Basement Membrane-Associated lncRNA Risk Model Predicts Prognosis and Guides Clinical Treatment in Clear Cell Renal Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collect and Identify lncRNAs Connected with Basement Membranes
2.2. Construction and Verification of a Basement Membrane-Associated lncRNA Risk Model
2.3. Creating and Validating Predicted Nomograms and Evaluating the Relationship between the Prognostic Signatures and Clinicopathological Features
2.4. PCA, Functional Enrichment, Tumor Immunity, Drug Sensitivity, and Mortality Analysis
2.5. Validation of Basement Membrane-Associated lncRNA Expression in Renal Cancer Cells
2.6. RNA Fish
2.7. Invasion Assay
3. Results
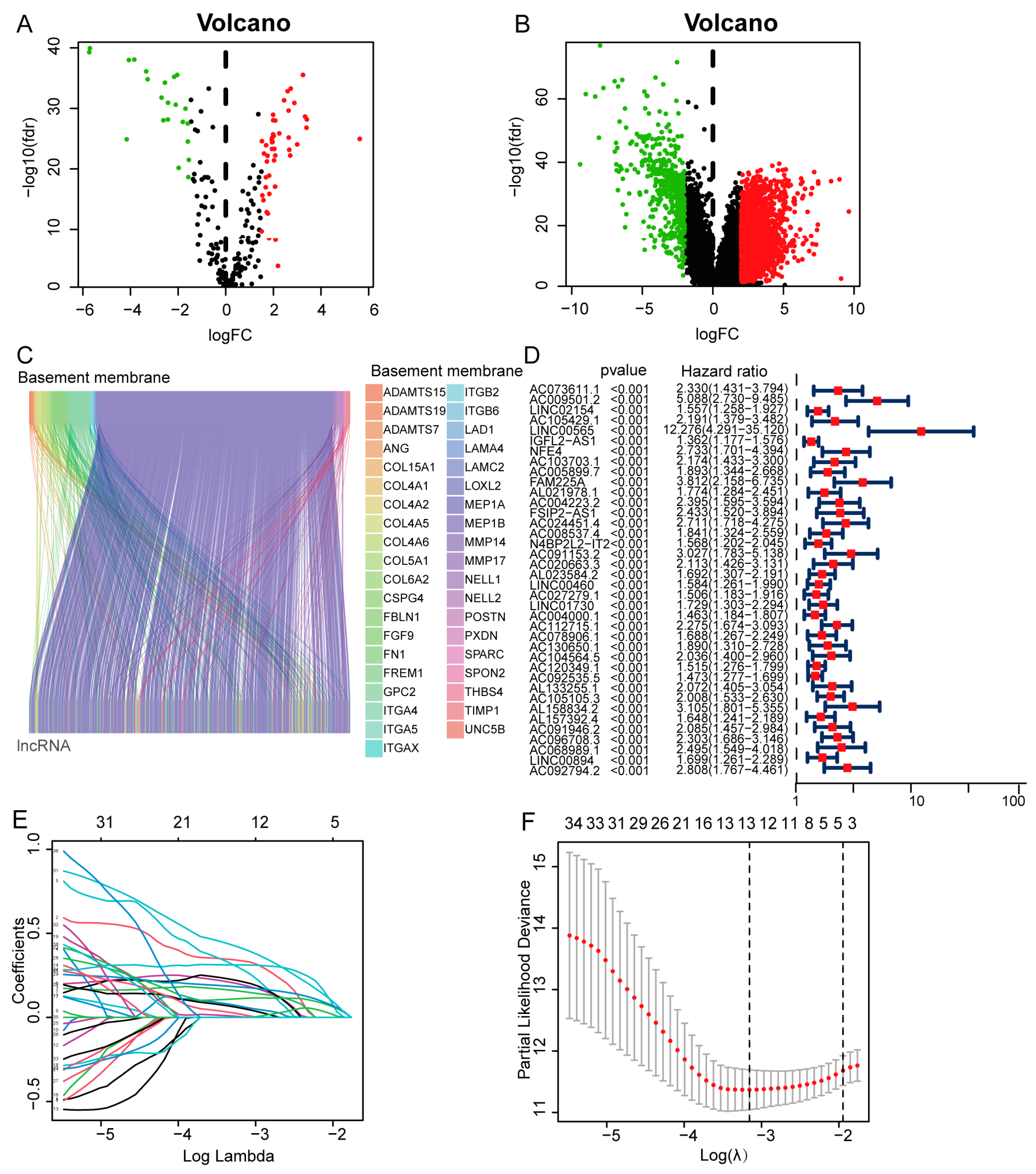
3.1. Construction of a Basement Membrane-Associated lncRNA Signature
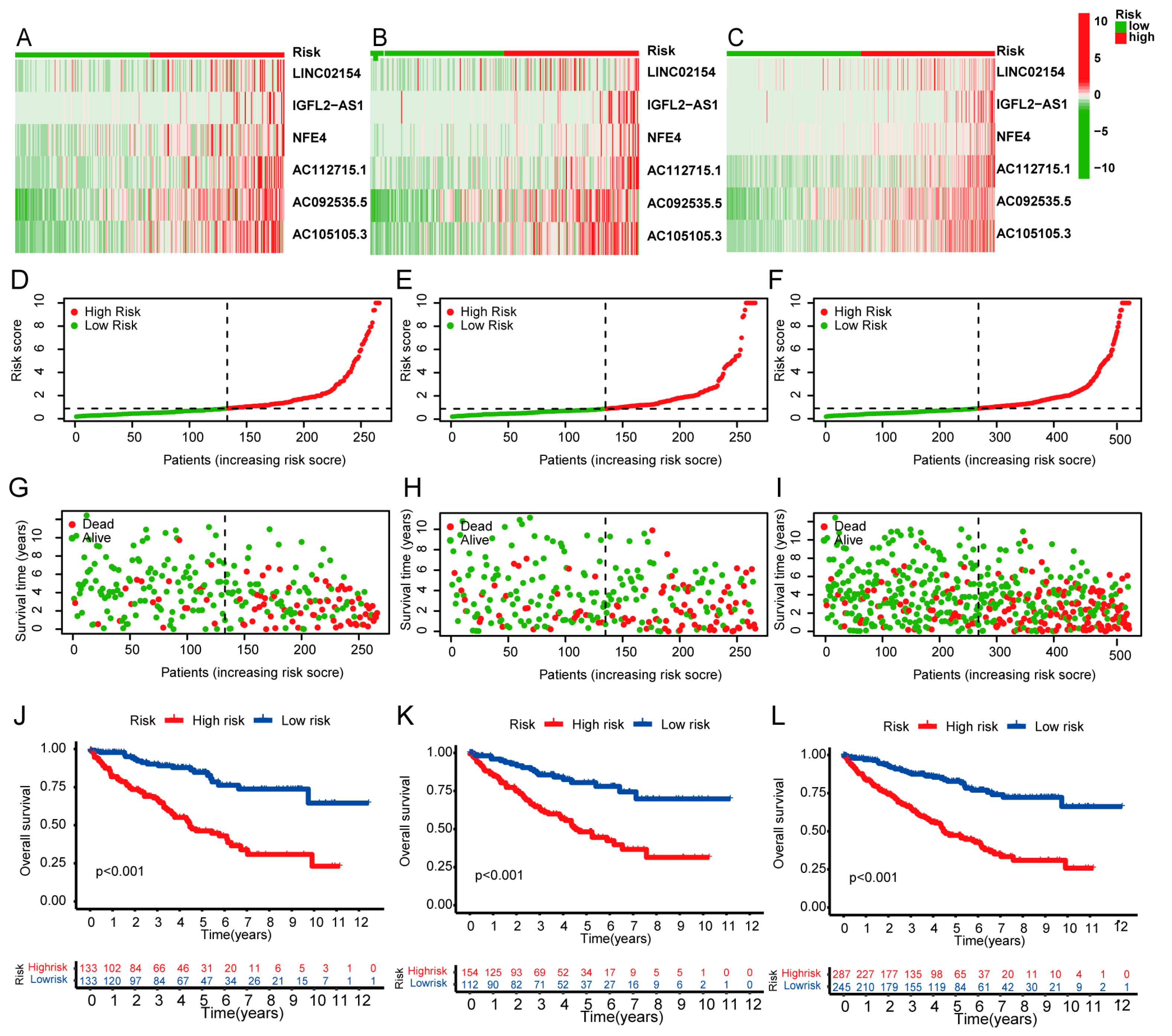
3.2. Expression and Survival Analysis of Prognostic Model lncRNAs
3.3. Survival Results and Multivariate Analysis
3.4. Independent Prognostic Value of the Risk Score
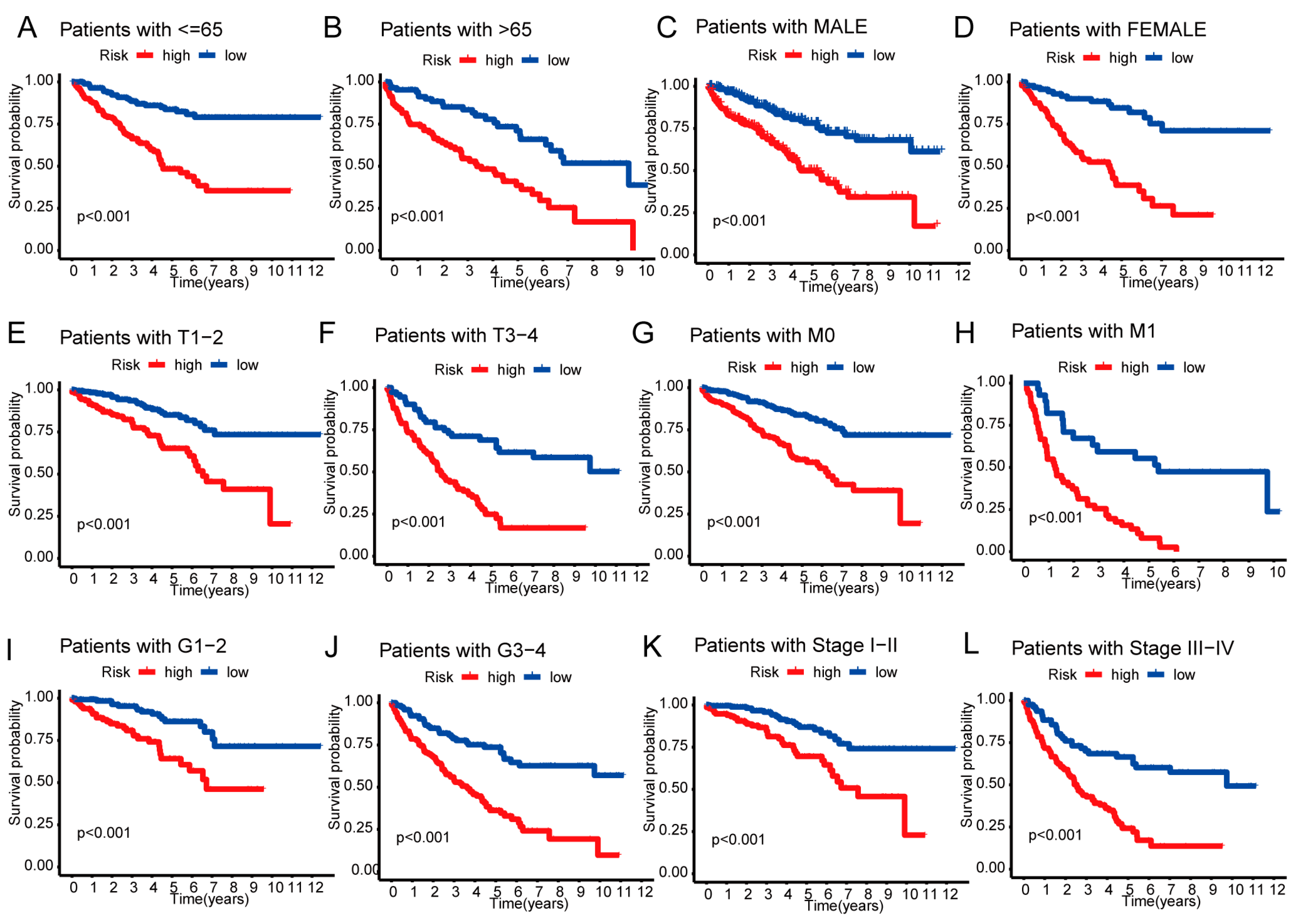
3.5. Nomogram and Heatmap of Clinical Factors
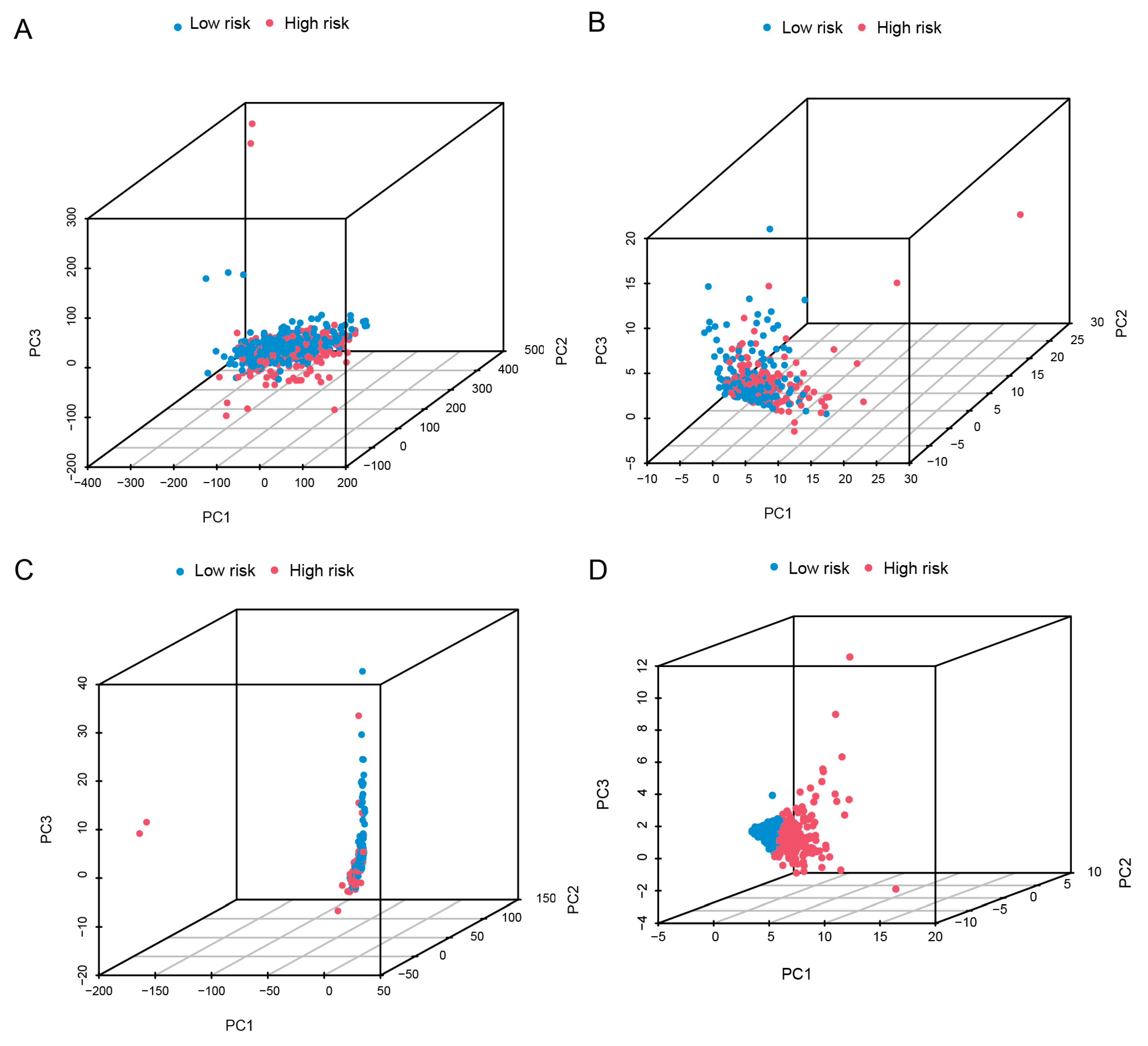
3.6. PCA and Enrichment Analysis
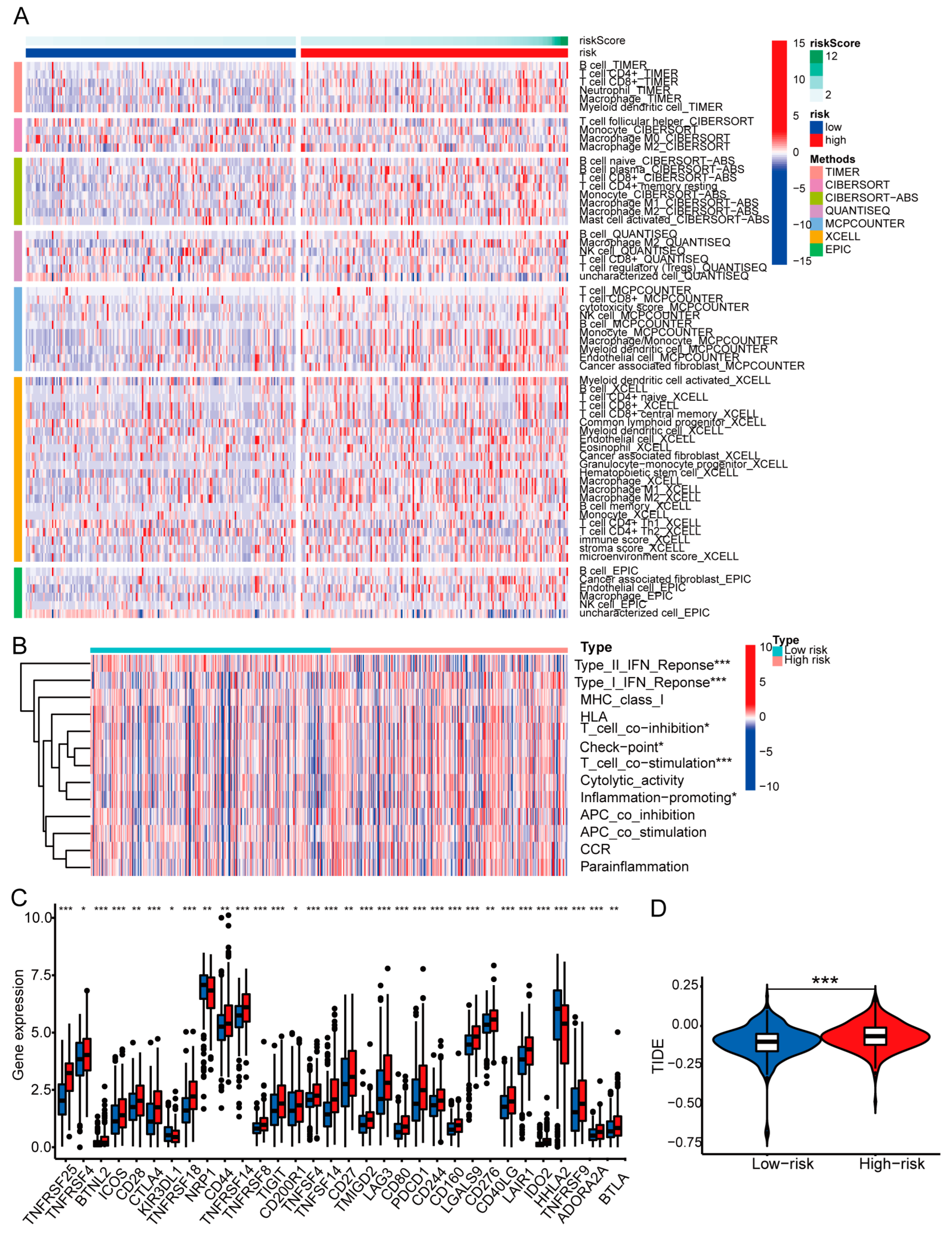
3.7. Examination of Immune Characteristics Using the Basement Membrane-Related lncRNA Signature
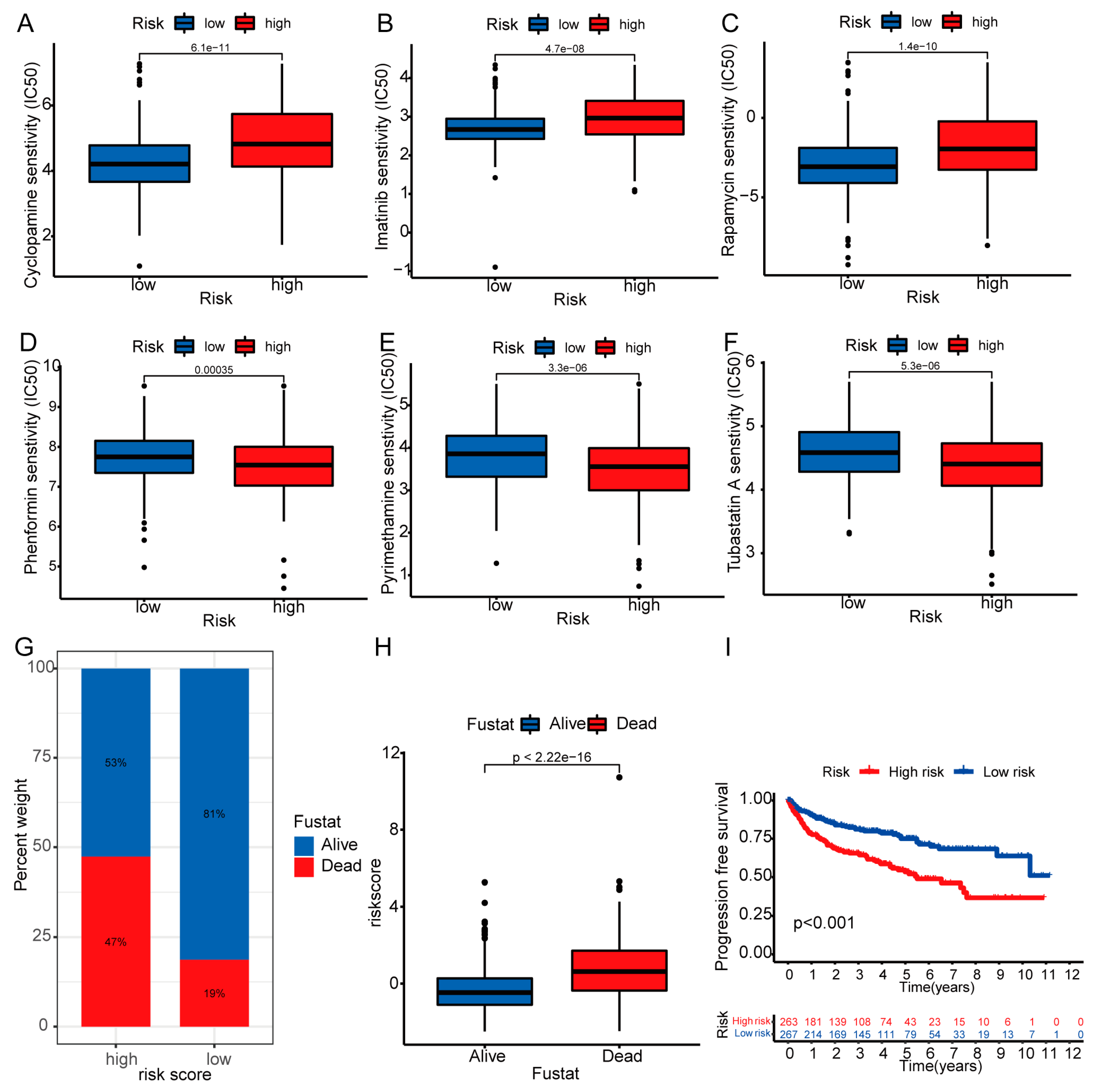
3.8. Therapeutic Drug Sensitivity and Mortality Rate
3.9. Expression of Risk Model lncRNAs
