Gene Set Enrichment Analysis and Genetic Experiment Reveal Changes in Cell Signaling Pathways Induced by α-Synuclein Overexpression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Acquisition
2.2. Differential Expression Matrix Acquisition
2.3. Acquisition of Heat Map and Volcano Map
2.4. GO and KEGG Analysis
2.5. Gene set Enrichment Analysis
2.6. Drosophila Culture, qRT-PCR Analysis and Immunostaining of Fly Brains
3. Results
3.1. Differentially Expressed Genes
3.2. Clustering of Differential Expression Profiles
3.3. Interaction Network of Differentially Expressed Genes
3.4. GO and KEGG Enrichment Analysis
3.5. Gene Set Enrichment Analysis
3.6. Reliability Analysis of Gene Enrichment
3.7. Knockdown of TNF-α Pathway Partially Rescues the DA Neurons Degeneration Caused by α-Syn Overexpression
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maroteaux, L.; Campanelli, J.T.; Scheller, R.H. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J. Neurosci. 1988, 8, 2804–2815. [Google Scholar] [CrossRef] [Green Version]
- Burre, J. The Synaptic Function of alpha-Synuclein. J. Parkinsons Dis. 2015, 5, 699–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lykkebo, S.; Jensen, P.H. Alpha-synuclein and presynaptic function: Implications for Parkinson’s disease. Neuromol. Med. 2002, 2, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Bekkers, J.M. Counting the number of releasable synaptic vesicles in a presynaptic terminal. Proc. Natl. Acad. Sci. USA 2009, 106, 2945–2950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polymeropoulos, M.H.; Lavedan, C.; Leroy, E.; Ide, S.E.; Dehejia, A.; Dutra, A.; Pike, B.; Root, H.; Rubenstein, J.; Boyer, R.; et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science 1997, 276, 2045–2047. [Google Scholar] [CrossRef] [Green Version]
