Comparative Efficacy of Metformin and Glimepiride in Modulating Pharmacological Network to Increase BDNF Levels and Benefit Type 2 Diabetes-Related Cognitive Impairment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Text Mining of T2DM-Associated Proteins/Genes
2.2. Drug Interactomes and Functional Enrichment
2.3. Critical Signals Transduction Process
2.4. Recruitment of the Study Individuals
2.5. RNA Isolation from Peripheral Blood Mononuclear Cells
2.6. cDNA Synthesis and Gene Expression Analysis
2.7. Estimation of Serum BDNF Levels
2.8. Statistical Analysis
3. Results
3.1. Text-Mining and T2DM Network
3.2. Drug Interactome Enrichment Analysis
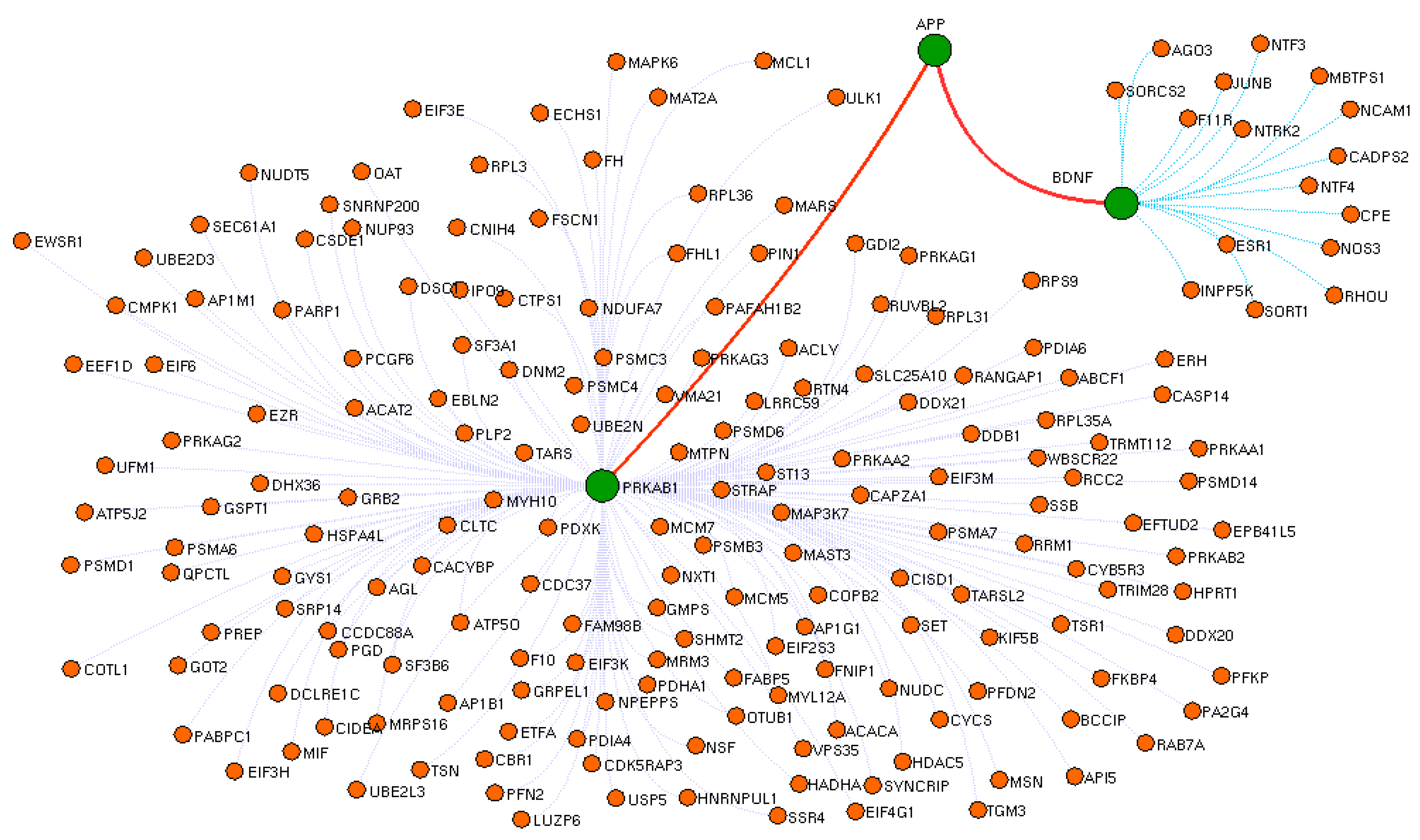
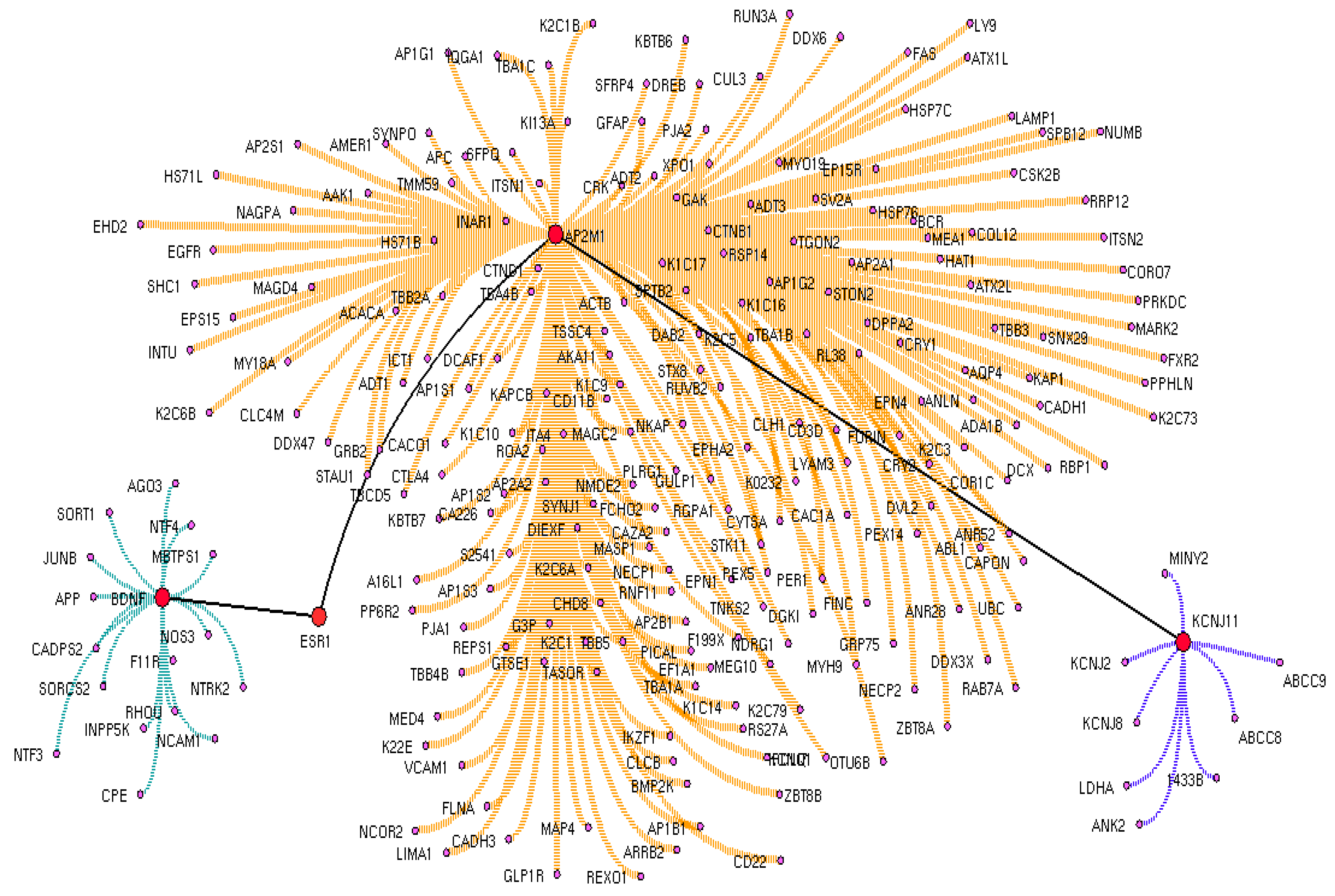
3.3. Crucial Protein in Drug Interactome
3.4. Gene Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of Type 2 Diabetes Mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Alam, S.; Hasan, M.K.; Neaz, S.; Hussain, N.; Hossain, M.F.; Rahman, T. Diabetes Mellitus: Insights from epidemiology, biochemistry, risk factors, diagnosis, complications and comprehensive management. Diabetology 2021, 2, 36–50. [Google Scholar] [CrossRef]
- Tyagi, A.; Pugazhenthi, S. Targeting Insulin Resistance to Treat Cognitive Dysfunction. Mol. Neurobiol. 2021, 58, 2672–2691. [Google Scholar] [CrossRef] [PubMed]
- De la Monte, M.S. Brain insulin resistance and deficiency as therapeutic targets in Alzheimer’s disease. Curr. Alzheimer Res. 2012, 9, 35–66. [Google Scholar] [CrossRef] [PubMed]
- Krentz, A.J.; Bailey, C.J. Oral antidiabetic agents. Drugs 2005, 65, 385–411. [Google Scholar] [CrossRef] [PubMed]
