Novel Serum Biomarkers for Patients with Allergic Asthma Phenotype
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Pulmonary Function Tests
2.2.1. Spirometry
2.2.2. Bronchial Reversibility Test
2.2.3. Methacholine Challenge Test
2.2.4. BAC with Dermatophagoides pteronyssinus
2.2.5. FeNO Test
2.3. Skin Prick Testing
2.4. Blood Tests
2.4.1. Complete Blood Count and Total IgE
2.4.2. Serum Levels of Selected Analytes Measurement
Enzyme-Linked Immunosorbent Assay (ELISA)
Luminex Assay
2.5. Confirmation of Participation in the Study
2.6. Statistical Analysis
3. Results
3.1. Study Subject Characteristics
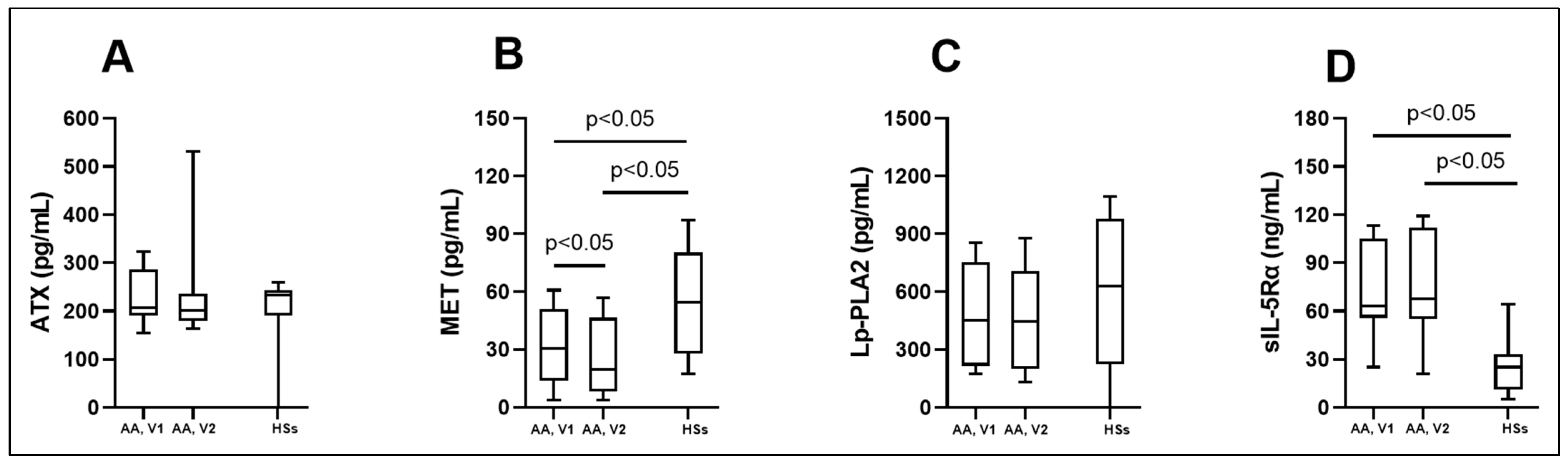
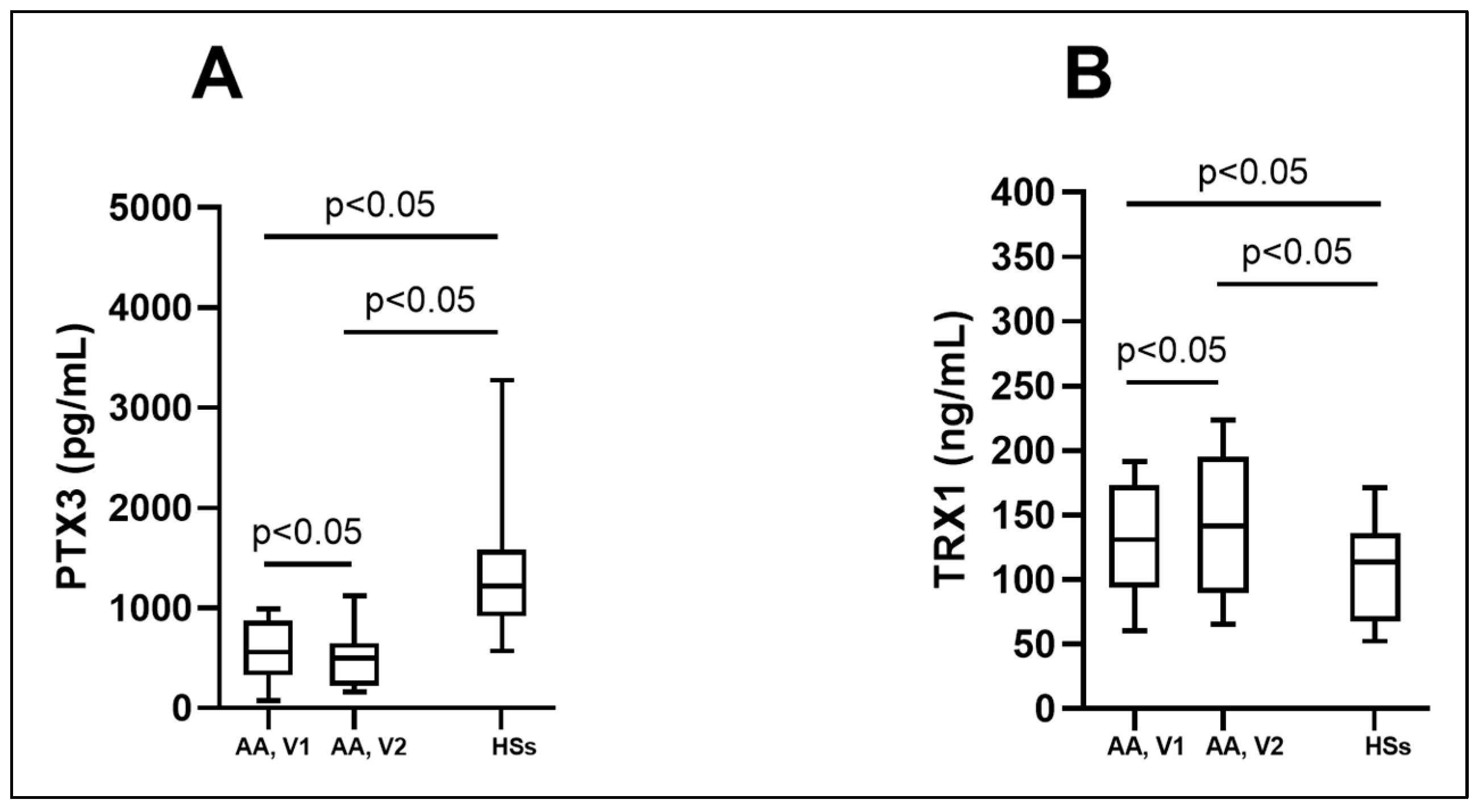
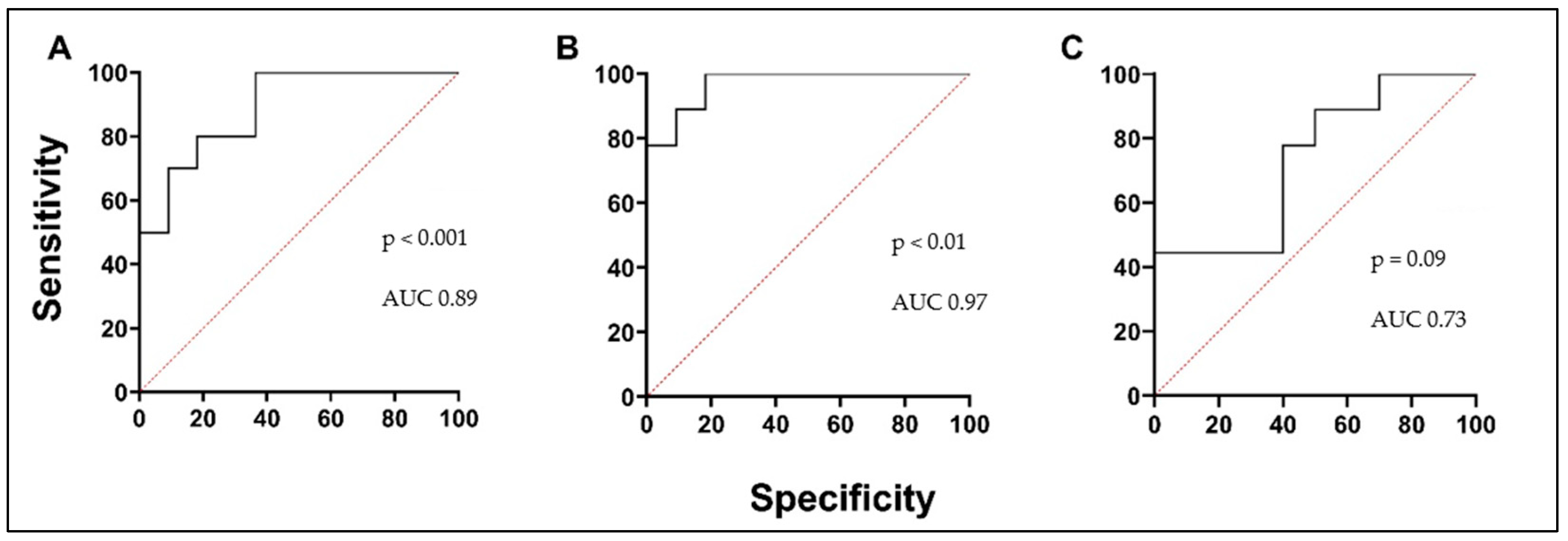
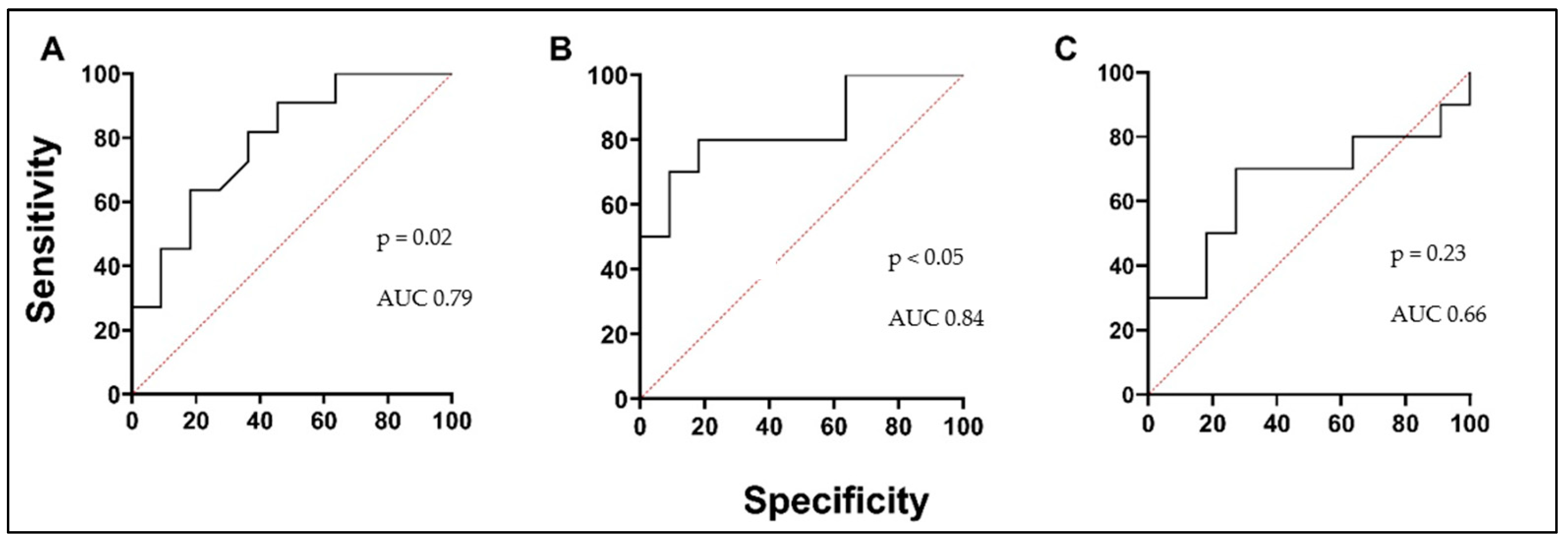
3.2. Biologically Active Chronic Inflammation-Related Substances as Biomarkers in Asthma
3.3. Blood Oxidative Stress Indicators as Biomarkers in Asthma
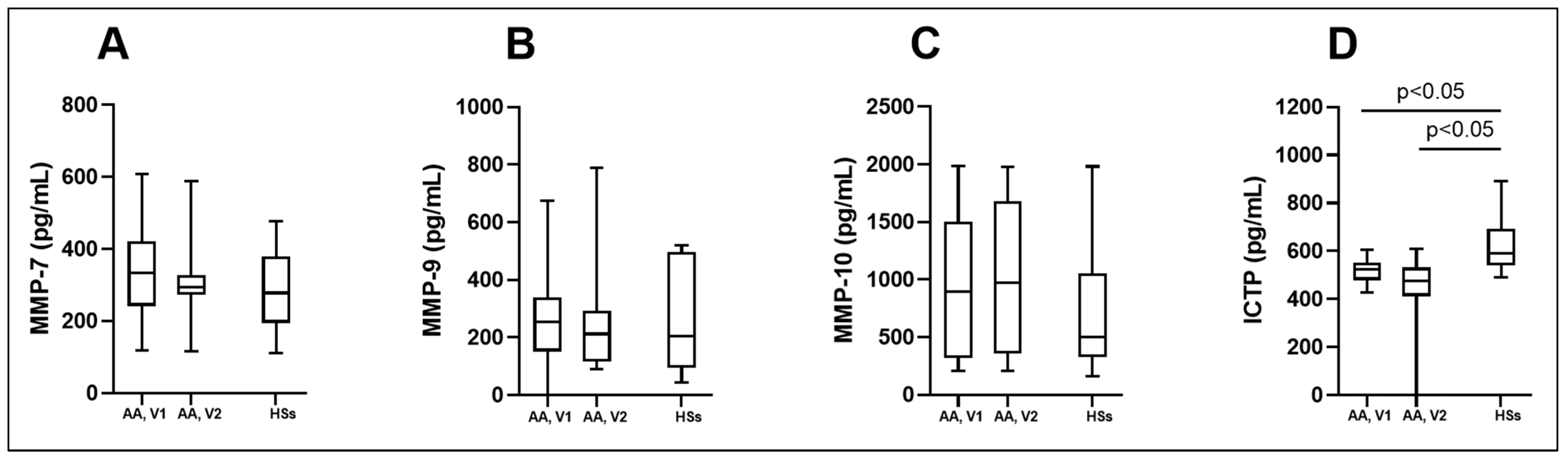
3.4. MMPs and ICTP Degradation Product as Biomarkers in Asthma
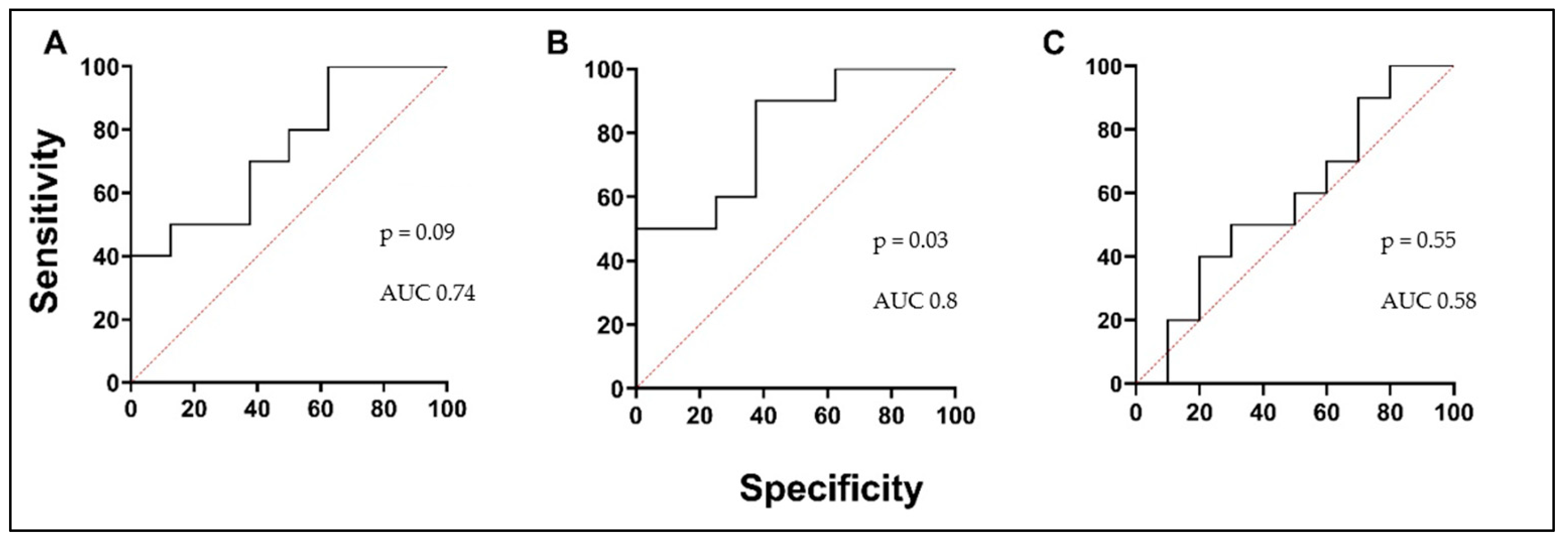
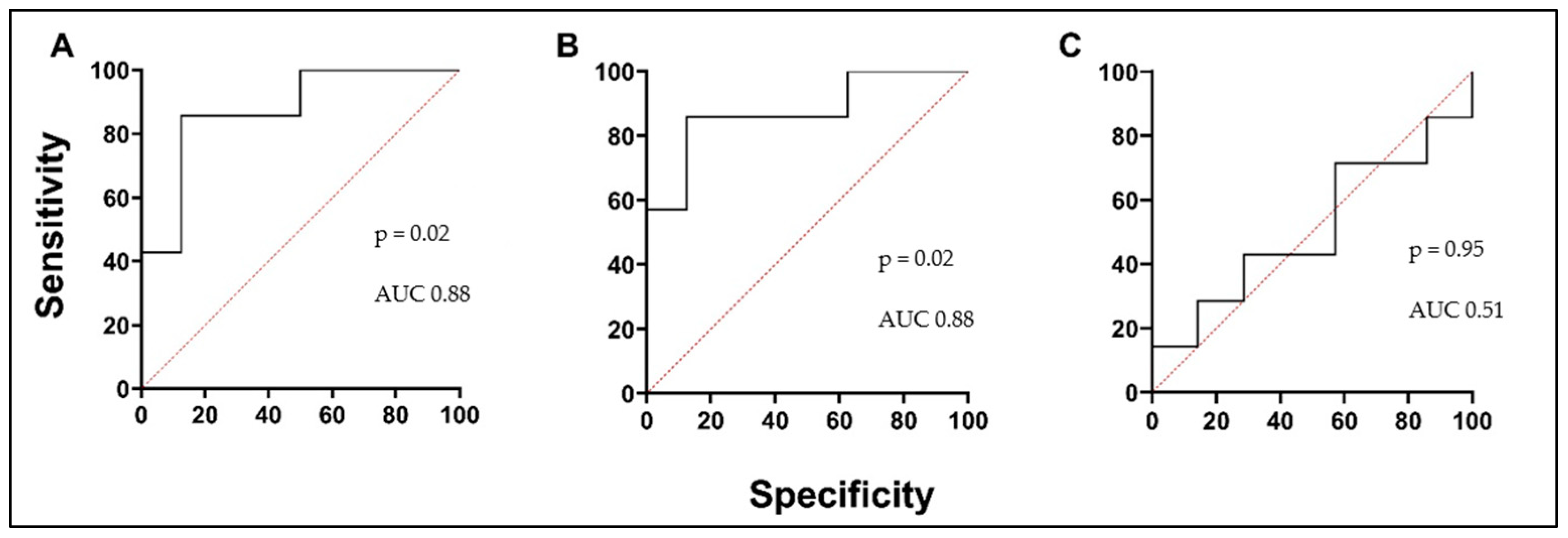
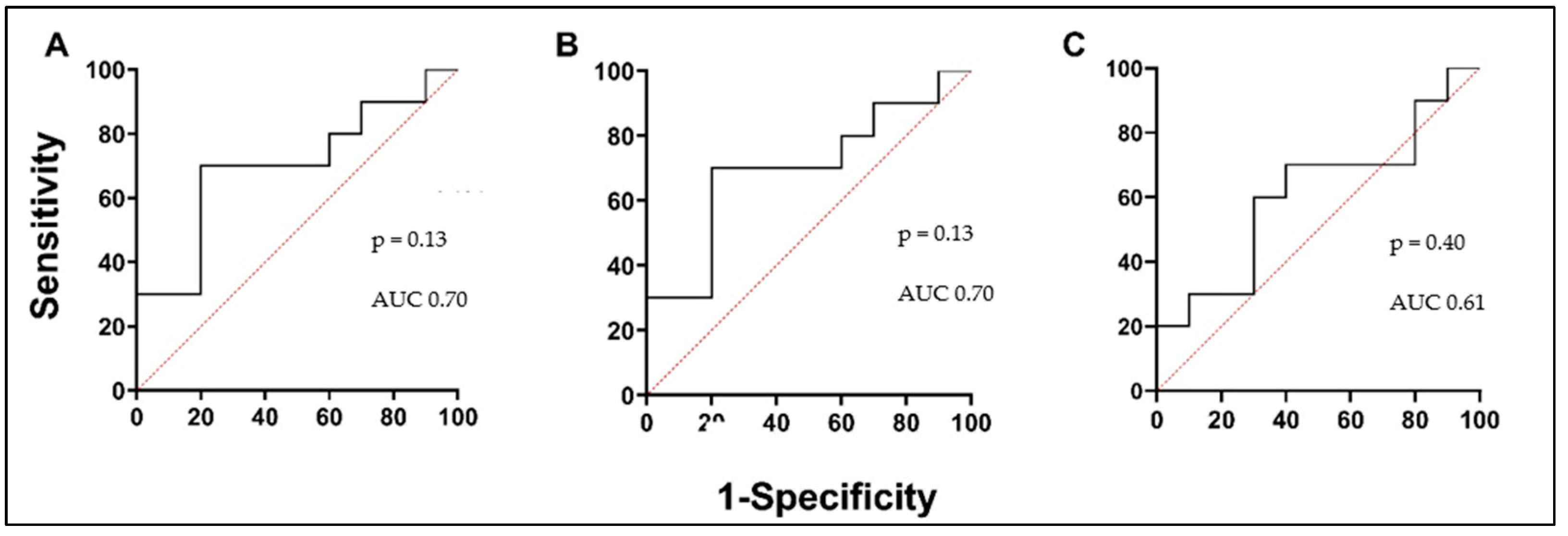
3.5. Biologically Active Substances in Correlation with Validated Asthma Biomarkers, Lung Function, and in Comparison with Each Other
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AA | allergic asthma |
| ATX | autotaxin |
| BAC | bronchial allergen challenge |
| ELISA | enzyme-linked immunosorbent assay |
| EOS | eosinophil |
| FeNO | fractional exhaled nitric oxide |
| FEV1 | forced expiratory volume in 1 s |
| HSs | healthy subjects |
| ICTP | I C-telopeptide of type I collagen |
| IgE | immunoglobulin E |
| IL-5 | interleukin-5 |
| Lp-PLA2 | lipoprotein-associated phospholipase A2 |
| LSMU | Lithuanian University of Health Sciences |
| MMP-10 | matrix metalloproteinase-10 |
| MMP-7 | matrix metalloproteinase-7 |
| MMP-9 | matrix metalloproteinase-9 |
| PTX3 | pentraxin 3 |
| ROC | receiver operating characteristic |
