RNA Modification in Inflammatory Bowel Diseases
Abstract
:1. Introduction
2. Modification-Dependent RNA Function
2.1. Methylation System as a Nucleotide Modification
2.2. RNA Modifications
3. Modification-Dependent Alterations in RNA Structure
3.1. Modification-Dependent RNA Structures
3.2. G-Quadruplexes as Therapeutic Targets in Gastrointestinal Oncogenes
4. RNA Modifications in IBD
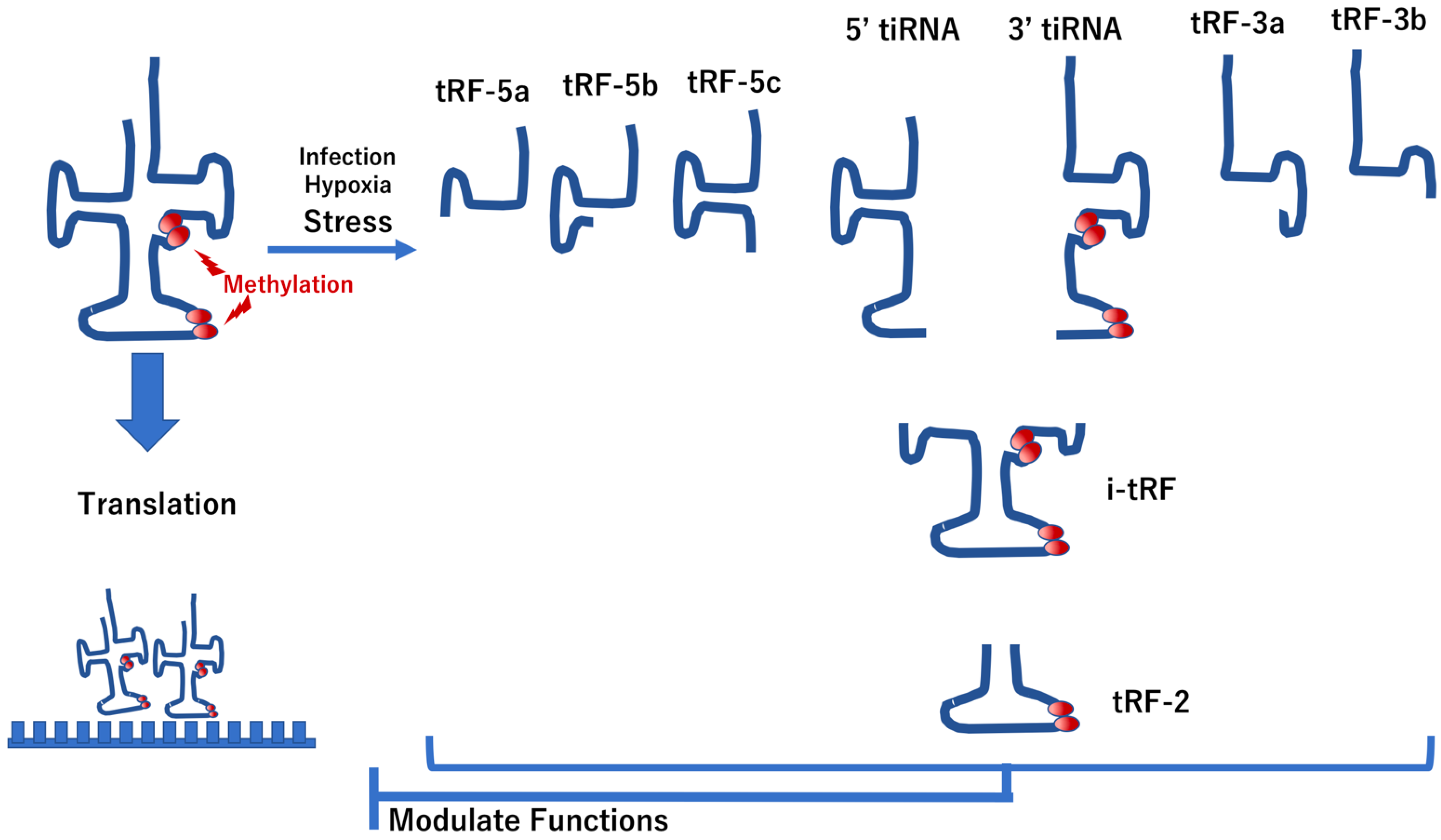
4.1. Modification of tRNAs in IBD
4.2. Modification of mRNA in IBD
4.3. Modification of ncRNAs in IBD
4.4. Modifications of miRNA in IBD
5. RNA Modifications That Regulate IBD Phenotypes
5.1. RNA Modification-Dependent Alterations in IBD Phenotypes
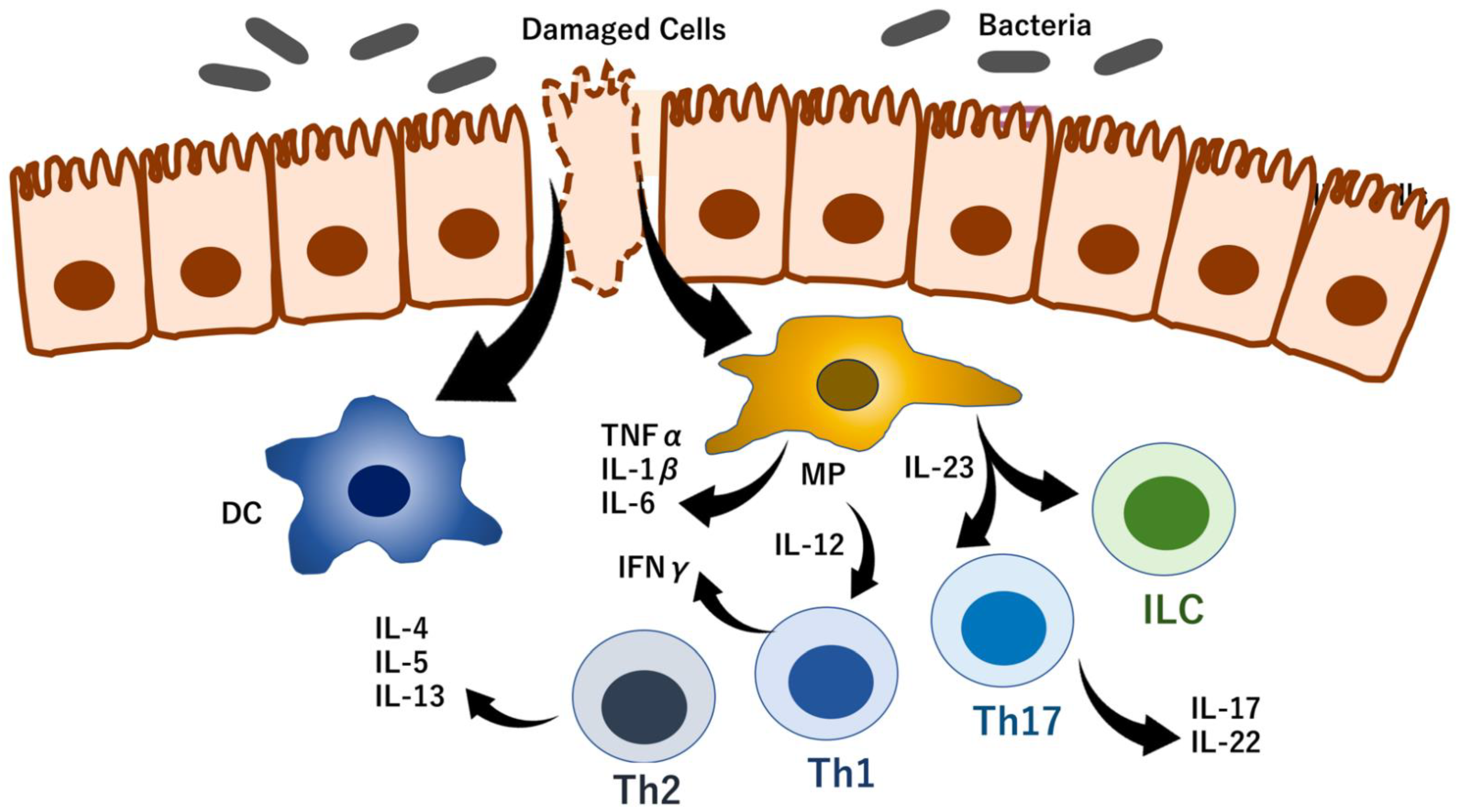
5.2. RNA Modification Control of T-Cell Homeostasis
5.3. RNA Modifications Control Immune Checkpoints
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xavier, R.J.; Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.N.; Benchimol, E.I.; Bitton, A.; Murthy, S.K.; Nguyen, G.C.; Lee, K.; Cooke-Lauder, J.; Kaplan, G.G. The impact of inflammatory bowel disease in Canada 2018: Extra-intestinal diseases in IBD. J. Can. Assoc. Gastroenterol. 2019, 2 (Suppl. 1), S73–S80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, W.T.; Feuerstein, J.D. Colorectal cancer surveillance in inflammatory bowel disease: Practice guidelines and recent developments. World J. Gastroenterol. 2019, 25, 4148–4157. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, G.R.; Rivas, M.A. Rare and common variant discovery in complex disease: The IBD case study. Hum. Mol. Genet. 2019, 28, R162–R169. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 205–217. [Google Scholar] [CrossRef]
- Kudelka, M.R.; Stowell, S.R.; Cummings, R.D.; Neish, A.S. Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 597–617. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef]
- Neurath, M.F. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 2019, 20, 970–979. [Google Scholar] [CrossRef]
- Kaplan, G.G. The global burden of IBD: From 2015 to 2025. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 720–727. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Johnson, T.B.; Coghill, R.D. Researches on pyrimidines. c111. the discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. J. Am. Chem. Soc. 1925, 47, 2838–2844. [Google Scholar] [CrossRef]
- Hotchkiss, R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948, 175, 315–332. [Google Scholar] [CrossRef]
- Wyatt, G.R. Occurrence of 5-methyl-cytosine in nucleic acids. Nature 1950, 166, 237–238. [Google Scholar] [CrossRef]
- Wyatt, G.R. The purine and pyrimidine composition of deoxypentose nucleic acids. Biochem. J. 1951, 48, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, G.R. Recognition and estimation of 5-methylcytosine in nucleic acids. Biochem. J. 1951, 48, 581–584. [Google Scholar] [CrossRef] [Green Version]
