Modulating TRPV4 Channel Activity in Pro-Inflammatory Macrophages within the 3D Tissue Analog
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Macrophage Cultivation
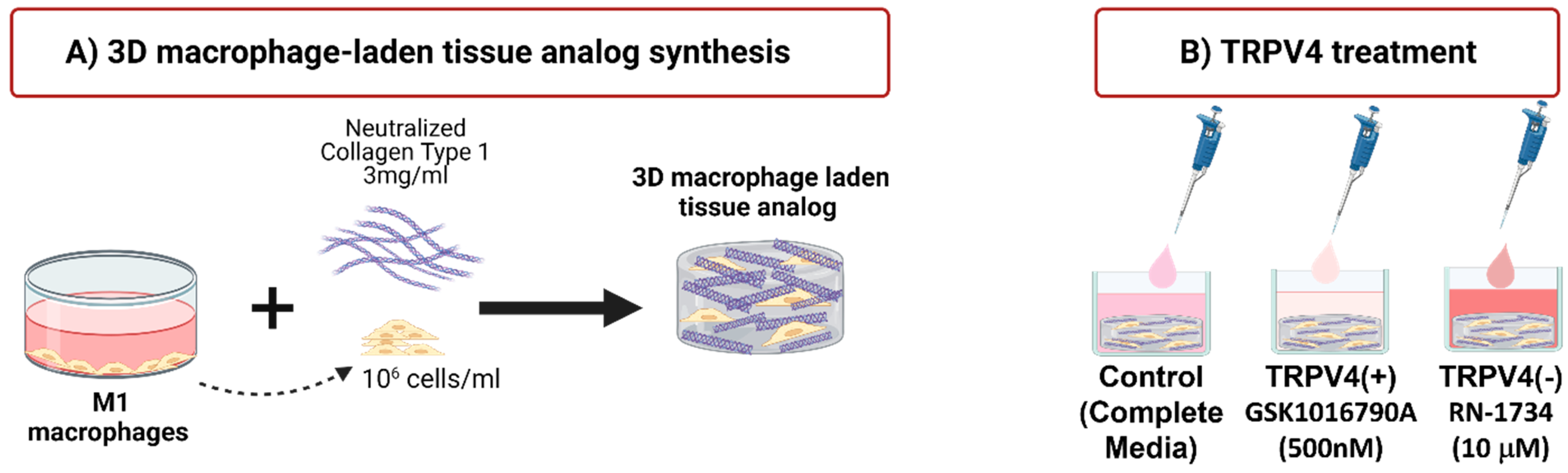
2.2. Treatment of Macrophages with TRPV4 Ion Channel Modulators
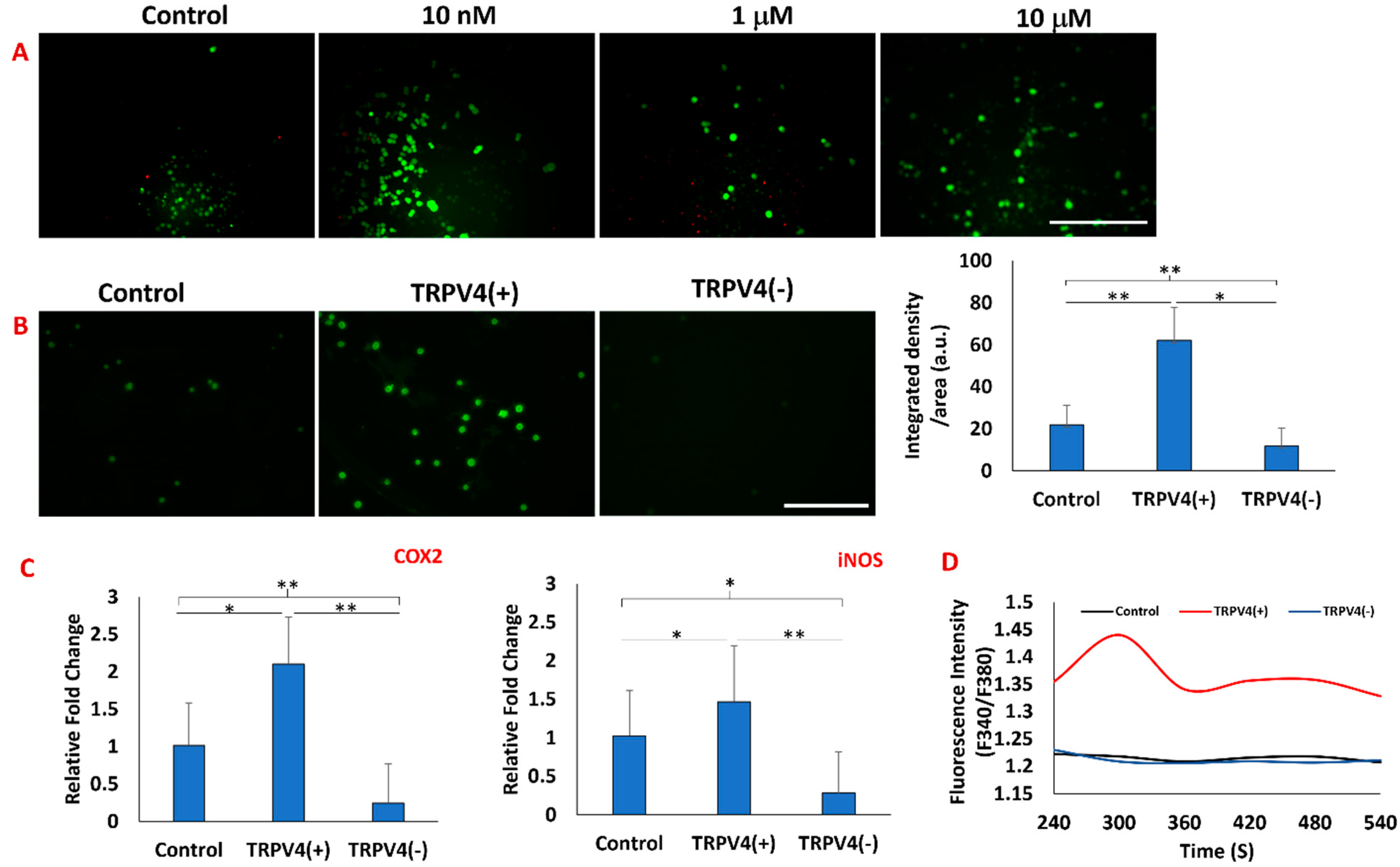
2.3. Confirmation of Activation/Deactivation of TRPV4 Channels upon TRPV4 Modulators using TRPV4 Antibody Staining and Intracellular Calcium Level
2.4. Synthesizing Macrophage-Laden 3D Tissue Matrix
2.5. Structural Changes in 3D M1-Laden Tissue Matrix upon TRPV4 Modulators
2.6. Identifying the Effect of TRPV4 Modulators on Macrophage Phenotypic Profile
2.7. Statistical Analysis
3. Results
3.1. Validating the Effectiveness of TRPV4 Agonist and Antagonist on Macrophages
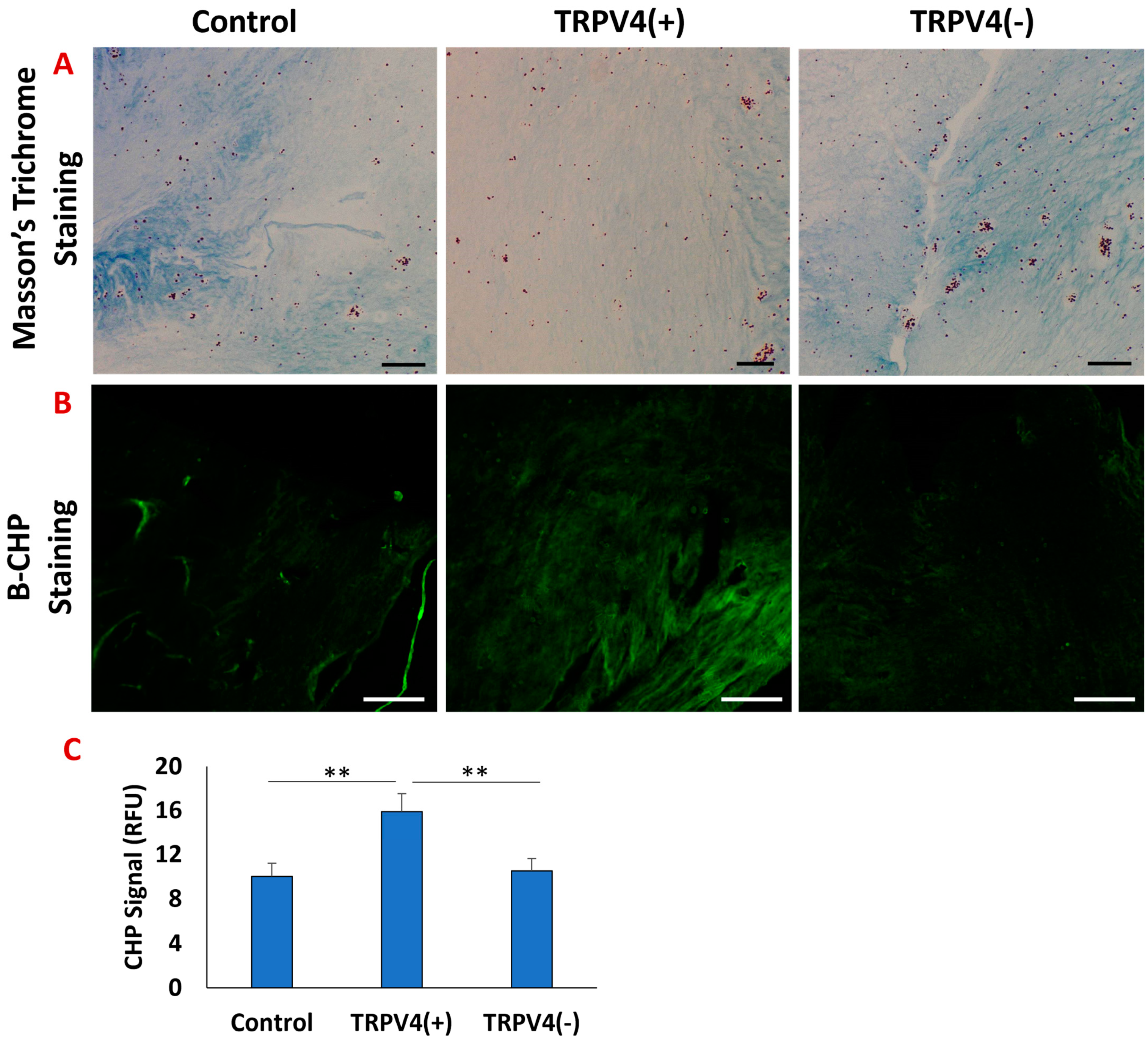
3.2. The Effect of Pharmacological Activation of TRPV4 Channel on Collagen Matrix Surrounding the Macrophages
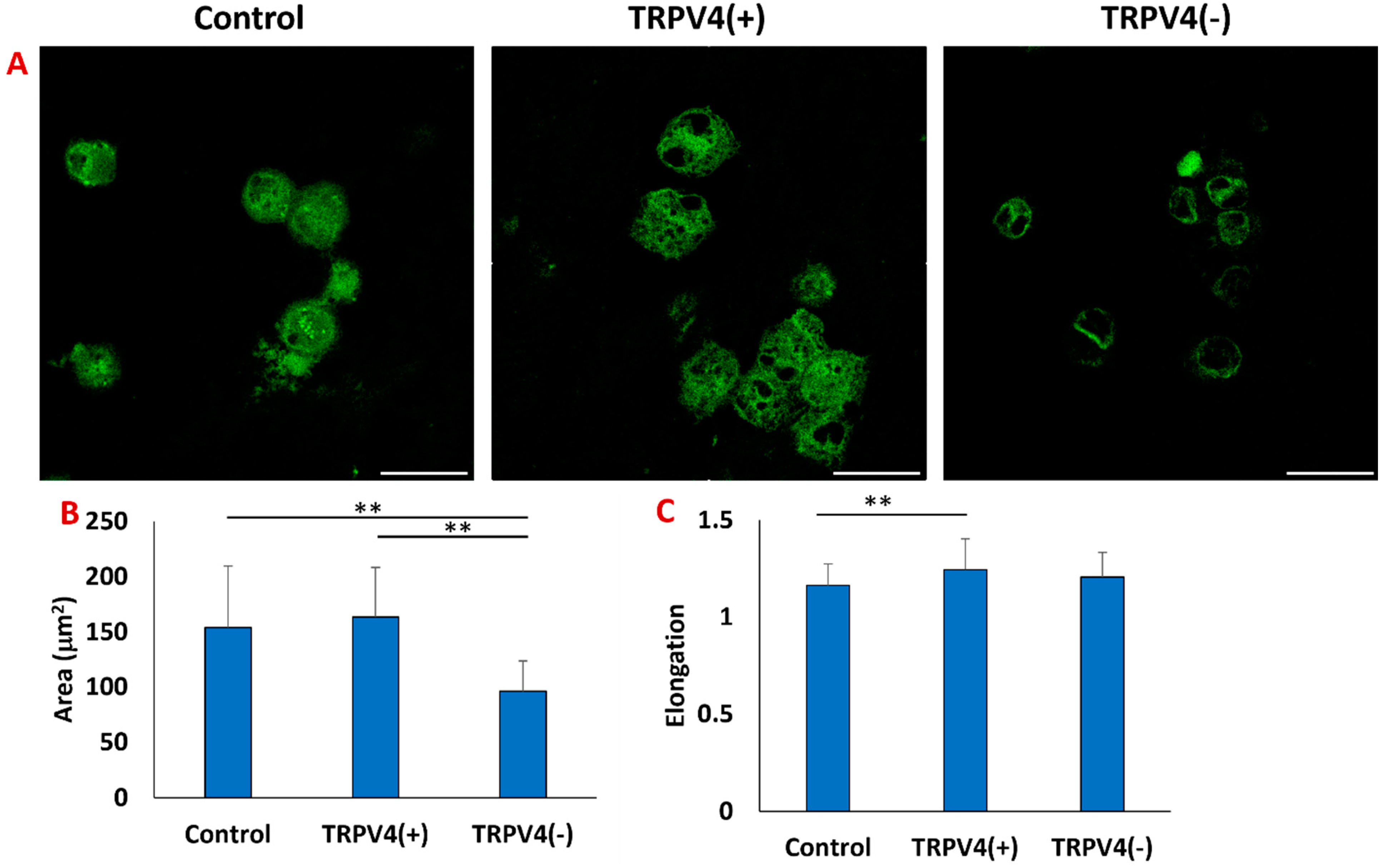
3.3. The Effect of Pharmacological Activation of TRPV4 Channels on the Cytoskeletal Structure of Macrophages within the 3D Collagen Matrix
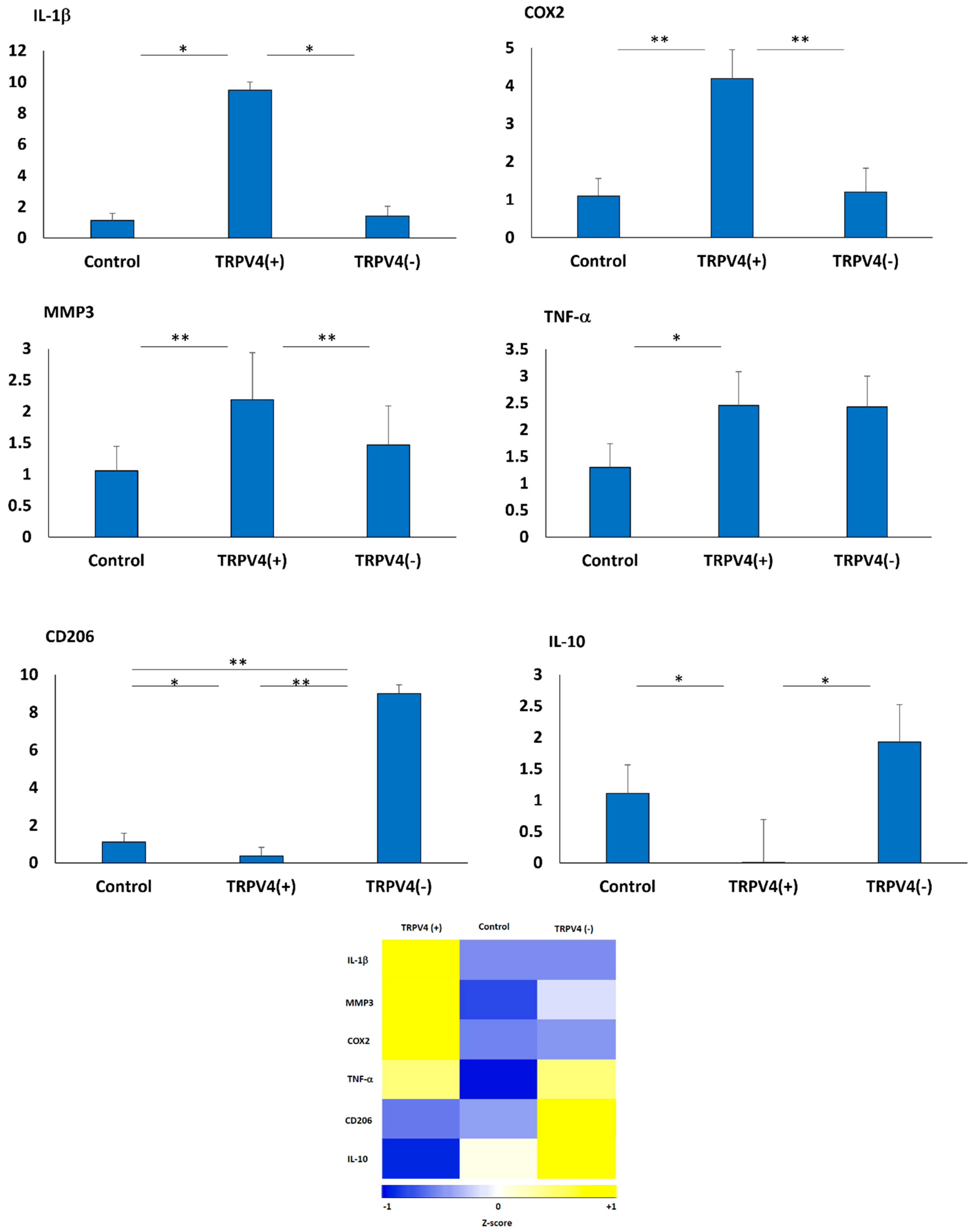
3.4. The Effect of Pharmacological Activation of TRPV4 Channel on Phenotypic Changes of M1 Macrophages within the 3D Collagen Matrix
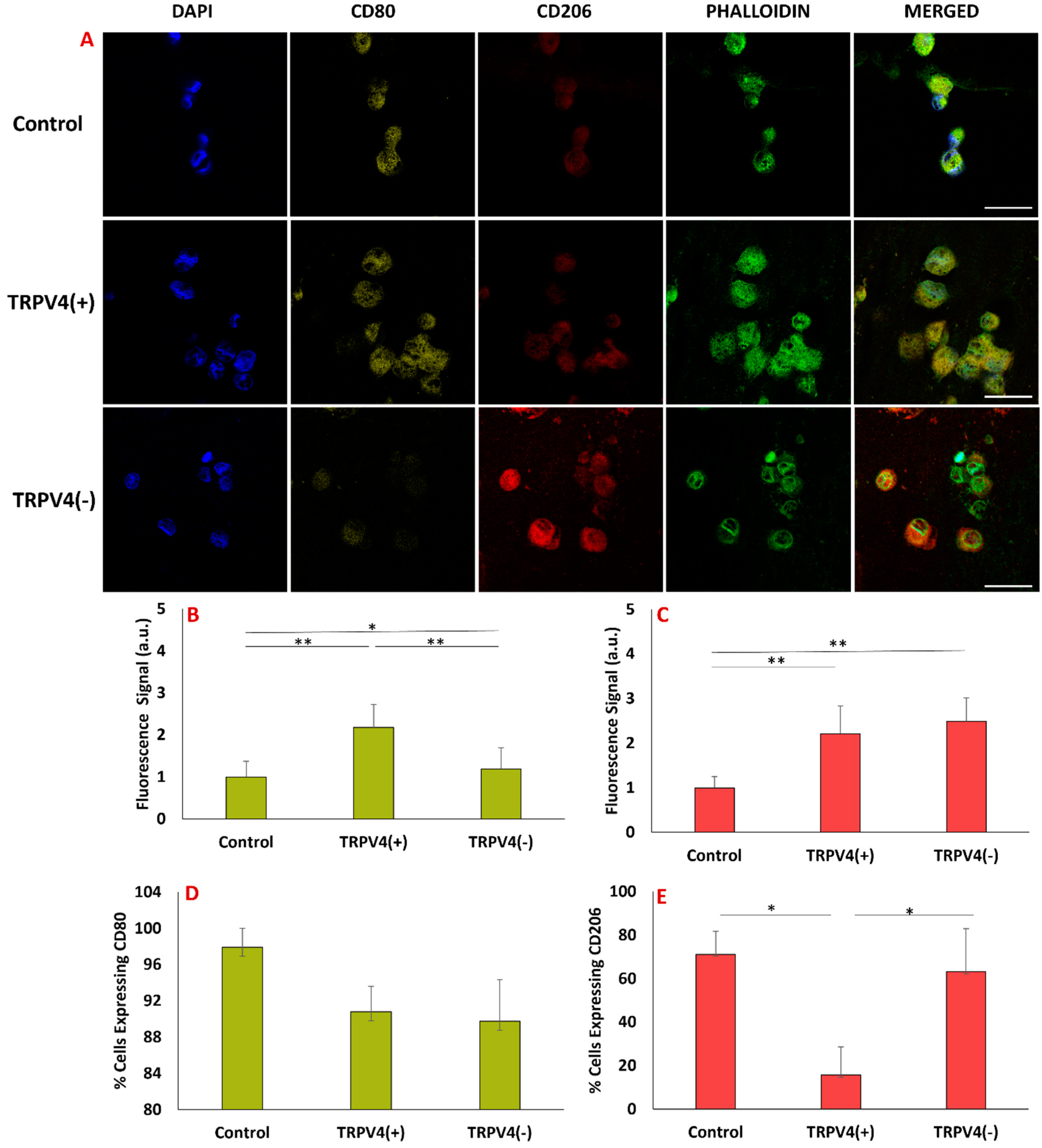
3.5. Pharmacological Activation/Blockage of TRPV4 Channel in M1 Macrophages Promotes Pro-Inflammatory/Anti-Inflammatory Surface Protein Markers in M1 Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| CCL-18 | Chemokine (C-C motif) ligand-18 |
| CD163 | Cluster of differentiation 163 |
| CD206 | Cluster of differentiation 206 |
| COX-2 | Cyclooxygenase-2 |
| ECM | Extracellular matrix |
| FBS | Fetal bovine serum |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| IL | Interleukin |
| IL-10 | Interleukin-10 |
| LPS | Lipid polysaccharide |
| M0 | Naïve macrophage |
| M1 | Pro-inflammatory macrophage |
| M2 | Anti-inflammatory macrophage |
| MMP | Matrix metalloproteinase |
| PBS | Phosphate buffered saline |
| PCR | Polymerase chain reaction |
| PMA | Phorbol 12-myristate 13-acetate |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| RFU | Relative fluorescence unit |
| RNA | Ribonucleic acid |
| RPMI | Roswell Park Memorial Institute |
| TNF-α | Tumor necrosis factor-α |
| U937 | Human myeloid cell line |