| sIL-5Rα | soluble interleukin 5 receptor subunit alpha |
| T2 | type 2 |
| TRX1 | thioredoxin 1 |
| V1 | experimental day |
| V2 | experimental day—24 h after V1. |
References
- Alsharairi, N.A. Antioxidant Intake and Biomarkers of Asthma in Relation to Smoking Status-A Review. Curr. Issues Mol. Biol. 2023, 45, 5099–5117. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.L.; Bacharier, L.B.; Bateman, E.; Boulet, L.P.; Brightling, C.; Buhl, R.; Brusselle, G.; Cruz, A.A.; Drazen, J.M.; Duijts, L.; et al. Key recommendations for primary care from the 2022 Global Initiative for Asthma (GINA) update. NPJ Prim. Care Respir. Med. 2023, 33, 7. [Google Scholar] [CrossRef] [PubMed]
- Papi, A.; Brightling, C.; Pedersen, S.E.; Reddel, H.K. Asthma. Lancet 2018, 391, 783–800. [Google Scholar] [CrossRef] [PubMed]
- Akar-Ghibril, N.; Casale, T.; Custovic, A.; Phipatanakul, W. Allergic Endotypes and Phenotypes of Asthma. J. Allergy Clin. Immunol. Pract. 2020, 8, 429–440. [Google Scholar] [CrossRef]
- Maspero, J.; Adir, Y.; Al-Ahmad, M.; Celis-Preciado, C.A.; Colodenco, F.D.; Giavina-Bianchi, P.; Lababidi, H.; Ledanois, O.; Mahoub, B.; Perng, D.-W.; et al. Type 2 inflammation in asthma and other airway diseases. ERJ Open Res. 2022, 8, 1–19. [Google Scholar] [CrossRef]
- Henry, E.K.; Inclan-Rico, J.M.; Siracusa, M.C. Type 2 cytokine responses: Regulating immunity to helminth parasites and allergic inflammation. Curr. Pharmacol. Rep. 2017, 3, 346–359. [Google Scholar] [CrossRef]
- Wan, X.C.; Woodruff, P.G. Biomarkers in Severe Asthma. Immunol. Allergy Clin. N. Am. 2016, 36, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Tiotiu, A. Biomarkers in asthma: State of the art. Asthma Res. Pract. 2018, 4, 10. [Google Scholar] [CrossRef]
- Puzzovio, P.G.; Levi-Schaffer, F. Latest Progresses in Allergic Diseases Biomarkers: Asthma and Atopic Dermatitis. Front. Pharmacol. 2021, 12, 747364. [Google Scholar] [CrossRef]