- Kruger, R.; Kuhn, W.; Muller, T.; Woitalla, D.; Graeber, M.; Kosel, S.; Przuntek, H.; Epplen, J.T.; Schols, L.; Riess, O. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat. Genet. 1998, 18, 106–108. [Google Scholar] [CrossRef] [PubMed]
- Nonnekes, J.; Post, B.; Tetrud, J.W.; Langston, J.W.; Bloem, B.R. MPTP-induced parkinsonism: An historical case series. Lancet Neurol. 2018, 17, 300–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhyre, T.R.; Boyd, J.T.; Hamill, R.W.; Maguire-Zeiss, K.A. Parkinson’s disease. Subcell Biochem. 2012, 65, 389–455. [Google Scholar] [CrossRef] [Green Version]
- Andrei, S. Biomarkers in Parkinson’s Disease; Humana: Totowa, NJ, USA, 2022; pp. 155–180. [Google Scholar] [CrossRef]
- Stefanis, L. Alpha-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2012, 2, a009399. [Google Scholar] [CrossRef]
- Ozansoy, M.; Basak, A.N. The central theme of Parkinson’s disease: Alpha-synuclein. Mol. Neurobiol. 2013, 47, 460–465. [Google Scholar] [CrossRef]
- Lücking, C. Alpha-synuclein and Parkinson’s disease. Cell. Mol. Life Sci. CMLS 2000, 57, 1894–1908. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Pu, J. Alpha-Synuclein in Parkinson’s Disease: From Pathogenetic Dysfunction to Potential Clinical Application. Parkinsons Dis. 2016, 2016, 1720621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Y.; Yu, J. Modeling Parkinson’s Disease in Drosophila: What Have We Learned for Dominant Traits? Front. Neurol. 