| List of Units and Symbols | |
| M | Molar |
| U | Unit |
| v/v | Volume per volume |
| w/v | Weight per volume |
| μ | Micro |
References
- Vincent, F.; AJ Duncton, M. TRPV4 agonists and antagonists. Curr. Top. Med. Chem. 2011, 11, 2216–2226. [Google Scholar] [CrossRef] [PubMed]
- Toft-Bertelsen, T.L.; MacAulay, N. TRPing to the point of clarity: Understanding the function of the complex TRPV4 ion channel. Cells 2021, 10, 165. [Google Scholar] [CrossRef] [PubMed]
- Chaigne, S.; Barbeau, S.; Ducret, T.; Guinamard, R.; Benoist, D. Pathophysiological Roles of the TRPV4 Channel in the Heart. Cells 2023, 12, 1654. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Anishkin, A.; Zinkevich, N.S.; Nishijima, Y.; Korishettar, A.; Wang, Z.; Fang, J.; Wilcox, D.A.; Zhang, D.X. Transient receptor potential vanilloid 4 (TRPV4) activation by arachidonic acid requires protein kinase A–mediated phosphorylation. J. Biol. Chem. 2018, 293, 5307–5322. [Google Scholar] [CrossRef] [PubMed]
- Grace, M.S.; Bonvini, S.J.; Belvisi, M.G.; McIntyre, P. Modulation of the TRPV4 ion channel as a therapeutic target for disease. Pharmacol. Ther. 2017, 177, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Nadezhdin, K.D.; Talyzina, I.A.; Parthasarathy, A.; Neuberger, A.; Zhang, D.X.; Sobolevsky, A.I. Structure of human TRPV4 in complex with GTPase RhoA. Nat. Commun. 2023, 14, 3733. [Google Scholar] [CrossRef]
- Baratchi, S.; Keov, P.; Darby, W.G.; Lai, A.; Khoshmanesh, K.; Thurgood, P.; Vahidi, P.; Ejendal, K.; McIntyre, P. The TRPV4 Agonist GSK1016790A Regulates the Membrane Expression of TRPV4 Channels. Front. Pharmacol. 2019, 10, 6. [Google Scholar] [CrossRef]
- Shin, S.H.; Lee, E.J.; Hyun, S.; Chun, J.; Kim, Y.; Kang, S.S. Phosphorylation on the Ser 824 residue of TRPV4 prefers to bind with F-actin than with microtubules to expand the cell surface area. Cell. Signal. 2012, 24, 641–651. [Google Scholar] [CrossRef]
- McNulty, A.L.; Leddy, H.A.; Liedtke, W.; Guilak, F. TRPV4 as a therapeutic target for joint diseases. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2015, 388, 437–450. [Google Scholar] [CrossRef]
- Michalick, L.; Kuebler, W.M. TRPV4—A Missing Link Between Mechanosensation and Immunity. Front. Immunol. 2020, 11, 413. [Google Scholar] [CrossRef]