- Rath, N.; Raje, N.; Rosenwasser, L. Immunoglobulin E as a Biomarker in Asthma. Immunol. Allergy Clin. N. Am. 2018, 38, 587–597. [Google Scholar] [CrossRef]
- Narendra, D.; Blixt, J.; Hanania, N.A. Immunological biomarkers in severe asthma. Semin. Immunol. 2019, 46, 101332. [Google Scholar] [CrossRef] [PubMed]
- Nixon, J.; Newbold, P.; Mustelin, T.; Anderson, G.P.; Kolbeck, R. Monoclonal antibody therapy for the treatment of asthma and chronic obstructive pulmonary disease with eosinophilic inflammation. Pharmacol. Ther. 2017, 169, 57–77. [Google Scholar] [CrossRef] [PubMed]
- Coumou, H.; Bel, E.H. Improving the diagnosis of eosinophilic asthma. Expert Rev. Respir. Med. 2016, 10, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Vijverberg, S.J.; Hilvering, B.; Raaijmakers, J.A.; Lammers, J.W.; Maitland-van der Zee, A.H.; Koenderman, L. Clinical utility of asthma biomarkers: From bench to bedside. Biologics 2013, 7, 199–210. [Google Scholar] [PubMed]
- Kelly, E.A.; Jarjour, N.N. Role of matrix metalloproteinases in asthma. Curr. Opin. Pulm. Med. 2003, 9, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Ronnow, S.R.; Sand, J.M.B.; Staunstrup, L.M.; Bahmer, T.; Wegmann, M.; Lunding, L.; Burgess, J.; Rabe, K.; Sorensen, G.L.; Fuchs, O.; et al. A serological biomarker of type I collagen degradation is related to a more severe, high neutrophilic, obese asthma subtype. Asthma Res. Pract. 2022, 8, 2. [Google Scholar] [CrossRef]
- Kuczia, P.; Mastalerz, L.; Potaczek, D.P.; Cybulska, A.; Zareba, L.; Bazan-Socha, S.; Undas, A. Increased activity of lipoprotein-associated phospholipase A(2) in non-severe asthma. Allergol. Int. 2019, 68, 450–455. [Google Scholar] [CrossRef]
- Park, G.Y.; Lee, Y.G.; Berdyshev, E.; Nyenhuis, S.; Du, J.; Fu, P.; Gorshkova, I.A.; Li, Y.; Chung, S.; Karpurapu, M.; et al. Autotaxin production of lysophosphatidic acid mediates allergic asthmatic inflammation. Am. J. Respir. Crit. Care Med. 2013, 188, 928–940. [Google Scholar] [CrossRef]