- Littlefield, J.W.; Dunn, D.B. The occurrence and distribution of thymine and three methylated-adenine bases in ribonucleic acids from several sources. Biochem. J. 1958, 70, 642–651. [Google Scholar] [CrossRef] [Green Version]
- Littlefield, J.W.; Dunn, D.B. Natural occurrence of thymine and three methylated adenine bases in several ribonucleic acids. Nature 1958, 181, 254–255. [Google Scholar] [CrossRef]
- Cantoni, G.L. The nature of the active methyl donor formed enzymatically from L-methionine and adenosinetriphosphate. J. Am. Chem. Soc. 1952, 74, 2942–2943. [Google Scholar] [CrossRef]
- Cantoni, G.L. Methylation of nicotinamide with soluble enzyme system from rat liver. J. Biol. Chem. 1951, 189, 203–216. [Google Scholar] [CrossRef]
- Kujundžić, R.N.; Prpić, M.; Đaković, N.; Dabelić, N.; Tomljanović, M.; Mojzeš, A.; Fröbe, A.; Trošelj, K.G. Nicotinamide N-methyltransferase in acquisition of stem cell properties and therapy resistance in cancer. Int. J. Mol. Sci. 2021, 22, 5681. [Google Scholar] [CrossRef]
- Pissios, P. Nicotinamide methyltransferase: More than a vitamin B3 clearance enzyme. Trends Endocrinol. Metab. 2017, 28, 340–353. [Google Scholar] [CrossRef] [Green Version]
- Ramsden, D.B.; Waring, R.H.; Barlow, D.J.; Parsons, R.B. Nicotinamide N-methyltransferase in health and cancer. Int. J. Tryptophan Res. 2017, 10, 1178646917691739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.M.; Long, H. Nicotinamide N-methyltransferase as a potential marker for cancer. Neoplasma 2018, 65, 656–663. [Google Scholar] [CrossRef]
- Li, J.; You, S.; Zhang, S.; Hu, Q.; Wang, F.; Chi, X.; Zhao, W.; Xie, C.; Zhang, C.; Yu, Y.; et al. Elevated N-methyltransferase expression induced by hepatic stellate cells contributes to the metastasis of hepatocellular carcinoma via regulation of the CD44v3 isoform. Mol. Oncol. 2019, 13, 1993–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wnorowski, A.; Wnorowska, S.; Kurzepa, J.; Parada-Turska, J. Alterations in kynurenine and NAD + salvage pathways during the successful treatment of inflammatory bowel disease suggest HCAR3 and NNMT as potential drug targets. Int. J. Mol. Sci. 2021, 22, 13497. [Google Scholar] [CrossRef] [PubMed]
- Ofusa, K.; Chijimatsu, R.; Ishii, H. Detection techniques for epitranscriptomic marks. Am. J. Physiol. Cell. Physiol. 2022, 322, C787–C793. [Google Scholar] [CrossRef]
- Konno, M.; Taniguchi, M.; Ishii, H. Significant epitranscriptomes in heterogeneous cancer. Cancer Sci. 2019, 110, 2318–2327. [Google Scholar] [CrossRef] [Green Version]