- Blahova, J.; Martiniakova, M.; Babikova, M.; Kovacova, V.; Mondockova, V.; Omelka, R. Pharmaceutical Drugs and Natural Therapeutic Products for the Treatment of Type 2 Diabetes Mellitus. Pharmaceuticals 2021, 14, 806. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, D.M.; Cha, B.S.; Park, T.S.; Kim, K.A.; Kim, D.L.; Chung, C.H.; Park, J.H.; Jang, H.C.; Choi, D.S. Efficacy of glimepiride/metformin fixed-dose combination vs metformin uptitration in type 2 diabetic patients inadequately controlled on low-dose metformin monotherapy: A randomized, open label, parallel group, multicenter study in Korea. J. Diabetes Investig. 2014, 5, 701–708. [Google Scholar] [CrossRef]
- Rojas, L.B.A.; Gomes, M.B. Metformin: An old but still the best treatment for type 2 diabetes. Diabetol. Metab. Syndr. 2013, 5, 6–15. [Google Scholar] [CrossRef]
- Basit, A.; Riaz, M.; Fawwad, A. Glimepiride: Evidence-based facts, trends, and observations. Vasc. Health Risk Manag. 2012, 8, 463. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Christian, K.; Lu, B. BDNF: A key regulator for protein synthesis-dependent LTP and long-term memory? Neurobiol. Learn. Mem. 2008, 89, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Manni, L.; Nikolova, V.; Vyagova, D.; Chaldakov, G.N.; Aloe, L. Reduced plasma levels of NGF and BDNF in patients with acute coronary syndromes. Int. J. Cardiol. 2005, 102, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Suwa, M.; Kishimoto, H.; Nofuji, Y.; Nakano, H.; Sasaki, H.; Radak, Z.; Kumagai, S. Serum brain-derived neurotrophic factor level is increased and associated with obesity in newly diagnosed female patients with type 2 diabetes mellitus. Metabolism 2006, 55, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Teillon, S.; Calderon, G.A.; Rios, M. Diminished diet-induced hyperglycemia and dyslipidemia and enhanced expression of PPARα and FGF21 in mice with hepatic ablation of brain-derived neurotropic factor. J. Endocrinol. 