- D’Aldebert, E.; Cenac, N.; Rousset, P.; Martin, L.; Rolland, C.; Chapman, K.; Selves, J.; Alric, L.; Vinel, J.; Vergnolle, N. Transient Receptor Potential Vanilloid 4 Activated Inflammatory Signals by Intestinal Epithelial Cells and Colitis in Mice. Gastroenterology 2011, 140, 275–285.e3. [Google Scholar] [CrossRef] [PubMed]
- Sumioka, T.; Iwanishi, H.; Yasuda, S.; Ichikawa, K.; Miyazima, M.; Kokado, M.; Okada, Y.; Saika, S. Loss of TRPV4 Cation Channel Inhibition of Macrophage Infiltration and Neovascularization in a Mouse Cornea. Lab. Investig. 2023, 103, 100061. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.-N.; Siddiqui, G.; Veldhuis, N.A.; Poole, D.P. Diverse Roles of TRPV4 in Macrophages: A Need for Unbiased Profiling. Front. Immunol. 2022, 12, 828115. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Cheng, Y.; Zhang, S.; Sun, X.; Wu, J. TRPV4-induced Müller cell gliosis and TNF-α elevation-mediated retinal ganglion cell apoptosis in glaucomatous rats via JAK2/STAT3/NF-κB pathway. J. Neuroinflammation 2021, 18, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhou, L.; An, D.; Xu, W.; Wu, C.; Sha, S.; Li, Y.; Zhu, Y.; Chen, A.; Du, Y.; et al. TRPV4-induced inflammatory response is involved in neuronal death in pilocarpine model of temporal lobe epilepsy in mice. Cell Death Dis. 2019, 10, 386. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.O.; Dalloneau, E.; Pérez-Berezo, M.-T.; Plata, C.; Wu, Y.; Guillon, A.; Morello, E.; Aimar, R.-F.; Potier-Cartereau, M.; Esnard, F.; et al. In vitro and in vivo evidence for an inflammatory role of the calcium channel TRPV4 in lung epithelium: Potential involvement in cystic fibrosis. Am. J. Physiol. Cell. Mol. Physiol. 2016, 311, L664–L675. [Google Scholar] [CrossRef]
- Ye, L.; Kleiner, S.; Wu, J.; Sah, R.; Gupta, R.K.; Banks, A.S.; Cohen, P.; Khandekar, M.J.; Boström, P.; Mepani, R.J.; et al. TRPV4 Is a Regulator of Adipose Oxidative Metabolism, Inflammation, and Energy Homeostasis. Cell 2012, 151, 96–110. [Google Scholar] [CrossRef]
- Dutta, B.; Arya, R.K.; Goswami, R.; Alharbi, M.O.; Sharma, S.; Rahaman, S.O. Role of macrophage TRPV4 in inflammation. Lab. Investig. 2020, 100, 178–185. [Google Scholar] [CrossRef]
- Chen, X.; Lu, W.; Lu, C.; Zhang, L.; Xu, F.; Dong, H. The CaSR/TRPV4 coupling mediates pro-inflammatory macrophage function. Acta Physiol. 2023, 237, e13926. [Google Scholar] [CrossRef]