- Konno, M.; Koseki, J.; Asai, A.; Yamagata, A.; Shimamura, T.; Motooka, D.; Okuzaki, D.; Kawamoto, K.; Mizushima, T.; Eguchi, H.; et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019, 10, 3888. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell. Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizawa, Y.; Konno, M.; Asai, A.; Koseki, J.; Kawamoto, K.; Miyoshim, N.; Takahashi, H.; Nishida, N.; Haraguchi, N.; Sakai, D.; et al. Oncogene c-Myc promotes epitranscriptome m6A reader YTHDF1 expression in colorectal cancer. Oncotarget 2017, 9, 7476–7486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Ascenzo, L.; Popova, A.M.; Abernathy, S.; Sheng, K.; Limbach, P.A.; Williamson, J.R. Pytheas: A software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry. Nat. Commun. 2022, 13, 2424. [Google Scholar] [CrossRef]
- Jeong, H.; Lee, Y.; Kim, J. Structural and functional characterization of TrmM in m6A modification of bacterial tRNA. Protein Sci. 2022, 31, e4319. [Google Scholar] [CrossRef]
- Ramasamy, S.; Mishra, S.; Sharma, S.; Parimalam, S.S.; Vaijayanthi, T.; Fujita, Y.; Kovi, B.; Sugiyama, H.; Pandian, G.N. An informatics approach to distinguish RNA modifications in nanopore direct RNA sequencing. Genom. Inform. 2022, 114, 110372. [Google Scholar] [CrossRef]
- Abebe, J.S.; Price, A.M.; Hayer, K.E.; Mohr, I.; Weitzman, M.D.; Wilson, A.C.; Depledge, D.P. DRUMMER-Rapid detection of RNA modifications through comparative nanopore sequencing. Bioinformatics 2022, btac274, 3113–3115. [Google Scholar] [CrossRef]
- Liu, H.; Begik, O.; Novoa, E.M. EpiNano: Detection of m6A RNA modifications using Oxford nanopore direct RNA sequencing. Method. Mol. Biol. 2021, 2298, 31–52. [Google Scholar] [CrossRef]
- Ohshiro, T.; Konno, M.; Asai, A.; Komoto, Y.; Yamagata, A.; Doki, Y.; Eguchi, H.; Ofusa, K.; Taniguchi, M.; Ishii, H. Single-molecule RNA sequencing for simultaneous detection of m6A and 5mC. Sci. Rep. 2021, 11, 19304. [Google Scholar] [CrossRef]
- Flati, T.; Gioiosa, S.; Spallanzani, N.; Tagliaferri, I.; Diroma, M.A.; Pesole, G.; Chillemi, G.; Picardi, E.; Castrignanò, T. HPC-REDItools: A novel HPC-aware tool for improved large scale RNA-editing analysis. BMC Bioinform. 2020, 21 (Suppl. 10), 353. [Google Scholar] [CrossRef]
- Xiong, H.; Liu, D.; Li, Q.; Lei, M.; Xu, L.; Wu, L.; Wang, Z.; Ren, S.; Li, W.; Xia, M.; et al. RED-ML: A novel, effective RNA editing detection method based on machine learning. Gigascience 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, D.; Weirick, T.; Dimmeler, S.; Uchida, S. RNAEditor: Easy detection of RNA editing events and the introduction of editing islands. Brief. Bioinform. 2017, 18, 993–1001. [Google Scholar] [CrossRef]