2010, 205, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, M.; Itakura, Y.; Tsuchida, A.; Nakagawa, T.; Taiji, M. Brain-derived neurotrophic factor (BDNF) prevents the development of diabetes in prediabetic mice. Biomed. Res. 2008, 29, 147–153. [Google Scholar] [CrossRef]
- Li, B.; Lang, N.; Cheng, Z.F. Serum Levels of Brain-Derived Neurotrophic Factor Are Associated with Diabetes Risk, Complications, and Obesity: A Cohort Study from Chinese Patients with Type 2 Diabetes. Mol. Neurobiol. 2016, 53, 5492–5499. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Nora, E.D.; Morieri, M.L.; Soavi, C.; Sanz, J.M.; Zurlo, A.; Fellin, R.; Zuliani, G. Brain-Derived Neurotrophic Factor Plasma Levels: Relationship with Dementia and Diabetes in the Elderly Population. J. Gerontol. Ser. A 2015, 70, 294–302. [Google Scholar] [CrossRef]
- Krallinger, M.; Valencia, A.; Hirschman, L. Linking genes to literature: Text mining, information extraction, and retrieval applications for biology. Genome Biol. 2008, 9, S8. [Google Scholar] [CrossRef]
- Pandi, M.-T.; van der Spek, P.J.; Koromina, M.; Patrinos, G.P. A Novel Text-Mining Approach for Retrieving Pharmacogenomics Associations from the Literature. Front. Pharmacol. 2020, 11, 602030. [Google Scholar] [CrossRef]
- Rani, J.; Shah, A.R.; Ramachandran, S. pubmed.mineR: An R package with text-mining algorithms to analyse PubMed abstracts. J. Biosci. 2015, 40, 671–682. [Google Scholar] [CrossRef]
- Available online: https://cran.r-project.org/web/packages/polmineR/vignettes/vignette.html (accessed on 14 January 2023).