- Shannon, A.H.; Elder, C.T.; Lu, G.; Su, G.; Mast, A.; Salmon, M.D.; Montgomery, W.G.; Spinosa, M.D.; Upchurch, G.R.; Sharma, A.K.; et al. Pharmacologic inhibition of transient receptor channel vanilloid 4 attenuates abdominal aortic aneurysm formation. FASEB J. 2020, 34, 9787–9801. [Google Scholar] [CrossRef]
- Hamanaka, K.; Jian, M.-Y.; Townsley, M.I.; King, J.A.; Liedtke, W.; Weber, D.S.; Eyal, F.G.; Clapp, M.M.; Parker, J.C.; Wang, T.; et al. TRPV4 channels augment macrophage activation and ventilator-induced lung injury. Am. J. Physiol. Cell. Mol. Physiol. 2010, 299, L353–L362. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Sun, Z.; Xu, X.; Lv, Z.; Li, J.; Wu, R.; Fei, Y.; Tan, G.; Liu, Z.; Liu, Y.; et al. Blocking TRPV4 Ameliorates Osteoarthritis by Inhibiting M1 Macrophage Polarization via the ROS/NLRP3 Signaling Pathway. Antioxidants 2022, 11, 2315. [Google Scholar] [CrossRef] [PubMed]
- Pairet, N.; Mang, S.; Fois, G.; Keck, M.; Kühnbach, M.; Gindele, J.; Frick, M.; Dietl, P.; Lamb, D.J. TRPV4 inhibition attenuates stretch-induced inflammatory cellular responses and lung barrier dysfunction during mechanical ventilation. PLoS ONE 2018, 13, e0196055. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Du, H.; Winer, D.A.; Clemente-Casares, X.; Tsai, S. Mechanosensing in macrophages and dendritic cells in steady-state and disease. Front. Cell Dev. Biol. 2022, 10, 1044729. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Shirai, K.; Miyajima, M.; Reinach, P.S.; Yamanaka, O.; Sumioka, T.; Kokado, M.; Tomoyose, K.; Saika, S. Loss of TRPV4 Function Suppresses Inflammatory Fibrosis Induced by Alkali-Burning Mouse Corneas. PLoS ONE 2016, 11, e0167200. [Google Scholar] [CrossRef] [PubMed]
- Scheraga, R.G.; Abraham, S.; Niese, K.A.; Southern, B.D.; Grove, L.M.; Hite, R.D.; McDonald, C.; Hamilton, T.A.; Olman, M.A. TRPV4 Mechanosensitive Ion Channel Regulates Lipopolysaccharide-Stimulated Macrophage Phagocytosis. J. Immunol. 2016, 196, 428–436. [Google Scholar] [CrossRef]
- Yoshizumi, M.; Tazawa, N.; Watanabe, C.; Mizoguchi, H. TRPV4 activation prevents lipopolysaccharide-induced painful bladder hypersensitivity in rats by regulating immune pathways. Front. Immunol. 2022, 13, 1080302. [Google Scholar] [CrossRef]
- Babaniamansour, P.; Jacho, D.; Teow, A.; Rabino, A.; Garcia-Mata, R.; Yildirim-Ayan, E. Macrophage Mechanoresponsiveness within 3D Tissue Matrix upon Mechanotherapy-Associated Strains. Tissue Eng. 2023, 29, 100516. [Google Scholar]