- Hauenschild, R.; Werner, S.; Tserovski, L.; Hildebrandt, A.; Motorin, Y.; Helm, M. CoverageAnalyzer (CAn): A Tool for inspection of modification signatures in RNA sequencing profiles. Biomolecules 2016, 6, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishio, M.; Tsukakoshi, K.; Ikebukuro, K. G-quadruplex: Flexible conformational changes by cations, pH, crowding and its applications to biosensing. Biosens. Bioelectron. 2021, 178, 113030. [Google Scholar] [CrossRef] [PubMed]
- Banco, M.T.; Ferré-D’Amaré, A.R. The emerging structural complexity of G-quadruplex RNAs. RNA 2021, 27, 390–402. [Google Scholar] [CrossRef]
- Miglietta, G.; Russo, M.; Capranico, G. G-quadruplex-R-loop interactions and the mechanism of anticancer G-quadruplex binders. Nucleic Acid. Res. 2020, 48, 11942–11957. [Google Scholar] [CrossRef]
- Long, W.; Zheng, B.-X.; Li, Y.; Huang, X.-H.; Lin, D.-M.; Chen, C.-C.; Hou, J.-Q.; Ou, T.-M.; Wong, W.-L.; Zhang, K.; et al. Rational design of small-molecules to recognize G-quadruplexes of c-MYC promoter and telomere and the evaluation of their in vivo antitumor activity against breast cancer. Nucleic Acid. Res. 2022, 50, 1829–1848. [Google Scholar] [CrossRef]
- Ferino, A.; Marquevielle, J.; Choudhary, H.; Cinque, G.; Robert, C.; Bourdoncle, A.; Picco, R.; Mergny, J.-L.; Salgado, G.F.; Xodo, L.E. hnRNPA1/UP1 unfolds KRAS G-Quadruplexes and feeds a regulatory axis controlling gene expression. ACS Omega 2021, 6, 34092–34106. [Google Scholar] [CrossRef]
- Chen, Y.; Shen, J. Mucosal immunity and tRNA, tRF, and tiRNA. J. Mol. Med. 2021, 99, 47–56. [Google Scholar] [CrossRef]
- Schimmel, P. The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell. Biol. 2018, 19, 45–58. [Google Scholar] [CrossRef]
- Zhu, L.; Ge, J.; Li, T.; Shen, Y.; Guo, J. tRNA-derived fragments and tRNA halves: The new players in cancers. Cancer Lett. 2019, 452, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, Z.; Sheng, J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes 2018, 9, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Hu, G.-F. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J. Cell. Physiol. 2012, 227, 2822–2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, D.M.; Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell. Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.-F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell. Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Husain, B.; Hesler, S.; Cole, J.L. Regulation of PKR by RNA: Formation of active and inactive dimers. Biochemistry 2015, 54, 6663–6672. [Google Scholar] [CrossRef] [Green Version]
- Mayo, C.B.; Wong, C.J.; Lopez, P.E.; Lary, J.W.; Cole, J.L. Activation of PKR by short stem-loop RNAs containing single-stranded arms. RNA 2016, 22, 1065–1075. [Google Scholar] [CrossRef] [Green Version]