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36 (Suppl. S1), D901–D906. [Google Scholar] [CrossRef]
- Brown, K.R.; Otasek, D.; Ali, M.; McGuffin, M.J.; Xie, W.; Devani, B.; van Toch, I.L.; Jurisica, I. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics 2009, 25, 3327–3329. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Donohue, D.; Hoke, A.; Miller, S.A.; Srinivasan, S.; Sowe, B.; Detwiler, L.; Lynch, J.; Levangie, M.; Hammamieh, R.; et al. Investigating gene expression profiles of whole blood and peripheral blood mononuclear cells using multiple collection and processing methods. PLoS ONE 2019, 14, e0225137. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; Conklin, D. NanoDrop microvolume quantitation of nucleic acids. J. Vis. Exp. 2010, 22, e2565. [Google Scholar]
- Krabbe, K.; Nielsen, A.R.; Krogh-Madsen, R.; Plomgaard, P.; Rasmussen, P.; Erikstrup, C.; Fischer, C.P.; Lindegaard, B.; Petersen, A.M.; Taudorf, S.; et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia 2007, 50, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Brunoni, A.R.; Lopes, M.; Fregni, F. A systematic review and meta-analysis of clinical studies on major depression and BDNF levels: Implications for the role of neuroplasticity in depression. Int. J. Neuropsychopharmacol. 2008, 11, 1169–1180. [Google Scholar] [CrossRef]
- Rahmani, F.; Saghazadeh, A.; Rahmani, M.; Teixeira, A.L.; Rezaei, N.; Aghamollaii, V.; Ardebili, H.E. Plasma levels of brain-derived neurotrophic factor in patients with Parkinson disease: A systematic review and meta-analysis. Brain Res. 2019, 1704, 127–136. [Google Scholar] [CrossRef]
- Ng, T.K.S.; Ho, C.S.H.; Tam, W.W.S.; Kua, E.H.; Ho, R.C.-M. Decreased serum brain-derived neurotrophic factor (BDNF) levels in patients with Alzheimer’s disease (AD): A systematic review and meta-analysis. Int. J. Mol. Sci. 2019, 20, 257. [Google Scholar] [CrossRef]
- Suliman, S.; Hemmings, S.M.J.; Seedat, S. Brain-Derived Neurotrophic Factor (BDNF) protein levels in anxiety disorders: Systematic review and meta-regression analysis. Front. Integr. Neurosci. 2013, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Mojtabavi, H.; Saghazadeh, A.; Van den Heuvel, L.; Bucker, J.; Rezaei, N. Peripheral blood levels of brain-derived neurotrophic factor in patients with post-traumatic stress disorder (PTSD): A systematic review and meta-analysis. PLoS ONE 2020, 15, e0241928. [Google Scholar] [CrossRef] [PubMed]
- Saghazadeh, A.; Rezaei, N. Brain-Derived Neurotrophic Factor Levels in Autism: A Systematic Review and Meta-Analysis. J. Autism Dev. Disord. 2017, 47, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Han, X.; Zhou, X.; Zhang, S.; Cai, X.; Zhang, L.; Li, Y.; Li, M.; Gong, S.; Ji, L. Brain derived neurotrophic factor in newly diagnosed diabetes and prediabetes. Mol. Cell. Endocrinol. 2016, 429, 106–113. [Google Scholar] [CrossRef]