2018, 9, 228. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Sango, K.; Nagai, Y. Roles of alpha-Synuclein and Disease-Associated Factors in Drosophila Models of Parkinson’s Disease. Int. J. Mol. Sci. 2022, 23, 1519. [Google Scholar] [CrossRef] [PubMed]
- Feany, M.B.; Bender, W.W. A Drosophila model of Parkinson’s disease. Nature 2000, 404, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Surguchov, A. Invertebrate Models Untangle the Mechanism of Neurodegeneration in Parkinson’s Disease. Cells 2021, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Pinho, R.; Paiva, I.; Jercic, K.G.; Fonseca-Ornelas, L.; Gerhardt, E.; Fahlbusch, C.; Garcia-Esparcia, P.; Kerimoglu, C.; Pavlou, M.A.S.; Villar-Pique, A.; et al. Nuclear localization and phosphorylation modulate pathological effects of alpha-synuclein. Hum. Mol. Genet. 2019, 28, 31–50. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Igaki, T.; Kanda, H.; Yamamoto-Goto, Y.; Kanuka, H.; Kuranaga, E.; Aigaki, T.; Miura, M. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002, 21, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Andersen, D.S.; Colombani, J.; Palmerini, V.; Chakrabandhu, K.; Boone, E.; Rothlisberger, M.; Toggweiler, J.; Basler, K.; Mapelli, M.; Hueber, A.O.; et al. The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature 2015, 522, 482–486. [Google Scholar] [CrossRef] [PubMed]