- Wiseman, D.H.; May, A.; Jolles, S.; Connor, P.; Powell, C.; Heeney, M.M.; Giardina, P.J.; Klaassen, R.J.; Chakraborty, P.; Geraghty, M.T.; et al. A novel syndrome of congenital sideroblastic anemia, B-cell immunodeficiency, periodic fevers, and developmental delay (SIFD). Blood 2013, 122, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Wilusz, J.E.; Whipple, J.M.; Phizicky, E.M.; Sharp, P.A. tRNAs marked with CCACCA are targeted for degradation. Science 2011, 334, 817–821. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Lee, I.; Lee, Y.S.; Bao, X. Small non-coding transfer RNA-derived RNA fragments (tRFs): Their biogenesis, function and implication in human diseases. Genom. Inform. 2015, 13, 94–101. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Yang, H.; Cheng, X.; Wang, D.; Fu, S.; Shen, W.; Zhang, Q.; Zhang, L.; Xue, Z.; Li, Y.; et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal Cancer. Cancer Res. 2017, 77, 3194–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.; Jin, C.; Wu, J.; Zhu, S.; Liu, Y.-J.; Chen, J. Guards at the gate: Physiological and pathological roles of tissue-resident innate lymphoid cells in the lung. Protein Cell 2017, 8, 878–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saikia, M.; Jobava, R.; Parisien, M.; Putnam, A.; Krokowski, D.; Gao, X.-H.; Guan, B.-J.; Yuan, Y.; Jankowsky, E.; Feng, Z.; et al. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 2014, 34, 2450–2463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebastian-delaCruz, M.; Olazagoitia-Garmendia, A.; Gonzalez-Moro, I.; Santin, I.; Garcia-Etxebarria, K.; Castellanos-Rubio, A. Implication of m6A mRNA methylation in susceptibility to inflammatory bowel disease. Epigenomes 2020, 4, 16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huo, C.; Liu, Y.; Su, R.; Zhao, Y.; Li, Y. Mechanism and disease association with a ubiquitin conjugating E2 enzyme: UBE2L3. Front. Immunol. 2022, 13, 793610. [Google Scholar] [CrossRef] [PubMed]
- Koepsell, H.; Lips, K.; Volk, C. Polyspecific organic cation transporters: Structure, function, physiological roles, and biopharmaceutical implications. Pharm. Res. 2007, 24, 1227–1251. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liu, Y.; Zhang, H.; Qiu, R.; Zhao, H.; Xin, Q.; Shan, S.; Dang, J.; Li, J.; Yang, Z.; et al. Evidence for genetic association of CARD9 and SNAPC4 with ankylosing spondylitis in a Chinese Han population. J. Rheumatol. 2014, 41, 318–324. [Google Scholar] [CrossRef]
- Nie, K.; Yi, J.; Yang, Y.; Deng, M.; Yang, Y.; Wang, T.; Chen, X.; Zhang, Z.; Wang, X. A broad m6A modification landscape in inflammatory bowel disease. Front. Cell. Dev. Biol. 2022, 9, 782636. [Google Scholar] [CrossRef]
- He, R.; Man, C.; Huang, J.; He, L.; Wang, X.; Lang, Y.; Fan, Y. Identification of RNA methylation-related lncRNAs signature for predicting hot and cold tumors and prognosis in colon cancer. Front. Genet. 2022, 13, 870945. [Google Scholar] [CrossRef]
