zhao, Y.; Sun, M.; Chen, D.; Lu, C.; Wang, Z.; Shi, X.; Zhang, Q.; et al. Detection of HPV DNA in esophageal cancer specimens from different regions and ethnic groups: A descriptive study. BMC Cancer 2010, 10, 19. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.S.; Lundine, D.; Leeman, J.E.; Higginson, D.S. Genomic Signatures in HPV-Associated Tumors. Viruses 2021, 13, 1998. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Han, H.; Zhang, F.; Wang, B.; Ma, W.; Wang, Y.; Sun, G.; Shi, M.; Ren, Y.; Cheng, Y. HPV infection in esophageal squamous cell carcinoma and its relationship to the prognosis of patients in northern China. Sci. World J. 2014, 2014, 804738. [Google Scholar] [CrossRef]
- Youssef, E.M.; Matsuda, T.; Takada, N.; Osugi, H.; Higashino, M.; Kinoshita, H.; Watanabe, T.; Katsura, Y.; Wanibuchi, H.; Fukushima, S. Prognostic significance of the MIB-1 proliferation index for patients with squamous cell carcinoma of the esophagus. Cancer 1995, 76, 358–366. [Google Scholar] [CrossRef]
- Natsugoe, S.; Nakashima, S.; Matsumoto, M.; Xiangming, C.; Okumura, H.; Kijima, F.; Ishigami, S.; Takebayashi, Y.; Baba, M.; Takao, S.; et al. Expression of p21WAF1/Cip1 in the p53-dependent pathway is related to prognosis in patients with advanced esophageal carcinoma. Clin. Cancer Res. 1999, 5, 2445–2449. [Google Scholar]
- Joshi, M.B.; Shirota, Y.; Danenberg, K.D.; Conlon, D.H.; Salonga, D.S.; Herndon, J.E., II; Danenberg, P.V.; Harpole, D.H., Jr. High gene expression of TS1, GSTP1, and ERCC1 are risk factors for survival in patients treated with trimodality therapy for esophageal cancer. Clin. Cancer Res. 2005, 11, 2215–2221. [Google Scholar] [CrossRef]
- Okumura, H.; Natsugoe, S.; Matsumoto, M.; Yokomakura, N.; Uchikado, Y.; Takatori, H.; Ishigami, S.; Takao, S.; Aikou, T. Predictive value of p53 and 14-3-3sigma for the effect of chemoradiation therapy on esophageal squamous cell carcinoma. J. Surg. Oncol. 2005, 91, 84–89. [Google Scholar] [CrossRef]
- Kishi, K.; Doki, Y.; Miyata, H.; Yano, M.; Yasuda, T.; Monden, M. Prediction of the response to chemoradiation and prognosis in oesophageal squamous cancer. Br. J. Surg. 2002, 89, 597–603. [Google Scholar] [CrossRef]
- Takeuchi, H.; Ozawa, S.; Ando, N.; Kitagawa, Y.; Ueda, M.; Kitajima, M. Cell-cycle regulators and the Ki-67 labeling index can predict the response to chemoradiotherapy and the survival of patients with locally advanced squamous cell carcinoma of the esophagus. Ann. Surg. Oncol. 2003, 10, 792–800. [Google Scholar] [CrossRef]
- Zhu, W.; You, Z.; Li, T.; Yu, C.; Tao, G.; Hu, M.; Chen, X. Correlation of hedgehog signal activation with chemoradiotherapy sensitivity and survival in esophageal squamous cell carcinomas. Jpn. J. Clin. Oncol. 2011, 41, 386–393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.-L.; Li, H.; Zhu, Y.-J.; Xu, G. The treatments and postoperative complications of esophageal cancer: A review. J. Cardiothorac. Surg. 2020, 15, 163. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Chen, J.; Huang, Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 2019, 1, 24. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, D.Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141. [Google Scholar] [CrossRef]