- Kanda, H.; Igaki, T.; Kanuka, H.; Yagi, T.; Miura, M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J. Biol. Chem. 2002, 277, 28372–28375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daubner, S.C.; Le, T.; Wang, S. Tyrosine hydroxylase and regulation of dopamine synthesis. Arch. Biochem. Biophys. 2011, 508, 1–12. [Google Scholar] [CrossRef]
- Sadaei, H.J.; Cordova-Palomera, A.; Lee, J.; Padmanabhan, J.; Chen, S.F.; Wineinger, N.E.; Dias, R.; Prilutsky, D.; Szalma, S.; Torkamani, A. Genetically-informed prediction of short-term Parkinson’s disease progression. NPJ Parkinsons Dis. 2022, 8, 143. [Google Scholar] [CrossRef]
- Ayajuddin, M.; Phom, L.; Koza, Z.; Modi, P.; Das, A.; Chaurasia, R.; Thepa, A.; Jamir, N.; Neikha, K.; Yenisetti, S.C. Adult health and transition stage-specific rotenone-mediated Drosophila model of Parkinson’s disease: Impact on late-onset neurodegenerative disease models. Front. Mol. Neurosci. 2022, 15, 896183. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Da Silva Santos, C.; Zavala, N.C.; Gans, N.; Patracuolla, D.; Fehrenbach, M.; Babcock, D.T. Characterizing dopaminergic neuron vulnerability using genome-wide analysis. Genetics 2021, 218, iyab081. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, J.; Chen, W.; Chen, Y.; Zhang, X.; Yang, M.; Xiao, M.; Ma, F.; Yao, Y.; Ye, M.; et al. Attenuation of epigenetic regulator SMARCA4 and ERK-ETS signaling suppresses aging-related dopaminergic degeneration. Aging Cell 2020, 19, e13210. [Google Scholar] [CrossRef]
- Park, J.; Lee, S.B.; Lee, S.; Kim, Y.; Song, S.; Kim, S.; Bae, E.; Kim, J.; Shong, M.; Kim, J.M.; et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 2006, 441, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, A.J.; Theodore, D.A.; Greene, J.C.; Benes, H.; Wes, P.D.; Pallanck, L.J. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 8024–8029. [Google Scholar] [CrossRef] [Green Version]