- Shen, J.; Tu, L.; Chen, D.; Tan, T.; Wang, Y.; Wang, S. TRPV4 channels stimulate Ca2+-induced Ca2+ release in mouse neurons and trigger endoplasmic reticulum stress after intracerebral hemorrhage. Brain Res. Bull. 2019, 146, 143–152. [Google Scholar] [CrossRef]
- Wasilewski, A.; Krajewska, U.; Owczarek, K.; Lewandowska, U.; Fichna, J. Fatty acid amide hydrolase (FAAH) inhibitor PF-3845 reduces viability, migration and invasiveness of human colon adenocarcinoma Colo-205 cell line: An in vitro study. Acta Biochim. Pol. 2017, 64, 519–525. [Google Scholar] [CrossRef]
- Chaigne, S.; Cardouat, G.; Louradour, J.; Vaillant, F.; Charron, S.; Sacher, F.; Ducret, T.; Guinamard, R.; Vigmond, E.; Hof, T. Transient receptor potential vanilloid 4 channel participates in mouse ventricular electrical activity. Am. J. Physiol. Circ. Physiol. 2021, 320, H1156–H1169. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Jie, P.; Tian, Y.; Chen, T.; Chen, L.; Chen, L. Transient Receptor Potential Vanilloid 4-Induced Modulation of Voltage-Gated Sodium Channels in Hippocampal Neurons. Mol. Neurobiol. 2016, 53, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Roebke, E.; Jacho, D.; Eby, O.; Aldoohan, S.; Elsamaloty, H.; Yildirim-Ayan, E. Injectable Cell-Laden Nanofibrous Matrix for Treating Annulus Fibrosus Defects in Porcine Model: An Organ Culture Study. Life 2022, 12, 1866. [Google Scholar] [CrossRef] [PubMed]
- Jacho, D.; Rabino, A.; Garcia-Mata, R.; Yildirim-Ayan, E. Mechanoresponsive regulation of fibroblast-to-myofibroblast transition in three-dimensional tissue analogues: Mechanical strain amplitude dependency of fibrosis. Sci. Rep. 2022, 12, 16832. [Google Scholar] [CrossRef] [PubMed]
- Bayrak, E.; Huri, P.Y. Engineering Musculoskeletal Tissue Interfaces. Front. Mater. 2018, 5, 24. [Google Scholar] [CrossRef]
- Hettinger, Z.R.; Wen, Y.; Peck, B.D.; Hamagata, K.; Confides, A.L.; Van Pelt, D.W.; A Harrison, D.; Miller, B.F.; Butterfield, T.A.; Dupont-Versteegden, E.E. Mechanotherapy Reprograms Aged Muscle Stromal Cells to Remodel the Extracellular Matrix during Recovery from Disuse. Function 2022, 3, zqac015. [Google Scholar] [CrossRef]
- Yilmaz, M.; Demir, E.; Firatli, Y.; Firatli, E.; Gürsoy, U.K. Tissue Levels of CD80, CD163 and CD206 and Their Ratios in Periodontal and Peri-Implant Health and Disease. Curr. Issues Mol. Biol. 2022, 44, 4704–4713. [Google Scholar] [CrossRef]