- Chai, X.-K.; Qi, W.; Zou, C.-Y.; He, C.-X.; Su, M.; Zhao, D.-Q. Potential prognostic value of a seven m6A-related lncRNAs signature and the correlative immune infiltration in colon adenocarcinoma. Front. Genet. 2021, 12, 774010. [Google Scholar] [CrossRef]
- Morin, R.D.; O’Connor, M.D.; Griffith, M.; Kuchenbauer, F.; Delaney, A.; Prabhu, A.-L.; Zhao, Y.; McDonald, H.; Zeng, T.; Hirst, M.; et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genom. Res. 2008, 18, 610–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomasello, L.; Distefano, R.; Nigita, G.; Croce, C.M. The MicroRNA family gets wider: The isomiRs classification and role. Front. Cell. Dev. Biol. 2021, 9, 668648. [Google Scholar] [CrossRef] [PubMed]
- Olejniczak, M.; Kotowska-Zimmer, A.; Krzyzosiak, W. Stress-induced changes in miRNA biogenesis and functioning. Cell. Mol. Life Sci. 2018, 75, 177–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Xu, H.-M.; Yang, M.-F.; Liang, Y.-J.; Peng, Q.-Z.; Zhang, Y.; Tian, C.-M.; Wang, L.-S.; Yao, J.; Nie, Y.-Q.; et al. New insights into the epigenetic regulation of inflammatory bowel disease. Front. Pharmacol. 2022, 13, 813659. [Google Scholar] [CrossRef]
- Zhernakova, A.; van Diemen, C.C.; Wijmenga, C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 2009, 10, 43–55. [Google Scholar] [CrossRef]
- Zhang, T.; Ding, C.; Chen, H.; Zhao, J.; Chen, Z.; Chen, B.; Mao, K.; Hao, Y.; Roulis, M.; Xu, H.; et al. m6A mRNA modification maintains colonic epithelial cell homeostasis via NF-κB-mediated antiapoptotic pathway. Sci. Adv. 2022, 8, eabl5723. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Huang, J.; Ocansey, D.K.W.; Xia, Y.; Zhao, Z.; Xu, Z.; Yan, Y.; Zhang, X.; Mao, F. The emerging clinical application of m6A RNA modification in inflammatory bowel disease and its associated colorectal cancer. J. Inflamm. Res. 2021, 14, 3289–3306. [Google Scholar] [CrossRef]
- Li, H.-B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef] [Green Version]
- Gendo, Y.; Matsumoto, T.; Kamiyama, N.; Saechue, B.; Fukuda, C.; Dewayani, A.; Hidano, S.; Noguchi, K.; Sonoda, A.; Ozaki, T.; et al. Dysbiosis of the gut microbiota on the inflammatory background due to lack of suppressor of cytokine signalling-1 in mice. Inflamm. Intest. Dis. 2019, 3, 145–154. [Google Scholar] [CrossRef]
- Dobie, R.; MacRae, V.E.; Pass, C.; Milne, E.M.; Ahmed, S.F.; Farquharson, C. Suppressor of cytokine signaling 2 (Socs2) deletion protects bone health of mice with DSS-induced inflammatory bowel disease. Dis. Model. Mech. 2018, 11, dmm028456. [Google Scholar] [CrossRef] [Green Version]