3.10. Expression of Risk Model lncRNAs in Renal Cancer Tissue and Functional Verification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vuong, L.; Kotecha, R.R.; Voss, M.H.; Hakimi, A.A. Tumor Microenvironment Dynamics in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2019, 9, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Delman, K.A. Introducing the “Virtual Tumor Board” series in CA: A Cancer Journal for Clinicians. CA Cancer J. Clin. 2020, 70, 77. [Google Scholar] [CrossRef] [PubMed]
- Choueiri, T.K.; Motzer, R.J. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2017, 376, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Lalani, A.A.; McGregor, B.A.; Albiges, L.; Choueiri, T.K.; Motzer, R.; Powles, T.; Wood, C.; Bex, A. Systemic Treatment of Metastatic Clear Cell Renal Cell Carcinoma in 2018: Current Paradigms, Use of Immunotherapy, and Future Directions. Eur. Urol. 2019, 75, 100–110. [Google Scholar] [CrossRef]
- Jonasch, E.; Walker, C.L.; Rathmell, W.K. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2021, 17, 245–261. [Google Scholar] [CrossRef]
- Napolitano, L.; Manfredi, C.; Cirillo, L.; Fusco, G.M.; Passaro, F.; Abate, M.; La Rocca, R.; Mastrangelo, F.; Spirito, L.; Pandolfo, S.D.; et al. Cytoreductive Nephrectomy and Metastatic Renal Cell Carcinoma: State of the Art and Future Perspectives. Medicina 2023, 59, 767. [Google Scholar] [CrossRef]
- Boissier, R.; Hevia, V.; Bruins, H.M.; Budde, K.; Figueiredo, A.; Lledo-Garcia, E.; Olsburgh, J.; Regele, H.; Taylor, C.F.; Zakri, R.H.; et al. The Risk of Tumour Recurrence in Patients Undergoing Renal Transplantation for End-stage Renal Disease after Previous Treatment for a Urological Cancer: A Systematic Review. Eur. Urol. 2018, 73, 94–108. [Google Scholar] [CrossRef]