- Abu El-Asrar, A.M.; Nawaz, M.I.; Siddiquei, M.M.; Al-Kharashi, A.S.; Kangave, D.; Mohammad, G. High-Mobility Group Box-1 Induces Decreased Brain-Derived Neurotrophic Factor-Mediated Neuroprotection in the Diabetic Retina. Mediat. Inflamm. 2013, 2013, 863036. [Google Scholar] [CrossRef]
- Garcia, M.E.; Lee, A.; Neuhaus, J.; Gonzalez, H.; To, T.M.; Haan, M.N. Diabetes Mellitus as a Risk Factor for Development of Depressive Symptoms in a Population-Based Cohort of Older Mexican Americans. J. Am. Geriatr. Soc. 2016, 64, 619–624. [Google Scholar] [CrossRef]
- Hasan, S.S.; Clavarino, A.M.; Dingle, K.; Mamun, A.A.; Kairuz, T. Diabetes Mellitus and the Risk of Depressive and Anxiety Disorders in Australian Women: A Longitudinal Study. J. Women’s Health 2015, 24, 889–898. [Google Scholar] [CrossRef]
- Rozanska, O.; Uruska, A.; Zozulinska-Ziolkiewicz, D. Brain-Derived Neurotrophic Factor and Diabetes. Int. J. Mol. Sci. 2020, 21, 841. [Google Scholar] [CrossRef]
- Kim, O.Y.; Song, J. The importance of BDNF and RAGE in diabetes-induced dementia. Pharmacol. Res. 2020, 160, 105083. [Google Scholar] [CrossRef]
- Singh, V.P.; Bali, A.; Singh, N.; Jaggi, A.S. Advanced Glycation End Products and Diabetic Complications. Korean J. Physiol. Pharmacol. 2014, 18, 1–14. [Google Scholar] [CrossRef]
- Hristova, M.G. Metabolic Syndrome and Neurotrophins: Effects of Metformin and Non-Steroidal Antiinflammatory Drug Treatment. Eurasian J. Med. 2011, 42, 141–145. [Google Scholar] [CrossRef]
- Yoo, D.Y.; Kim, W.; Nam, S.M.; Yoo, K.Y.; Lee, C.H.; Choi, J.H.; Won, M.H.; Hwang, I.K.; Yoon, Y.S. Reduced cell proliferation and neuroblast differentiation in the dentate gyrus of high fat diet-fed mice are ameliorated by metformin and glimepiride treatment. Neurochem. Res. 2011, 36, 2401–2408. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.; Jain, P.D.; Ghumatkar, P.J.; Tambe, R.; Sathaye, S. Neuroprotective effect of metformin in MPTP-induced Parkinson’s disease in mice. Neuroscience 2014, 277, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Allard, J.S.; Perez, E.J.; Fukui, K.; Carpenter, P.; Ingram, D.K.; de Cabo, R. Prolonged metformin treatment leads to reduced transcription of Nrf2 and neurotrophic factors without cognitive impairment in older C57BL/6J mice. Behav. Brain Res. 2016, 301, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, J.; Yu, H.; Chen, Y.; Wang, Q.; Xiang, L. Effect of metformin on Schwann cells under hypoxia condition. Int. J. Clin. Exp. Pathol. 2015, 8, 6748–6755. [Google Scholar]
- Sola, D.; Rossi, L.; Schianca, G.P.; Maffioli, P.; Bigliocca, M.; Mella, R.; Corlianò, F.; Fra, G.P.; Bartoli, E.; Derosa, G. Sulfonylureas and their use in clinical practice. Arch. Med. Sci. 2015, 11, 840–848. [Google Scholar] [CrossRef]
- Butterfield, D.A. Oxidative stress in neurodegenerative disorders. Antioxid. Redox Signal 2006, 8, 1971–1973. [Google Scholar] [CrossRef]
- Markesbery, W.R. Oxidative Stress Hypothesis in Alzheimer’s Disease. Free Radic. Biol. Med. 1997, 23, 134–147. [Google Scholar] [CrossRef]