- Koussih, L.; Atoui, S.; Tliba, O.; Gounni, A.S. New Insights on the Role of pentraxin-3 in Allergic Asthma. Front. Allergy 2021, 2, 678023. [Google Scholar] [CrossRef]
- Januskevicius, A.; Jurkeviciute, E.; Janulaityte, I.; Kalinauskaite-Zukauske, V.; Miliauskas, S.; Malakauskas, K. Blood Eosinophils Subtypes and Their Survivability in Asthma Patients. Cells 2020, 9, 1248. [Google Scholar] [CrossRef]
- Kalinauskaite-Zukauske, V.; Januskevicius, A.; Janulaityte, I.; Miliauskas, S.; Malakauskas, K. Expression of eosinophil beta chain-signaling cytokines receptors, outer-membrane integrins, and type 2 inflammation biomarkers in severe non-allergic eosinophilic asthma. BMC Pulm. Med. 2019, 19, 158. [Google Scholar] [CrossRef] [PubMed]
- Kalinauskaite-Zukauske, V.; Janulaityte, I.; Januskevicius, A.; Malakauskas, K. Serum levels of epithelial-derived mediators and interleukin-4/interleukin-13 signaling after bronchial challenge with Dermatophagoides pteronyssinus in patients with allergic asthma. Scand. J. Immunol. 2019, 90, e12820. [Google Scholar] [CrossRef] [PubMed]
- Georas, S.N. LPA and Autotaxin: Potential Drug Targets in Asthma? Cell Biochem. Biophys. 2021, 79, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhong, M.; Zhan, D.; Zhang, Y.; Liu, S. Roles of the HGF/Met signaling in head and neck squamous cell carcinoma: Focus on tumor immunity (Review). Oncol. Rep. 2020, 44, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Correll, K.; Zemans, R.L.; Leslie, C.C.; Murphy, R.C.; Mason, R.J. Influenza induces IL-8 and GM-CSF secretion by human alveolar epithelial cells through HGF/c-Met and TGF-alpha/EGFR signaling. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L1178–L1188. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Ueki, S.; Konno, Y.; Ito, W.; Takeda, M.; Nakamura, Y.; Nishikawa, J.; Moritoki, Y.; Omokawa, A.; Saga, T.; et al. The effect of hepatocyte growth factor on secretory functions in human eosinophils. Cytokine 2016, 88, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.S.; Pavlidis, S.; Zhu, J.; Loza, M.; Baribaud, F.; Rowe, A.; Pandis, I.; Gibeon, D.; Hoda, U.; Sousa, A.; et al. Contribution of airway eosinophils in airway wall remodeling in asthma: Role of MMP-10 and MET. Allergy 2019, 74, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Principe, S.; Porsbjerg, C.; Bolm Ditlev, S.; Kjaersgaard Klein, D.; Golebski, K.; Dyhre-Petersen, N.; van Dijk, Y.E.; van Bragt, J.J.; Dankelman, L.L.; Dahlen, S.; et al. Treating severe asthma: Targeting the IL-5 pathway. Clin. Exp. Allergy 2021, 51, 992–1005. [Google Scholar] [CrossRef]
- Fernandez-Botran, R. Soluble cytokine receptors: Novel immunotherapeutic agents. Expert Opin. Investig. Drugs 2000, 9, 497–514. [Google Scholar] [CrossRef]
- Wilson, T.M.; Maric, I.; Shukla, J.; Brown, M.; Santos, C.; Simakova, O.; Khouri, P.; Fay, M.P.; Kozich, A.; Kolbeck, R.; et al. IL-5 receptor alpha levels in patients with marked eosinophilia or mastocytosis. J. Allergy Clin. Immunol. 2011, 128, 1086–1092.e3. [Google Scholar] [CrossRef]
- Blumchen, K.; Kallinich, T.; Hamelmann, E. Interleukin-5: A novel target for asthma therapy. Expert Opin. Biol. Ther. 2001, 1, 433–453. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, J.; Wang, C.; Fukunaga, A.; Li, S.; Yodoi, J.; Tian, H. Thioredoxin-1: A Promising Target for the Treatment of Allergic Diseases. Front. Immunol. 2022, 13, 883116. [Google Scholar] [CrossRef] [PubMed]
- Dozor, A.J. The role of oxidative stress in the pathogenesis and treatment of asthma. Ann. N. Y. Acad. Sci. 2010, 1203, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Doni, A.; Garlanda, C.; Mantovani, A. Innate immunity, hemostasis and matrix remodeling: PTX3 as a link. Semin. Immunol. 2016, 28, 570–577. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, S.; Qu, X.; Liu, F.; Wang, C. Pentraxin 3 as a prognostic biomarker in patients with systemic inflammation or infection. Mediat. Inflamm. 