- Yurchenco, P.D. Basement membranes: Cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 2011, 3, a004911. [Google Scholar] [CrossRef]
- Chang, J.; Chaudhuri, O. Beyond proteases: Basement membrane mechanics and cancer invasion. J. Cell Biol. 2019, 218, 2456–2469. [Google Scholar] [CrossRef]
- Fidler, A.L.; Darris, C.E.; Chetyrkin, S.V.; Pedchenko, V.K.; Boudko, S.P.; Brown, K.L.; Gray Jerome, W.; Hudson, J.K.; Rokas, A.; Hudson, B.G. Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues. eLife 2017, 6, e24176. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Dang, H.X.; Lim, D.A.; Feng, F.Y.; Maher, C.A. Long noncoding RNAs in cancer metastasis. Nat. Rev. Cancer 2021, 21, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Pandey, G.K.; Kanduri, C. Long Non-Coding RNAs: Tools for Understanding and Targeting Cancer Pathways. Cancers 2022, 14, 4760. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.F.; Farooqi, A.A.; Peng, S.Y.; Yu, T.J.; Dahms, H.U.; Lee, C.H.; Tang, J.Y.; Wang, S.C.; Shu, C.W.; Chang, H.W. Regulatory effects of noncoding RNAs on the interplay of oxidative stress and autophagy in cancer malignancy and therapy. Semin. Cancer Biol. 2022, 83, 269–282. [Google Scholar] [CrossRef]
- Shan, G.; Huang, T.; Tang, T. Long non-coding RNA MEG8 induced by PLAG1 promotes clear cell renal cell carcinoma through the miR-495-3p/G3BP1 axis. Pathol. Res. Pract. 2022, 229, 153734. [Google Scholar] [CrossRef]