- Trinh, K.; Moore, K.; Wes, P.D.; Muchowski, P.J.; Dey, J.; Andrews, L.; Pallanck, L.J. Induction of the phase II detoxification pathway suppresses neuron loss in Drosophila models of Parkinson’s disease. J. Neurosci. 2008, 28, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.X.; Cheng, X.Y.; Jin, M.; Cao, Y.L.; Yang, Y.P.; Wang, J.D.; Li, Q.; Wang, F.; Hu, L.F.; Liu, C.F. TNF compromises lysosome acidification and reduces alpha-synuclein degradation via autophagy in dopaminergic cells. Exp. Neurol. 2015, 271, 112–121. [Google Scholar] [CrossRef]
- Sznejder-Pacholek, A.; Joniec-Maciejak, I.; Wawer, A.; Ciesielska, A.; Mirowska-Guzel, D. The effect of alpha-synuclein on gliosis and IL-1alpha, TNFalpha, IFNgamma, TGFbeta expression in murine brain. Pharmacol. Rep. 2017, 69, 242–251. [Google Scholar] [CrossRef]
- Palmerini, V.; Monzani, S.; Laurichesse, Q.; Loudhaief, R.; Mari, S.; Cecatiello, V.; Olieric, V.; Pasqualato, S.; Colombani, J.; Andersen, D.S.; et al. Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions. Nat. Commun. 2021, 12, 2070. [Google Scholar] [CrossRef]
- Ludtmann, M.H.; Angelova, P.R.; Ninkina, N.N.; Gandhi, S.; Buchman, V.L.; Abramov, A.Y. Monomeric Alpha-Synuclein Exerts a Physiological Role on Brain ATP Synthase. J. Neurosci. 2016, 36, 10510–10521. [Google Scholar] [CrossRef] [Green Version]