- Isozaki, Y.; Hoshino, I.; Akutsu, Y.; Hanari, N.; Mori, M.; Nishimori, T.; Murakami, K.; Akanuma, N.; Takeshita, N.; Maruyama, T.; et al. Usefulness of microRNA-375 as a prognostic and therapeutic tool in esophageal squamous cell carcinoma. Int. J. Oncol. 2015, 46, 1059–1066. [Google Scholar] [CrossRef]
- Zhang, J.; Lieu, Y.K.; Ali, A.M.; Penson, A.; Reggio, K.S.; Rabadan, R.; Raza, A.; Mukherjee, S.; Manley, J.L. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc. Natl. Acad. Sci. USA 2015, 112, E4726–E4734. [Google Scholar] [CrossRef]
- Castanotto, D.; Stein, C.A. Antisense oligonucleotides in cancer. Curr. Opin. Oncol. 2014, 26, 584–589. [Google Scholar] [CrossRef]




| Method | Sample Number | Number of Non-Silent MUTATIONS/ Tumor | Ref. |
|---|---|---|---|
| Whole exome sequencing | 12 | 83 | [18] |
| Whole exome and transcriptome sequencing | 20 exomes and 119 transcriptomes | 59 | [19] |
| Whole exome sequencing | 113 exomes | 82 | [20] |
| Whole genome and exome sequencing | 14 genomes, 90 exomes | 104 | [21] |
| Gene Symbol | Chromosomal Location | Nonsynonymous Mutation | Copy Number Loss | Copy Number Gain | Ref. |
|---|---|---|---|---|---|
| TP53 | 17p13.1 | 92 | 0 | 0 | [18] |
| 83 | 0 | 1 | [20] | ||
| 60 | 0 | 0 | [19] | ||
| 93 | 0 | 0 | [20] | ||
| 88 | 0 | 0 | [15] | ||
| NOTCH1 | 9q34.3 | 33 | 0 | 0 | [18] |
| 9 | 0 | 0 | [20] | ||
| 8 | 0 | 0 | [19] | ||
| 14 | 0 | 0 | [20] | ||
| 19 | 0 | 0 | [15] | ||
| PIK3CA | 3q26.3 | 0 | 0 | 0 | [18] |
| 5 | 4 | 0 | [20] | ||
| 7 | 10 | 0 | [19] | ||
| 9 | 2 | 0 | [20] | ||
| 17 | 0 | 0 | [15] | ||
| CDKN2A | 9p21.3 | 8 | 0 | 0 | [18] |
| 5 | 0 | 44 | [20] | ||
| 3 | 0 | 33 | [19] | ||
| 8 | 0 | 12 | [20] | ||
| 8 | 0 | 64 | [15] | ||
| CCND1 | 11q13 | 0 | 0 | 0 | [18] |
| 0 | 46 | 0 | [23] | ||
| 0 | 46 | 0 | [19] | ||
| 0 | 33 | 0 | [20] | ||
| 0 | 64 | 0 | [15] | ||
| FAT1 | 4q35.2 | 8 | 0 | 0 | [18] |
| 5 | 0 | 0 | [23] | ||
| 12 | 0 | 0 | [19] | ||
| 11 | 0 | 0 | [20] | ||
| 15 | 0 | 0 | [15] |
| miRNA | Type of Marker | Host Gene or Chromosome Location | Description | Ref |
|---|---|---|---|---|
| Upregulated | ||||
| Changes are a consequence of OSCC | ||||
| 142-3p | Prognostic | GRCh37 | Indicates poor prognosis. | [49] |
| 181b | Prognostic | SALL4 | Indicates poor prognosis. | [47] |
| 223 | Prognostic | Xq12 | Indicates poor prognosis. | [50] |
| 146b | Prognostic | 10. | Indicates poor prognosis. | [51] |
| Changes contribute to OSCC development ad progression | ||||
| 20b | Diagnostic | - | Targets RB1 and TP53. | [52] |
| 21 | Diagnostic: and prognostic | TMEM49 | Found in serum in serum and plasma indicates poor prognosis. Target for inhibition. | [53] |
| 23 | Prognostic | 19. | Indicates poor prognosis. | [54] |
| 25 | Diagnostic | MCM7 | Plays a role in metastasis. Target for inhibition. | [55] |
| 34b | Diagnostic | 11q23.1 | Oncogenic role in OSCC. Target for inhibition. | [56] |
| 96 | Prognostic | MIRN183-MIRN96-MIRN182 cluster 7q32.2 | Indicates poor prognosis. Target for inhibition. | [57] |
| 128b | Prognostic | 3p22 | Indicates poor prognosis. Target for inhibition. | [58] |
| 129 | Diagnostic and prognostic | 11p11.2 | Indicates poor prognosis. | [59] |
| 130b | Diagnostic | 2q11.21-q11.22. | Promotes angiogenesis. Target for inhibition. | [60] |
| 138 | Diagnostic | [61,62] | ||