- Cui, S.-J.; Yang, F.-J.; Wang, X.-D.; Mao, Z.-B.; Gu, Y. Mechanical overload induces TMJ disc degeneration via TRPV4 activation. Oral Dis. 2023, Online ahead of print. [Google Scholar] [CrossRef]
- Rayees, S.; Joshi, J.C.; Tauseef, M.; Anwar, M.; Baweja, S.; Rochford, I.; Joshi, B.; Hollenberg, M.D.; Reddy, S.P.; Mehta, D. PAR2-Mediated cAMP Generation Suppresses TRPV4-Dependent Ca2+ Signaling in Alveolar Macrophages to Resolve TLR4-Induced Inflammation. Cell Rep. 2019, 27, 793–805.e4. [Google Scholar] [CrossRef]
- Pelegrin, P.; Surprenant, A. Dynamics of macrophage polarization reveal new mechanism to inhibit IL-1β release through pyrophosphates. EMBO J. 2009, 28, 2114–2127. [Google Scholar] [CrossRef]
- McWhorter, F.Y.; Davis, C.T.; Liu, W.F. Physical and mechanical regulation of macrophage phenotype and function. Cell. Mol. Life Sci. 2015, 72, 1303–1316. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, X.; Wang, L.; Wang, Y.; Dong, F.; Wu, J.; Qu, X.; Liu, Y.; Liu, Z.; Fan, H.; et al. TRPV4 Inhibition Improved Myelination and Reduced Glia Reactivity and Inflammation in a Cuprizone-Induced Mouse Model of Demyelination. Front. Cell. Neurosci. 2018, 12, 392. [Google Scholar] [CrossRef] [PubMed]
- Dragoni, S.; Guerra, G.; Pla, A.F.; Bertoni, G.; Rappa, A.; Poletto, V.; Bottino, C.; Aronica, A.; Lodola, F.; Cinelli, M.P.; et al. A functional transient receptor potential vanilloid 4 (TRPV4) channel is expressed in human endothelial progenitor cells. J. Cell. Physiol. 2015, 230, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Alenmyr, L.; Uller, L.; Greiff, L.; Högestätt, E.D.; Zygmunt, P.M. TRPV 4-Mediated Calcium Influx and Ciliary Activity in Human Native Airway Epithelial Cells. Basic Clin. Pharmacol. Toxicol. 2014, 114, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Chen, Z.; Zhao, M.; Zu, S.; Zhao, S.; Wang, S.; Zhang, X. Cell Deformation at the Air-Liquid Interface Evokes Intracellular Ca2+ Increase and ATP Release in Cultured Rat Urothelial Cells. Front. Physiol. 2021, 12, 631022. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.; Merth, M.; Sharma, S.; Alharbi, M.O.; Aranda-Espinoza, H.; Zhu, X.; Rahaman, S.O. TRPV4 calcium-permeable channel is a novel regulator of oxidized LDL-induced macrophage foam cell formation. Free. Radic. Biol. Med. 2017, 110, 142–150. [Google Scholar] [CrossRef]