- Bauché, D.; Joyce-Shaikh, B.; Fong, J.; Villarino, A.V.; Ku, K.S.; Jain, R.; Lee, Y.C.; Annamalai, L.; Yearley, J.H.; Cua, D.J. IL-23 and IL-2 activation of STAT5 is required for optimal IL-22 production in ILC3s during colitis. Sci. Immunol. 2020, 5, eaav1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Hui, H.; Agrawal, K.; Kang, Y.; Li, N.; Tang, R.; Yuan, J.; Rana, T.M. m6 A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020, 39, e104514. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Kang, Y.; Wang, L.; Huff, S.; Tang, R.; Hui, H.; Agrawal, K.; Gonzalez, G.M.; Wang, Y.; Patel, S.P.; et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 20159–20170. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nagpal, R.; Kumar, A.; Ashraf, M.U.; Bae, Y.-S. Immunotherapeutic potential of m6A-modifiers and microRNAs in controlling acute myeloid leukaemia. Biomedicines 2021, 9, 690. [Google Scholar] [CrossRef]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef]
- Moroz-Omori, E.V.; Huang, D.; Bedi, R.K.; Cheriyamkunnel, S.J.; Bochenkova, E.; Dolbois, A.; Rzeczkowski, M.D.; Li, Y.; Wiedmer, L.; Caflisch, A. METTL3 inhibitors for epitranscriptomic modulation of cellular processes. ChemMedChem 2021, 16, 3035–3043. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell 2020, 38, 79–96.e11. [Google Scholar] [CrossRef]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-molecule targeting of oncogenic fto demethylase in acute myeloid leukemia. Cancer Cell 2019, 35, 677–691.e10. [Google Scholar] [CrossRef] [Green Version]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6 A/MYC/CEBPA signaling. Cell 2018, 172, 90–105.e23. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Kennedy, S.; Hajian, T.; Gibson, E.; Seitova, A.; Xu, C.; Arrowsmith, C.H.; Vedadi, M. A radioactivity-based assay for screening human m6a-rna methyltransferase, mettl3-mettl14 complex, and demethylase alkbh5. J. Biomol. Screen. 2016, 21, 290–297. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Kinne, H.E.; Milligan, R.D.; Washburn, L.J.; Olsen, M.; Lucci, A. Important role of fto in the survival of rare panresistant triple-negative inflammatory breast cancer cells facing a severe metabolic challenge. PLoS ONE 2016, 11, e0159072. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yan, J.; Li, Q.; Li, J.; Gong, S.; Zhou, H.; Gan, J.; Jiang, H.; Jia, G.-F.; Luo, C.; et al. Meclofenamic acid selectively inhibits fto demethylation of m6a over alkbh5. Nucleic Acid. Res. 2015, 43, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Fong, C.Y.; Gilan, O.; Lam, E.Y.N.; Rubin, A.F.; Ftouni, S.; Tyler, D.; Stanley, K.; Sinha, D.; Yeh, P.; Morison, J.; et al. Bet inhibitor resistance emerges from leukaemia stem cells. Nature 2015, 525, 538–542. [Google Scholar] [CrossRef] [PubMed]