| 151 | Diagnostic | FAK | Oncogenic. | [45] |
| 330 | Diagnostic | 724063 | Oncogene. Target for inhibition. | [63] |
| 373 | Diagnostic | Linked to MIRN371 and MIRN372 | Promotes migration and invasion. Target for inhibition. | [64] |
| Down-regulated | ||||
| Changes are a consequence of OSCC | ||||
| 27b | Prognostic | 9q22, | Indicates poor prognosis. | [54] |
| 103 | Prognostic | Indicates good prognosis. | [65] | |
| 34c | Diagnostic | 11q23.1 | Tumor suppressor. | [59] |
| 140 | Diagnostic | 16q22. | Downregulated. | [66] |
| 143 | Predictive and prognostic | 5q33 | Indicates a poor prognosis and predicts nonresponse to treatment. | [67] |
| 145 | Diagnostic and predictive | 5q33.1 | Predicts poor response to treatment. | [67] |
| 205 | Diagnostic and Predictive | 1q32.2 | Downregulation serves as diagnostic marker. Upregulation predicts poor response to treatment. | [65] |
| 518b | Prognostic | 19q13.42 | Indicates poor prognosis. | [68] |
| Changes contribute to OSCC development ad progression | ||||
| 26a | Prognostic | CTDSPL | Indicates poor prognosis. Possible therapy through mimics. | [69] |
| 29c | Diagnostic | 1q32.2 | Leads to increased proliferation. Possible therapy through mimics. | [70] |
| 30a-3p | Diagnostic | 6q13 | Downregulation leads to increased proliferation. | [71] |
| 31 | Prognostic and Diagnostic | 9p21.3 | Found in serum indicates poor prognosis. | [72] |
| 92a | Prognostic and predictive | C13ORF25 | Indicates good prognosis and predicts nonresponse to treatment. | [73] |
| 99a | Diagnostic | MIR99AHG | Tumor suppressor. Possible therapy through mimics. | [74] |
| 100 | Diagnostic | MIR100HG | Downregulated. Possible therapy through mimics. | [75] |
| 106a | Prognostic and predictive | Xq26.2 | Indicates poor prognosis and predicts nonresponse to treatment. | [76] |
| 107 | Prognostic | 10 | Indicates poor prognosis. Possible therapy through mimics. | [77] |
| 133a | Diagnostic | MIB1 | Tumor suppressor. | [78] |
| 133b | Diagnostic | LINCMD1 | Tumor suppressor. | [78] |
| 148a | Prognostic and predictive | 7p15.2 | Downregulation is an indicator of poor prognosis and predicts lack of response to treatment. | [79] |
| 150 | Prognostic | 19q13.33 | Downregulation is associated with poor prognosis. | [80] |
| 203 | Diagnostic | 14q32.33 | Tumor suppressor. Possible therapy through mimics. | [81] |
| 296 | Prognostic | MIRN296 | Downregulation indicts poor prognosis. Possible therapy through mimics. | [82] |
| 340 | Diagnostic | 16q11 | Acts as a tumor suppressor. Possible therapy through mimics. | [83] |
| 375 | Plasma Diagnostic, prognostic | DLK1 and DIO3 | Decreased level in plasma indicates poor prognosis. Possible therapy through mimics. | [84] |
| Let-7d | Diagnostic | 387247 | Blocks EMT transition low levels indicates poor prognosis. | [85] |
| Gene | Function | Splicing | Ref. |
|---|---|---|---|
| Cyclin D1 | Proliferation | Cyclin D1b levels increased. | [99] |
| FIR | Splicing, apoptosis, and transcription | Increased expression of isoforms lacking exon 2. | [100] |
| FGF | inhibits proliferation | Splice variants of FGF-2 and variant b increased in cancer. | [101] |
| GHRHR | Growth hormone receptor | Splice variant 1 levels increase. | [102] |