2014, 2014, 421429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shan, L.; Koussih, L.; Redhu, N.S.; Halayko, A.J.; Chakir, J.; Gounni, A.S. Pentraxin 3 (PTX3) expression in allergic asthmatic airways: Role in airway smooth muscle migration and chemokine production. PLoS ONE 2012, 7, e34965. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Lee, H.S.; Sol, I.S.; Kim, M.N.; Hong, J.Y.; Lee, K.E.; Kim, Y.H.; Kim, K.W.; Sohn, M.H.; Kim, K.-E. Sputum pentraxin 3 as a candidate to assess airway inflammation and remodeling in childhood asthma. Medicine 2016, 95, e5677. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tang, K.; Wang, M.; Yang, Q.; Xu, Y.; Wang, J.; Zhao, J.; Xie, J. Pentraxin levels in non-eosinophilic versus eosinophilic asthma. Clin. Exp. Allergy. 2018, 48, 981–989. [Google Scholar] [CrossRef]
- Hoshino, T.; Okamoto, M.; Takei, S.; Sakazaki, Y.; Iwanaga, T.; Aizawa, H. Redox-regulated mechanisms in asthma. Antioxid. Redox Signal. 2008, 10, 769–783. [Google Scholar] [CrossRef]
- Imaoka, H.; Hoshino, T.; Takei, S.; Sakazaki, Y.; Kinoshita, T.; Okamoto, M.; Kawayama, T.; Yodoi, J.; Kato, S.; Iwanaga, T.; et al. Effects of thioredoxin on established airway remodeling in a chronic antigen exposure asthma model. Biochem. Biophys. Res. Commun. 2007, 360, 525–530. [Google Scholar] [CrossRef]
- Hanschmann, E.M.; Berndt, C.; Hecker, C.; Garn, H.; Bertrams, W.; Lillig, C.H.; Hudemann, C. Glutaredoxin 2 Reduces Asthma-Like Acute Airway Inflammation in Mice. Front. Immunol. 2020, 11, 561724. [Google Scholar] [CrossRef] [PubMed]
- Ito, W.; Kobayashi, N.; Takeda, M.; Ueki, S.; Kayaba, H.; Nakamura, H.; Yodoi, J.; Chihara, J. Thioredoxin in allergic inflammation. Int. Arch. Allergy Immunol. 2011, 155 (Suppl. 1), 142–146. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Nakamura, H.; Adachi, T.; Sannohe, S.; Oyamada, H.; Kayaba, H.; Yodoi, J.; Chihara, J. Elevated serum levels of thioredoxin in patients with acute exacerbation of asthma. Immunol. Lett. 2003, 86, 199–205. [Google Scholar] [CrossRef] [PubMed]