| Tool | Description | Merits and Applications | References |
|---|---|---|---|
| IndoC | “Trace” and “current signal intensity” of potentially modified sites are compared |
| [36] |
| DRUMMER | A rapid detection of RNA modifications through comparative nanopore sequencing |
| [37] |
| EpiNano | Detection of m6A RNA modifications using Oxford nanopore direct RNA sequencing |
| [38] |
| Quantum Sequencer System | Sequencing and mapping tool of RNA base modifications in microRNAs, such as m6A or m5C |
| [39] |
| HPC-REDItools | A novel HPC-aware tool for improved large-scale RNA-editing analysis |
| [40] |
| RED-ML | An effective RNA-editing detection method based on machine learning |
| [41] |
| RNAEditor | An easy method for the detection of RNA-editing events and the introduction of editing islands |
| [42] |
| CoverageAnalyzer (CAn) | A tool for inspection of modification signatures in RNA sequencing profiles |
| [43] |
| Regents | Target(s) and Action | Diseases | Expected Trials | References |
|---|---|---|---|---|
| STM2457 | Inhibitor of METTL3 (and METTL14) | Myeloid leukemia | Phase I | [85] |
| UZH1 | METTL3 | Leukemia | SC | [86] |
| CS1, CS2 | FTO inhibitor | Leukemia and solid tumors | SC | [87] |
| FB23 | FTO inhibitor | Leukemia | SC | [88] |
| R-2-hydroxyglutarate (R-2HG) | FTO inhibitor | Glioma and leukemia | SC | [89] |
| S-adenosyl-homocysteine | Inhibitor of METTL3 and METTL14 | Metabolic diseases | Phase I | [90] |
| MO-I-500 | FTO inhibitor | Breast cancer | SC | [91] |
| Meclofenamic acid | FTO inhibitor | Glioblastoma | Phase I | [92] |
| I-BET | Bromodomain and extra-terminal (BET) protein inhibitors | Leukemia | Phase I | [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakayama, M.; Ozato, Y.; Tsuji, Y.; Arao, Y.; Otsuka, C.; Hamano, Y.; Sumi, G.; Ofusa, K.; Uchida, S.; Vecchione, A.; et al. RNA Modification in Inflammatory Bowel Diseases. Biomedicines 2022, 10, 1695. https://doi.org/10.3390/biomedicines10071695
Nakayama M, Ozato Y, Tsuji Y, Arao Y, Otsuka C, Hamano Y, Sumi G, Ofusa K, Uchida S, Vecchione A, et al. RNA Modification in Inflammatory Bowel Diseases. Biomedicines. 2022; 10(7):1695. https://doi.org/10.3390/biomedicines10071695
Chicago/Turabian StyleNakayama, Mika, Yuki Ozato, Yoshiko Tsuji, Yasuko Arao, Chihiro Otsuka, Yumiko Hamano, Genzo Sumi, Ken Ofusa, Shizuka Uchida, Andrea Vecchione, and et al. 2022. "RNA Modification in Inflammatory Bowel Diseases" Biomedicines 10, no. 7: 1695. https://doi.org/10.3390/biomedicines10071695
APA StyleNakayama, M., Ozato, Y., Tsuji, Y., Arao, Y., Otsuka, C., Hamano, Y., Sumi, G., Ofusa, K., Uchida, S., Vecchione, A., & Ishii, H. (2022). RNA Modification in Inflammatory Bowel Diseases. Biomedicines, 10(7), 1695. https://doi.org/10.3390/biomedicines10071695