- Saedi, E.; Gheini, M.R.; Faiz, F.; Arami, M.A. Diabetes mellitus and cognitive impairments. World J. Diabetes 2016, 7, 412–422. [Google Scholar] [CrossRef]
- Gregorio, K.C.; Laurindo, C.P.; Machado, U.F. Estrogen and glycemic homeostasis: The fundamental role of nuclear estrogen receptors ESR1/ESR2 in glucose transporter GLUT4 regulation. Cells 2021, 10, 99. [Google Scholar] [CrossRef]
- Faustini-Fustini, M.; Rochira, V.; Carani, C. Oestrogen deficiency in men: Where are we today? Eur. J. Endocrinol. 1999, 140, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Belgorosky, A.; Guercio, G.; Pepe, C.; Saraco, N.; Rivarola, M. Genetic and Clinical Spectrum of Aromatase Deficiency in Infancy, Childhood and Adolescence. Horm. Res. Paediatr. 2009, 72, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Sundermann, E.E.; Maki, P.M.; Bishop, J.R. A review of estrogen receptor alpha gene (ESR1) polymorphisms, mood, and cognition. Menopause 2010, 17, 874–886. [Google Scholar] [CrossRef] [PubMed]




| Clinical Characteristics of Participants | Group 1: Untreated T2DM | Group 2: Metformin | Group 3: Glimepiride | Group 4: Healthy Control |
|---|---|---|---|---|
| Age (years) | 43 ± 6 | 40 ± 3 | 41 ± 6 | 39 ± 8 |
| Gender (Male/Female) | 18/12 | 15/15 | 13/17 | 20/10 |
| HbA1C(%) | 6.9 ± 0.7 | 7.2 ± 0.8 | 7.9 ± 0.2 | 5.2 ± 0.3 |
| Onset (months) | 2 ± 0.5 | 12 ± 2 | 11 ± 4 | -- |
| Drug Dosage per day | -- | Metformin-500 mg | Glimepiride-2 mg | -- |
| Gene | Orientation | Primer |
|---|---|---|
| APP | FORWARD | TGGCCAACATGATTAGTGAACC |
| REVERSE | AAGATGGCATGAGAGCATCGT | |
| GAPDH | FORWARD | GCCACATCGCTCAGACACC |
| REVERSE | AATCCGTTGACTCCGACCTTC | |
| AP2M1 | FORWARD | TCTCGGCTTGTCCTAACACAG |
| REVERSE | CTTACAGCCGCTCTCCTCTG | |
| ESR1 | FORWARD | CTCTTAGCTATGCCTGGGGC |
| REVERSE | GGTGGATGTTTACTCCGCCA | |
| BDNF | FORWARD | GATGCTGCAAACATGTCCATGAG |
| REVERSE | TTTTGTCTGCCGCCGTTACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anirudhan, A.; Ahmad, S.F.; Emran, T.B.; Angulo-Bejarano, P.I.; Sharma, A.; Ahmed, S.S.S.J. Comparative Efficacy of Metformin and Glimepiride in Modulating Pharmacological Network to Increase BDNF Levels and Benefit Type 2 Diabetes-Related Cognitive Impairment. Biomedicines 2023, 11, 2939. https://doi.org/10.3390/biomedicines11112939
Anirudhan A, Ahmad SF, Emran TB, Angulo-Bejarano PI, Sharma A, Ahmed SSSJ. Comparative Efficacy of Metformin and Glimepiride in Modulating Pharmacological Network to Increase BDNF Levels and Benefit Type 2 Diabetes-Related Cognitive Impairment. Biomedicines. 2023; 11(11):2939. https://doi.org/10.3390/biomedicines11112939
Chicago/Turabian StyleAnirudhan, Athira, Sheikh F. Ahmad, Talha Bin Emran, Paola Isabel Angulo-Bejarano, Ashutosh Sharma, and Shiek S. S. J. Ahmed. 2023. "Comparative Efficacy of Metformin and Glimepiride in Modulating Pharmacological Network to Increase BDNF Levels and Benefit Type 2 Diabetes-Related Cognitive Impairment" Biomedicines 11, no. 11: 2939. https://doi.org/10.3390/biomedicines11112939