- Grassi, D.; Howard, S.; Zhou, M.; Diaz-Perez, N.; Urban, N.T.; Guerrero-Given, D.; Kamasawa, N.; Volpicelli-Daley, L.A.; LoGrasso, P.; Lasmezas, C.I. Identification of a highly neurotoxic alpha-synuclein species inducing mitochondrial damage and mitophagy in Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2018, 115, E2634–E2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.S.; Kim, Y.M.; Junn, E.; Lee, G.; Park, K.H.; Tanaka, M.; Ronchetti, R.D.; Quezado, M.M.; Mouradian, M.M. Cell cycle aberrations by alpha-synuclein over-expression and cyclin B immunoreactivity in Lewy bodies. Neurobiol. Aging 2003, 24, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.L.; Song, L.K.; Yuan, Y.H.; Zhang, Y.; Han, N.; Gao, K.; Chen, N.H. The nuclear accumulation of alpha-synuclein is mediated by importin alpha and promotes neurotoxicity by accelerating the cell cycle. Neuropharmacology 2014, 82, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Jellinger, K.A. Multiple System Atrophy: An Oligodendroglioneural Synucleinopathy1. J. Alzheimers Dis. 2018, 62, 1141–1179. [Google Scholar] [CrossRef] [Green Version]
- Valera, E.; Masliah, E. The neuropathology of multiple system atrophy and its therapeutic implications. Auton. Neurosci. 2018, 211, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Surmeier, D.J. Determinants of dopaminergic neuron loss in Parkinson’s disease. FEBS J. 2018, 285, 3657–3668. [Google Scholar] [CrossRef]
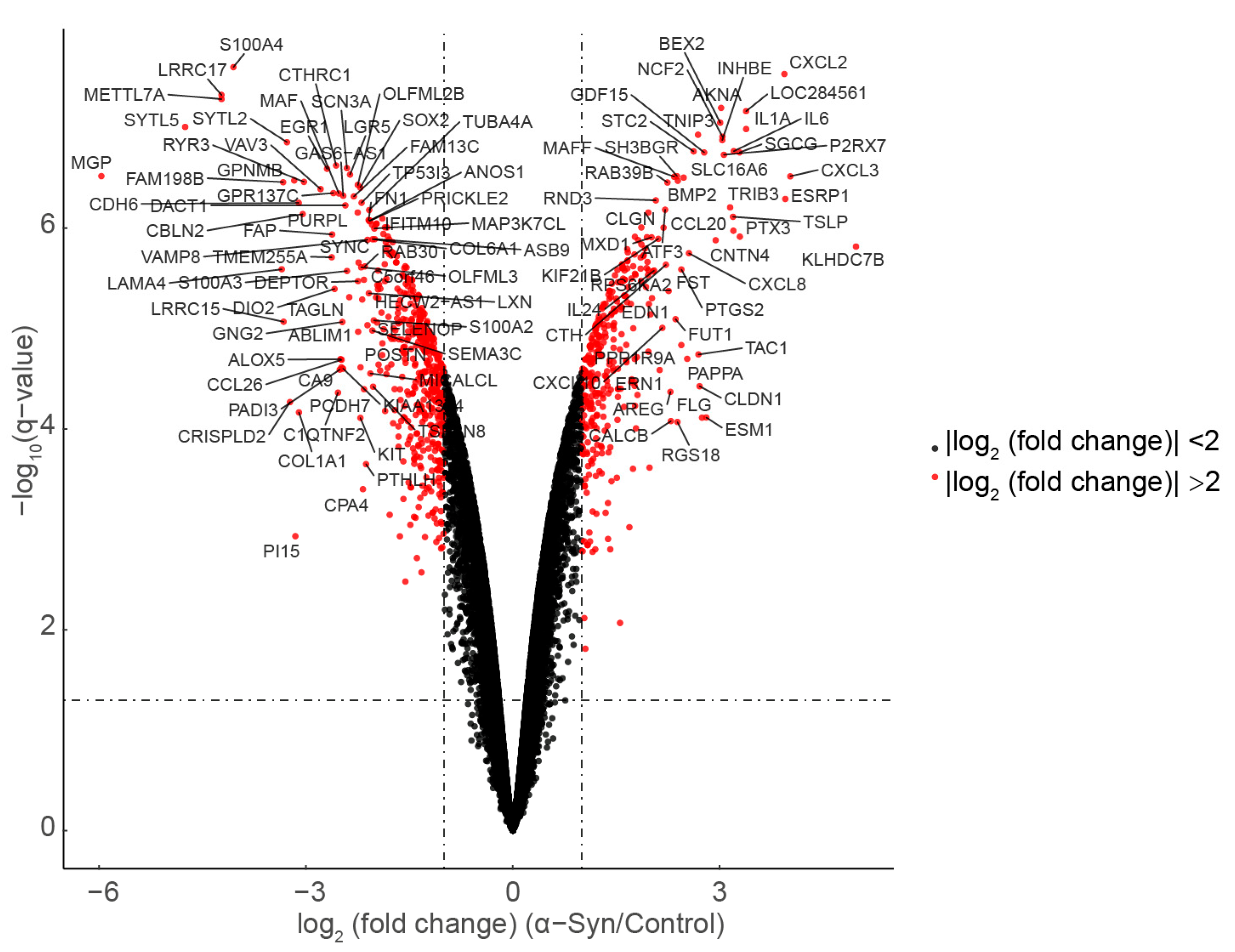

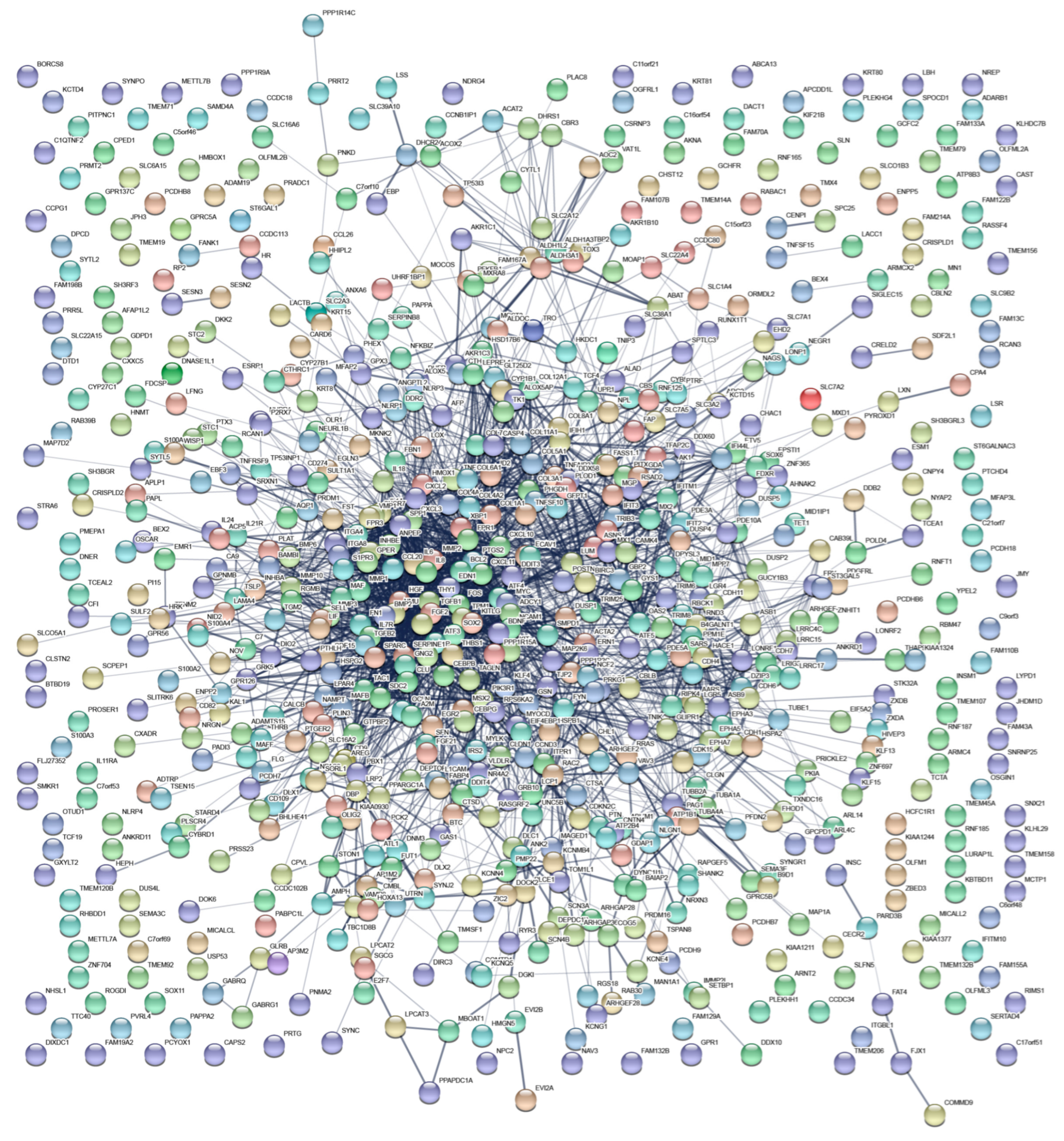
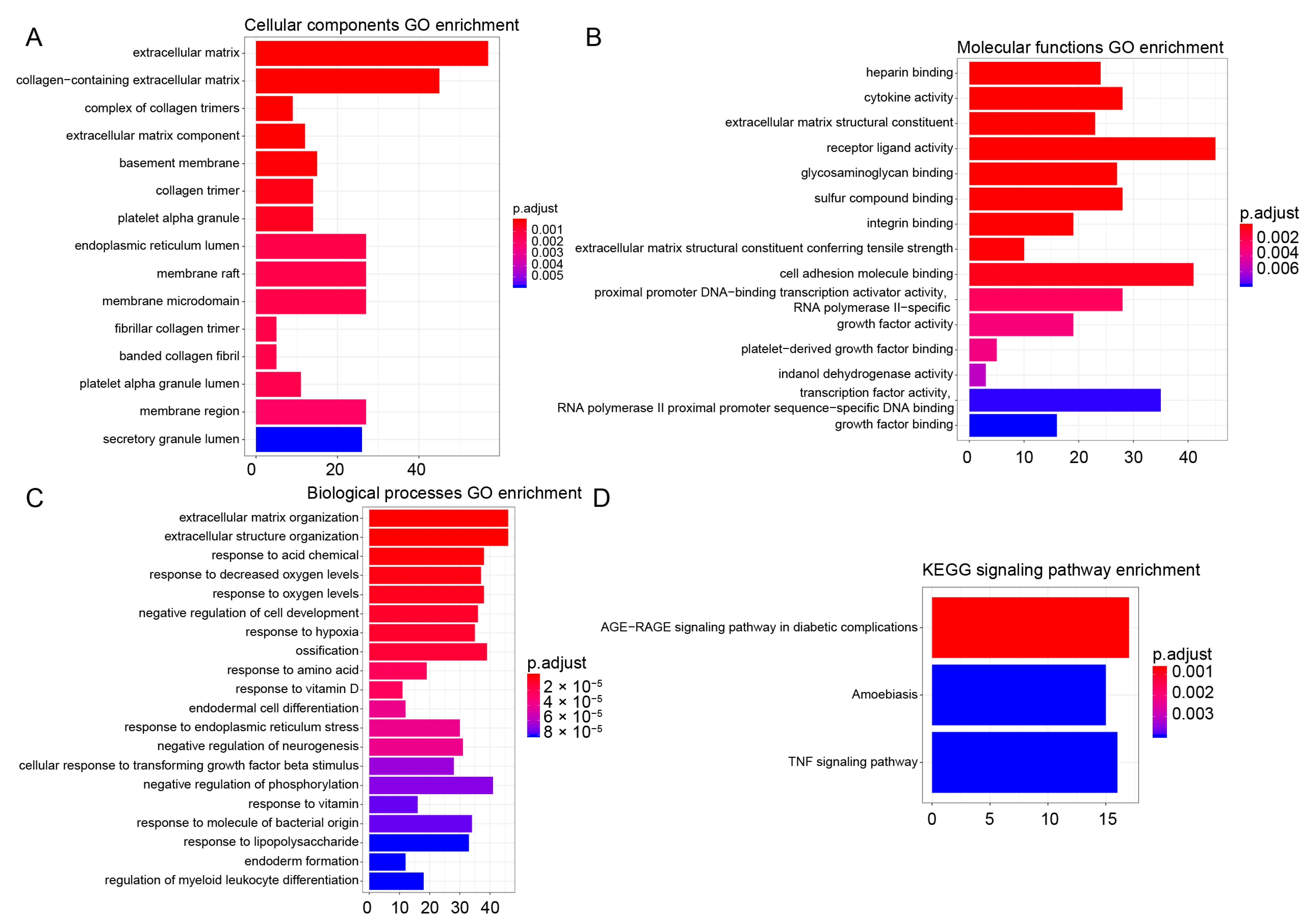
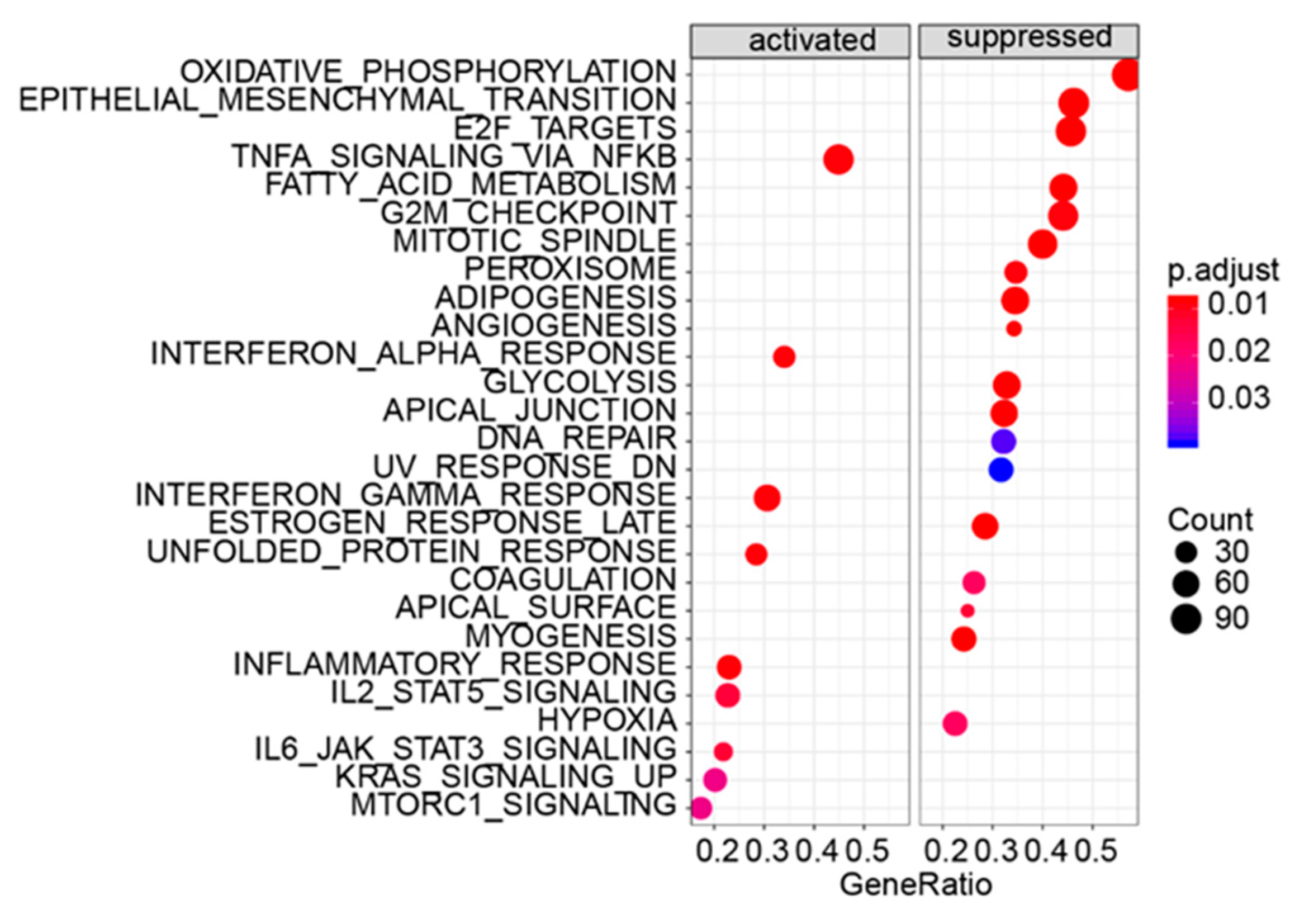


| Gene Sets | Gene Symbol of Enrichment Genes in NCBI Database |
|---|---|