- Zhang, Z.; Fu, X.; Gao, Y.; Nie, Z. LINC01535 Attenuates ccRCC Progression through Regulation of the miR-146b-5p/TRIM2 Axis and Inactivation of the PI3K/Akt Pathway. J. Oncol. 2022, 2022, 2153337. [Google Scholar] [CrossRef]
- Barik, G.K.; Sahay, O.; Behera, A.; Naik, D.; Kalita, B. Keep your eyes peeled for long noncoding RNAs: Explaining their boundless role in cancer metastasis, drug resistance, and clinical application. Biochim. Biophys Acta Rev. Cancer 2021, 1876, 188612. [Google Scholar] [CrossRef]
- Jayadev, R.; Morais, M.; Ellingford, J.M.; Srinivasan, S.; Naylor, R.W.; Lawless, C.; Li, A.S.; Ingham, J.F.; Hastie, E.; Chi, Q.; et al. A basement membrane discovery pipeline uncovers network complexity, regulators, and human disease associations. Sci. Adv. 2022, 8, eabn2265. [Google Scholar] [CrossRef]
- Fang, C.; Liu, S.; Feng, K.; Huang, C.; Zhang, Y.; Wang, J.; Lin, H.; Wang, J.; Zhong, C. Ferroptosis-related lncRNA signature predicts the prognosis and immune microenvironment of hepatocellular carcinoma. Sci. Rep. 2022, 12, 6642. [Google Scholar] [CrossRef]
- Morell-Quadreny, L.; Rubio, J.; Lopez-Guerrero, J.A.; Casanova, J.; Ramos, D.; Iborra, I.; Solsona, E.; Llombart-Bosch, A. Disruption of basement membrane, extracellular matrix metalloproteinases and E-cadherin in renal-cell carcinoma. Anticancer Res. 2003, 23, 5005–5010. [Google Scholar] [PubMed]