- Habib, N.; Pasha, M.A.; Tang, D.D. Current Understanding of Asthma Pathogenesis and Biomarkers. Cells 2022, 11, 2764. [Google Scholar] [CrossRef] [PubMed]
- Grzela, K.; Litwiniuk, M.; Zagorska, W.; Grzela, T. Airway Remodeling in Chronic Obstructive Pulmonary Disease and Asthma: The Role of Matrix Metalloproteinase-9. Arch. Immunol. Ther. Exp. 2016, 64, 47–55. [Google Scholar] [CrossRef]
- Nomura, A.; Uchida, Y.; Sakamoto, T.; Ishii, Y.; Masuyama, K.; Morishima, Y.; Hirano, K.; Sekizawa, K. Increases in collagen type I synthesis in asthma: The role of eosinophils and transforming growth factor-beta. Clin. Exp. Allergy 2002, 32, 860–865. [Google Scholar] [CrossRef]
- Burgess, J.K.; Mauad, T.; Tjin, G.; Karlsson, J.C.; Westergren-Thorsson, G. The extracellular matrix—The under-recognized element in lung disease? J. Pathol. 2016, 240, 397–409. [Google Scholar] [CrossRef]
- Halwani, R.; Al-Muhsen, S.; Al-Jahdali, H.; Hamid, Q. Role of transforming growth factor-beta in airway remodeling in asthma. Am. J. Respir. Cell Mol. Biol. 2011, 44, 127–133. [Google Scholar] [CrossRef]
| Inclusion Criteria | Exclusion Criteria | |
|---|---|---|
| AA Group | HSs | |
| Diagnosed with AA Free of inhaled steroids at least 1 month Positive skin prick test for at least Dermatophagoides pteronyssinus allergen Positive methacholine challenge or bronchial reversibility test | No symptoms of respiratory disease No clinical allergy symptoms All skin prick tests negative | Age < 18 years Smoker Active airway infection ≤ 1 months prior to study Other clinically significant non-controlled disease |
| Screening Visit | V1 | V2 * |
|---|---|---|
| Inclusion and exclusion criteria | CBC | CBC |
| Written informed consent | Total IgE | Total IgE |
| Spirometry | FeNO measurement | FeNO measurement |
| Methacholine challenge test/bronchial reversibility test * | Spirometry | Spirometry |
| Skin prick testing | BAC with Dermatophagoides pteronyssinus allergen * |