| TNF-α_SIGNALING_VIA_NFKB | AREG/ATF3/BCL2A1/BIRC3/BMP2/BTG1/CCL20/CD69/CD83/CEBPB/CEBPD/CSF2/CXCL10/CXC/CXCL2/CXCL3/DENND5A/DRAM1/DUSP1/DUSP4/DUSP5/EDN1/EIF1/FJX1/FOSL1/GADD45A/GCH/HBEGF/IER3/IFIH1/IFIT2/IL15RA/IL1A/IL6/IL6ST/INHBA/IRF1/IRS2/KDM6B/KLF4/KLF9/KYNU/LDLR/LIF/MAFF/MCL1/MSC/MXD1/MYC/NAMPT/NFIL3/NFKB1/NFKB2/NFKBIA/NFKBIE/NR4A2/OLR1/PHLDA1/PNRC1/PPP1R15A/PTGS2/PTX3/RCAN1/REL/RELA/RELB/RIGI/RIPK2/SAT1/SERPINB2/SERPINB8/SIK1/SLC16A6/SLC2A3/SOCS3/SOD2/SQSTM1/TGIF1/TIPARP/TNFAIP3/TNFAIP6/TNFRSF9/TNIP1/TRIB1/TSC22D1/VEGFA/YRDC/ZBTB10 |
| OXIDATIVE_PHOSPHORYLATION | ACAA1/ACAA2/ACADM/ACADSB/ACADVL/ATP1B1/ATP5F1B/ATP5F1C/ATP5F1D/ATP5F1E /ATP5MC1/ATP5MC3/ATP5ME/ATP5MG/ATP5PB/ATP5PD/ATP5PF/ATP5PO/ATP6AP1/ATP6V0B/ATP6V0C/ATP6V0E1/ATP6V1D/ATP6V1E1/ATP6V1F/ATP6V1H/BAX/BDH2/CASP7/COX4I1/COX5A/COX5B/COX6A1/COX6B1/COX6C/COX7A2/COX7B/COX8A/CS/CYB5A/CYB5R3/DECR1/DLAT/ECH1/ECHS1/ETFA/ETFB/ETFDH/FDX1/FXN/GPI/GPX4/HSD17B10/HTRA2/IDH2/IDH3A/IDH3G ISCA1/LDHA/MDH1/MGST3/MPC1/MRPL15/MRPL34/MRPL35/MRPS11/MRPS12/MTRF1/NDUFA2/NDUFA3/NDUFA4/NDUFA5/NDUFA6/NDUFA7/NDUFA8/NDUFB1/NDUFB2/NDUFB3/NDUFB5/NDUFB6/NDUFB7/NDUFB8/NDUFS3/NDUFS6/NNT/OAT/OGDH/PDHB/PHYH/POLR2F/PRDX3/RETSAT/SDHB/SDHD/SLC25A11/SLC25A12/SLC25A20/SUCLA2/SUCLG1/TCIRG1/TIMM10/TIMM13/TIMM8B/TOMM22/UQCR10/UQCR11/UQCRB/UQCRC1/UQCRFS1/UQCRQ/VDAC1/VDAC3 |
| EPITHELIAL_MESENCHYMAL_TRANSTION | ACTA2/ANPEP/APLP1/BASP1/BMP1/CALD1/CALU/CAP2/CAPG/CD59/CDH11/CDH6/COL11A1/COL12A1/COL16A1/COL1A1/COL3A1/COL4A1/COL4A2/COL5A1/COL7A1/CRLF1/CTHRC1/DCN/DPYSL3/ECM1/EDIL3/EMP3/FAP/FAS/FBLN5/FBN1/FBN2/FLNA/FN1/FSTL3/FUCA1/GAS1/GEM/GLIPR1/GPC1/HTRA1/ID2/IGFBP2/ITGA5/ITGAV/ITGB3/ITGB5/LAMA1/LAMA2/LAMC1/LGALS1/LOXL1/LRRC15/LUM/MATN2/MATN3/MGP/MMP2/MYLK/NID2/P3H1/PCOLCE/PDGFRB/PDLIM4/PLOD1/PLOD2/PLOD3/PMEPA1/PMP22/POSTN/PTHLH/RHOB/SDC1/SERPINE1/SFRP1/SGCB/SNAI2/SPARC/SPOCK1/SPP1/TAGLN/TGFB1/TGFBI/TGM2/THBS1/THY1/TIMP1/TNFRSF11B/TPM1/TPM4/VCAN |
| E2F_TARGETS | ASF1B/ATAD2/AURKA/BIRC5/BRMS1L/BUB1B/CBX5/CCNB2/CCNE1/CCP110/CDCA3/CDCA8/CDK1/CDK4/CDKN1A/CDKN2C/CDKN3/CENPE/CENPM/CKS2/DCLRE1B/DCTPP1/DLGAP5/DNMT1/DONSON/DSCC1/DUT/E2F8/EED/ESPL1/GINS1/GINS4/H2AX/HELLS/HMMR/JPT1/KIF22/KIF4A/KPNA2/LIG1/LMNB1/MAD2L1/MCM3/MCM4/MKI67/MSH2/MXD3/MYBL2/NCAPD2/NME1/PAICS/PCNA/PDS5B/PLK1/PLK4/PNN/POLA2/POLD1/POLD2/PPM1D/PRIM2/PRKDC/PRPS1/RACGAP1/RAD51AP1/RANBP1/RBBP7/RFC2/RFC3/RNASEH2A/RPA3/RRM2/SMC1A/SPAG5/SPC24/SPC25/STMN1/TACC3/TCF19/TIMELESS/TK1/TOP2A/TRIP13/TUBB/TUBG1/UBE2T/UBR7/UNG/WEE1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Wen, D.; Yuan, Y.; Chen, W. Gene Set Enrichment Analysis and Genetic Experiment Reveal Changes in Cell Signaling Pathways Induced by α-Synuclein Overexpression. Biomedicines 2023, 11, 263. https://doi.org/10.3390/biomedicines11020263
Huang Y, Wen D, Yuan Y, Chen W. Gene Set Enrichment Analysis and Genetic Experiment Reveal Changes in Cell Signaling Pathways Induced by α-Synuclein Overexpression. Biomedicines. 2023; 11(2):263. https://doi.org/10.3390/biomedicines11020263
Chicago/Turabian StyleHuang, Yusong, Dongjing Wen, Yao Yuan, and Wenfeng Chen. 2023. "Gene Set Enrichment Analysis and Genetic Experiment Reveal Changes in Cell Signaling Pathways Induced by α-Synuclein Overexpression" Biomedicines 11, no. 2: 263. https://doi.org/10.3390/biomedicines11020263