- Majo, S.; Courtois, S.; Souleyreau, W.; Bikfalvi, A.; Auguste, P. Impact of Extracellular Matrix Components to Renal Cell Carcinoma Behavior. Front. Oncol. 2020, 10, 625. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lu, H.; Tao, D.; Fan, M.; Zhuang, Q.; Xing, Z.; Chen, Z.; He, X. Membrane type-2 matrix metalloproteinases improve the progression of renal cell cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 10618–10626. [Google Scholar] [PubMed]
- Zhou, J.; Zhang, Y.; Li, S.; Zhou, Q.; Lu, Y.; Shi, J.; Liu, J.; Wu, Q.; Zhou, S. Dendrobium nobile Lindl. alkaloids-mediated protection against CCl(4-)induced liver mitochondrial oxidative damage is dependent on the activation of Nrf2 signaling pathway. BioMedicine 2020, 129, 110351. [Google Scholar] [CrossRef]
- Zhao, P.; Deng, Y.; Wu, Y.; Guo, Q.; Zhou, L.; Yang, X.; Wang, C. Long noncoding RNA SNHG6 promotes carcinogenesis by enhancing YBX1-mediated translation of HIF1alpha in clear cell renal cell carcinoma. FASEB J. 2021, 35, e21160. [Google Scholar] [CrossRef]
- Li, J.K.; Chen, C.; Liu, J.Y.; Shi, J.Z.; Liu, S.P.; Liu, B.; Wu, D.S.; Fang, Z.Y.; Bao, Y.; Jiang, M.M.; et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol. Cancer 2017, 16, 111. [Google Scholar] [CrossRef]
- Song, C.; Xiong, Y.; Liao, W.; Meng, L.; Yang, S. Long noncoding RNA ATB participates in the development of renal cell carcinoma by downregulating p53 via binding to DNMT1. J. Cell. Physiol. 2019, 234, 12910–12917. [Google Scholar] [CrossRef]
- Yue, H.; Wu, K.; Liu, K.; Gou, L.; Huang, A.; Tang, H. LINC02154 promotes the proliferation and metastasis of hepatocellular carcinoma by enhancing SPC24 promoter activity and activating the PI3K-AKT signaling pathway. Cell. Oncol. 2022, 45, 447–462. [Google Scholar] [CrossRef]