- Jaudon, F.; Chiacchiaretta, M.; Albini, M.; Ferroni, S.; Benfenati, F.; Cesca, F. Kidins220/ARMS controls astrocyte calcium signaling and neuron–astrocyte communication. Cell Death Differ. 2020, 27, 1505–1519. [Google Scholar] [CrossRef]
- Fu, S.; Meng, H.; Inamdar, S.; Das, B.; Gupta, H.; Wang, W.; Thompson, C.L.; Knight, M.M. Activation of TRPV4 by mechanical, osmotic or pharmaceutical stimulation is anti-inflammatory blocking IL-1β mediated articular cartilage matrix destruction. Osteoarthr. Cartil. 2021, 29, 89–99. [Google Scholar] [CrossRef]
- Ji, C.; McCulloch, C.A. TRPV4 integrates matrix mechanosensing with Ca2+ signaling to regulate extracellular matrix remodeling. FEBS J. 2021, 288, 5867–5887. [Google Scholar] [CrossRef]
- Xiao, L.; Majumdar, R.; Dai, J.; Li, Y.; Xie, L.; Shen, F.H.; Jin, L.; Li, X. Molecular Detection and Assessment of Intervertebral Disc Degeneration via a Collagen Hybridizing Peptide. ACS Biomater. Sci. Eng. 2019, 5, 1661–1667. [Google Scholar] [CrossRef]
- Hwang, J.; Huang, Y.; Burwell, T.J.; Peterson, N.C.; Connor, J.; Weiss, S.J.; Yu, S.M.; Li, Y. In Situ Imaging of Tissue Remodeling with Collagen Hybridizing Peptides. ACS Nano 2017, 11, 9825–9835. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Liu, J.; Li, Y.; Zhang, T.; Wang, F.; Chang, L.; Li, C.; Ge, X.; Zuo, T.; Lu, S.; et al. Damaged collagen detected by collagen hybridizing peptide as efficient diagnosis marker for early hepatic fibrosis. Biochim. et Biophys. Acta (BBA)—Gene Regul. Mech. 2023, 1866, 194928. [Google Scholar] [CrossRef] [PubMed]
- Goswami, C.; Kuhn, J.; Heppenstall, P.A.; Hucho, T. Importance of Non-Selective Cation Channel TRPV4 Interaction with Cytoskeleton and Their Reciprocal Regulations in Cultured Cells. PLoS ONE 2010, 5, e11654. [Google Scholar] [CrossRef] [PubMed]
- Bagnell, A.M.; Sumner, C.J.; McCray, B.A. TRPV4: A trigger of pathological RhoA activation in neurological disease. BioEssays 2022, 44, e2100288. [Google Scholar] [CrossRef]
- Phuong, T.T.T.; Redmon, S.N.; Yarishkin, O.; Winter, J.M.; Li, D.Y.; Križaj, D. Calcium influx through TRPV4 channels modulates the adherens contacts between retinal microvascular endothelial cells. J. Physiol. 2017, 595, 6869–6885. [Google Scholar] [CrossRef]
- Lee, H.-P.; Stowers, R.; Chaudhuri, O. Volume expansion and TRPV4 activation regulate stem cell fate in three-dimensional microenvironments. Nat. Commun. 2019, 10, 529. [Google Scholar] [CrossRef]