| LCN2, NGAL | Inhibits proteolysis | Expression of NGAL-2 and NGAL-3 increased. | [103] |
| LOXL2 | ECM remodeling | LOXL2Δ72, which lacks 72 promotes greater cell migration. | [104] |
| MAGE-A10 | Development | Additional exons 3A and 3B. | [105] |
| MUC1 | Cell adhesion | MUC1/C, D, and Z are expressed at higher levels. | [106] |
| PHF6 | Transcriptional regulation | Splice variants retaining introns overexpressed. | [93] |
| SRSF5 | Splicing factor | Different splice variants have different splicing regulatory functions. | [93] |
| TCF4 | WNT signaling | Unique isoforms isolated from various cancers. | [93] |
| Gene Mutations/Expression | |
|---|---|
| Methylation status | CDKN2A/p16INK4a |
| Somatic mutations present in OAC, gastric cancer and OSCC | TP53, PIK3CA, CDKN2A, CCND1, ARID1A, KRAS, APC, PTEN, SMAD4, NFE2L2, CDH1 and FAT1 |
| Mutations unique or more common in OSCC | KMT2D, SETD2,CHEK2, FBXW7,NOTCH1, RB1, CDKN2A, BAP1, FOXO3 and MSH6 |
| Increased gene expression | PD-L1, OPN, ORAOV2 and FAP |
| Altered signaling pathways | FGF2-FGFR3, EGFR/AKT/S6 (EGF- EGFR), TGFβ- TGFβR3, Wnt pathway, PI3K pathway |
| Splicing isoforms | Cyclin D1 (Cyclin D1b), FIR (isoforms lacking exon 2), FGF (variant b), GHRHR (variant 1), NGAL (NGAL-2 and NGAL-3), LOXL2, MAGE-A10 (isoform with exons 3A and 3B), MUC1 (C, D and Z isoforms), PHF6(intron retaining isoforms), SRSF5, TCF4 |
| miRNA | |
| Upregulated biomarker miRNAs | miR-1269, miR 142-3p, miR 181b, miR 223, miR 146b, miR 20b, miR 23, miR 129, miR 138, miR 151 |
| Upregulated miRNAs that can be targeted by an inhibitor | miR 21, miR 25, miR 34b, miR 96, miR 128b, miR 130b, miR 330, miR 373 |
| Downregulated biomarker miRNAs | miR 27b, miR 103, miR 34c, miR 140, miR 143, miR 145, miR 205, miR 518b, miR 30a-3p, miR 31, miR 92a, miR 106a, miR 133a, miR 133b, miR 148a, miR 150, Let-7d |
| Downregulated miRNAs that can be supplemented with mimics for treatment | miR 26a, miR 29c, miR 99a, miR 100, miR 107, miR 203, miR 296, miR 340, miR 375 |
| Diagnosis and screening miRNA polymorphism in South African populations | miR 3184 (rs6505162) overlap of two oppositely orientated miRNAs, miR314 and miR413. (NSRP1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mbatha, S.; Hull, R.; Dlamini, Z. Exploiting the Molecular Basis of Oesophageal Cancer for Targeted Therapies and Biomarkers for Drug Response: Guiding Clinical Decision-Making. Biomedicines 2022, 10, 2359. https://doi.org/10.3390/biomedicines10102359
Mbatha S, Hull R, Dlamini Z. Exploiting the Molecular Basis of Oesophageal Cancer for Targeted Therapies and Biomarkers for Drug Response: Guiding Clinical Decision-Making. Biomedicines. 2022; 10(10):2359. https://doi.org/10.3390/biomedicines10102359
Chicago/Turabian StyleMbatha, Sikhumbuzo, Rodney Hull, and Zodwa Dlamini. 2022. "Exploiting the Molecular Basis of Oesophageal Cancer for Targeted Therapies and Biomarkers for Drug Response: Guiding Clinical Decision-Making" Biomedicines 10, no. 10: 2359. https://doi.org/10.3390/biomedicines10102359
APA StyleMbatha, S., Hull, R., & Dlamini, Z. (2022). Exploiting the Molecular Basis of Oesophageal Cancer for Targeted Therapies and Biomarkers for Drug Response: Guiding Clinical Decision-Making. Biomedicines, 10(10), 2359. https://doi.org/10.3390/biomedicines10102359