- Zhang, G.; Fan, E.; Zhong, Q.; Feng, G.; Shuai, Y.; Wu, M.; Chen, Q.; Gou, X. Identification and potential mechanisms of a 4-lncRNA signature that predicts prognosis in patients with laryngeal cancer. Hum. Genom. 2019, 13, 36. [Google Scholar] [CrossRef]
- Liu, L.; Zhuang, M.; Tu, X.H.; Li, C.C.; Liu, H.H.; Wang, J. Bioinformatics analysis of markers based on m(6) A related to prognosis combined with immune invasion of renal clear cell carcinoma. Cell Biol. Int. 2023, 47, 260–272. [Google Scholar] [CrossRef]
- Shen, J.; Wang, L.; Bi, J. Bioinformatics analysis and experimental validation of cuproptosis-related lncRNA LINC02154 in clear cell renal cell carcinoma. BMC Cancer 2023, 23, 160. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, S.; Tan, L.; Li, H.; Liu, J.; Zhang, S. IGFL2-AS1 facilitates tongue squamous cell carcinoma progression via Wnt/beta-catenin signaling pathway. Oral Dis. 2023, 29, 469–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shi, Y.; Chen, C.H.; Wen, Y.; Zhou, Z.; Yang, C.; Sun, J.; Du, G.; Wu, J.; Mao, X.; et al. KLF5-induced lncRNA IGFL2-AS1 promotes basal-like breast cancer cell growth and survival by upregulating the expression of IGFL1. Cancer Lett. 2021, 515, 49–62. [Google Scholar] [CrossRef]
- Tracy, K.M.; Tye, C.E.; Page, N.A.; Fritz, A.J.; Stein, J.L.; Lian, J.B.; Stein, G.S. Selective expression of long non-coding RNAs in a breast cancer cell progression model. J. Cell. Physiol. 2018, 233, 1291–1299. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, Y.; Pu, Y.S.; Cui, M.L.; Mao, Z.J.; Li, Z.Z.; He, L.; Wu, M.; Wang, J.H. LncRNA IGFL2-AS1 functions as a ceRNA in regulating ARPP19 through competitive binding to miR-802 in gastric cancer. Mol. Carcinog. 2020, 59, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Cen, X.; Huang, Y.; Lu, Z.; Shao, W.; Zhuo, C.; Bao, C.; Feng, S.; Wei, C.; Tang, X.; Cen, L.; et al. LncRNA IGFL2-AS1 Promotes the Proliferation, Migration, and Invasion of Colon Cancer Cells and is Associated with Patient Prognosis. Cancer Manag. Res. 2021, 13, 5957–5968. [Google Scholar] [CrossRef]
- Cheng, B.; Xie, M.; Zhou, Y.; Li, T.; Liu, W.; Yu, W.; Jia, M.; Yu, S.; Chen, L.; Dai, R.; et al. Vascular mimicry induced by m(6)A mediated IGFL2-AS1/AR axis contributes to pazopanib resistance in clear cell renal cell carcinoma. Cell Death Discov. 2023, 9, 121. [Google Scholar] [CrossRef]
- Pan, Y.; Lu, X.; Shu, G.; Cen, J.; Lu, J.; Zhou, M.; Huang, K.; Dong, J.; Li, J.; Lin, H.; et al. Extracellular Vesicle-Mediated Transfer of LncRNA IGFL2-AS1 Confers Sunitinib Resistance in Renal Cell Carcinoma. Cancer Res. 2023, 83, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Wang, L.; Zhang, H.; Liang, C.; Li, B. Identification of a 5-Gene Signature Predicting Progression and Prognosis of Clear Cell Renal Cell Carcinoma. Med. Sci. Monit. 2019, 25, 4401–4413. [Google Scholar] [CrossRef]