- Atsumi, Y.; Toriyama, M.; Kato, H.; Nakamura, M.; Morita, A.; Takaishi, M.; Saito, K.; Tanaka, M.; Okada, F.; Tominaga, M.; et al. Anti-Inflammatory Role of TRPV4 in Human Macrophages. ImmunoHorizons 2023, 7, 81–96. [Google Scholar] [CrossRef]
- Scheraga, R.G.; Perelas, A.; Abraham, S.; Grove, L.; Southern, B.D.; Crish, J.; Olman, M. TRPV4 Mediates the Macrophage Phagocytic and Cytokine Response to Leptin. J. Immunol. 2019, 202, 58.4. [Google Scholar] [CrossRef]
- Teow, A. Mechanoresponsive Naïve Human Macrophage Polarization within a 3D Collagen Matrix; University of Toledo: Toledo, OH, USA, 2021. [Google Scholar]
- Shortridge, C.; Akbari Fakhrabadi, E.; Wuescher, L.M.; Worth, R.G.; Liberatore, M.W.; Yildirim-Ayan, E. Impact of digestive inflammatory environment and genipin crosslinking on immunomodulatory capacity of injectable musculoskeletal tissue scaffold. Int. J. Mol. Sci. 2021, 22, 1134. [Google Scholar] [CrossRef]
- Li, H.; Sze, S.; Tong, Y.; Ng, T. Production of Th1-and Th2-dependent cytokines induced by the Chinese medicine herb, Rhodiola algida, on human peripheral blood monocytes. J. Ethnopharmacol. 2009, 123, 257–266. [Google Scholar] [CrossRef]
- Jacho, D.; Babaniamansour, P.; Osorio, R.; Toledano, M.; Rabino, A.; Garcia-Mata, R.; Yildirim-Ayan, E. Deciphering the Cell-Specific Effect of Osteoblast-Macrophage Crosstalk in Periodontitis. Tissue Eng. Part A 2023, 29, 579–593. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babaniamansour, P.; Jacho, D.; Niedzielski, S.; Rabino, A.; Garcia-Mata, R.; Yildirim-Ayan, E. Modulating TRPV4 Channel Activity in Pro-Inflammatory Macrophages within the 3D Tissue Analog. Biomedicines 2024, 12, 230. https://doi.org/10.3390/biomedicines12010230
Babaniamansour P, Jacho D, Niedzielski S, Rabino A, Garcia-Mata R, Yildirim-Ayan E. Modulating TRPV4 Channel Activity in Pro-Inflammatory Macrophages within the 3D Tissue Analog. Biomedicines. 2024; 12(1):230. https://doi.org/10.3390/biomedicines12010230
Chicago/Turabian StyleBabaniamansour, Parto, Diego Jacho, Skyler Niedzielski, Agustin Rabino, Rafael Garcia-Mata, and Eda Yildirim-Ayan. 2024. "Modulating TRPV4 Channel Activity in Pro-Inflammatory Macrophages within the 3D Tissue Analog" Biomedicines 12, no. 1: 230. https://doi.org/10.3390/biomedicines12010230