- Peng, D.; Fu, M.; Wang, M.; Wei, Y.; Wei, X. Targeting TGF-beta signal transduction for fibrosis and cancer therapy. Mol. Cancer 2022, 21, 104. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, M.M.; Zhang, G.G.; Yang, J.; Chen, K.S.; Xu, W.W.; Li, B. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduct. Target. Ther. 2021, 6, 425. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020, 20, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Morad, G.; Helmink, B.A.; Sharma, P.; Wargo, J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2021, 184, 5309–5337. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Kuang, Q.; Peng, M.; Yang, K.; Luo, P. Basement Membrane-Associated lncRNA Risk Model Predicts Prognosis and Guides Clinical Treatment in Clear Cell Renal Cell Carcinoma. Biomedicines 2023, 11, 2635. https://doi.org/10.3390/biomedicines11102635
Li X, Kuang Q, Peng M, Yang K, Luo P. Basement Membrane-Associated lncRNA Risk Model Predicts Prognosis and Guides Clinical Treatment in Clear Cell Renal Cell Carcinoma. Biomedicines. 2023; 11(10):2635. https://doi.org/10.3390/biomedicines11102635
Chicago/Turabian StyleLi, Xinxin, Qihui Kuang, Min Peng, Kang Yang, and Pengcheng Luo. 2023. "Basement Membrane-Associated lncRNA Risk Model Predicts Prognosis and Guides Clinical Treatment in Clear Cell Renal Cell Carcinoma" Biomedicines 11, no. 10: 2635. https://doi.org/10.3390/biomedicines11102635
APA StyleLi, X., Kuang, Q., Peng, M., Yang, K., & Luo, P. (2023). Basement Membrane-Associated lncRNA Risk Model Predicts Prognosis and Guides Clinical Treatment in Clear Cell Renal Cell Carcinoma. Biomedicines, 11(10), 2635. https://doi.org/10.3390/biomedicines11102635