| Circulating Inflammatory Biomarkers | Blood Oxidative Stress Markers | Extracellular Matrix Proteinases and Degradation Product |
|---|---|---|
| ATX | MMP-7 | |
| MET | PTX-3 | MMP-9 |
| Lp-PLA2 | TRX1 | MMP-10 |
| sIL-5Rα | ICTP |
| Characteristic | AA Patients | HSs | |
|---|---|---|---|
| Number, n | 20 | 16 | |
| Sex, M/F | 11/9 | 6/10 | |
| Age, years | 22 (19; 35) | 27 (25; 30) | |
| BMI, kg/m2 | 22.4 (21.1; 28.7) | 22.3 (20.1; 24.2) | |
| Asthma control test, score | 21 (12; 23) | NA | |
| PD20M, mg | 0.27 (0.10; 0.45) | NA | |
| PD20A, HEP/mL (range) | 10 (1; 10) | NA | |
| V1 | V2 | ||
| FEV1, L | 3.75 (3.05; 3.94) # | 3.41 (2.63; 3.88) | 3.66 (3.21; 4.64) |
| FEV1, % of predicted | 87 (79; 102) * # | 81 (65; 95) * | 97 (89; 108) |
| FeNO, ppb | 42 (23; 70) * # | 64 (37; 108) * | 13 (9; 18) |
| Blood EOS count, × 109/L | 0.31 (0.19; 0.44) * # | 0.41 (0.32; 0.57) * | 0.10 (0.07; 0.13) |
| Total IgE, IU/mL | 295 (122; 640) * | 377 (104; 670) * | 13 (4; 44) |
| Biologically Active Substances | AA Patients | HSs | |
|---|---|---|---|
| V1 | V2 | ||
| ATX (pg/mL) | 206.8 (190.9; 286.3) | 201.5 (180.3; 235.5) | 233.3 (190.9; 243.9) |
| MET (pg/mL) | 30.5 (14.0; 51.1) * # | 19.8 (8.0; 46.6) * | 54.3 (28.0; 80.3) |
| Lp-PLA2 (pg/mL) | 451.4 (216.4; 755.1) | 445.5 (199.9; 706.6) | 629.3 (224.9; 980.8) |
| sIL-5Rα (ng/mL) | 63.4 (55.9; 105.4) * | 67.9 (54.9; 112.1) * | 25.2 (11.3; 33.2) |
| PTX3 (pg/mL) | 560.1 (326.1; 875.2) * # | 498.9 (226.5; 651.1) * | 1217.6 (919.8; 1580.1) |
| TRX1 (ng/mL) | 131.2 (93.8; 173.6) * # | 142.0 (89.4; 195.6) * | 113.9 (67.4; 136.1) |
| MMP-7 (pg/mL) | 332.9 (241.5; 421.3) | 295.1 (272.1; 327.7) | 278.3 (195.3; 379.2) |
| MMP-9 (pg/mL) | 253.7 (151.4; 339.9) | 211.9 (116.9; 293.7) | 204.2 (94.8; 498.0) |
| MMP-10 (pg/mL) | 896.1 (318.9; 1501.0) | 971.0 (355.7; 1682.6) | 502.7 (324.0; 1052.5) |
| ICTP (pg/mL) | 523.0 (478.0; 552.0) * | 476.3 (411.6; 531.4) * | 589.0 (540.0; 693.0) |
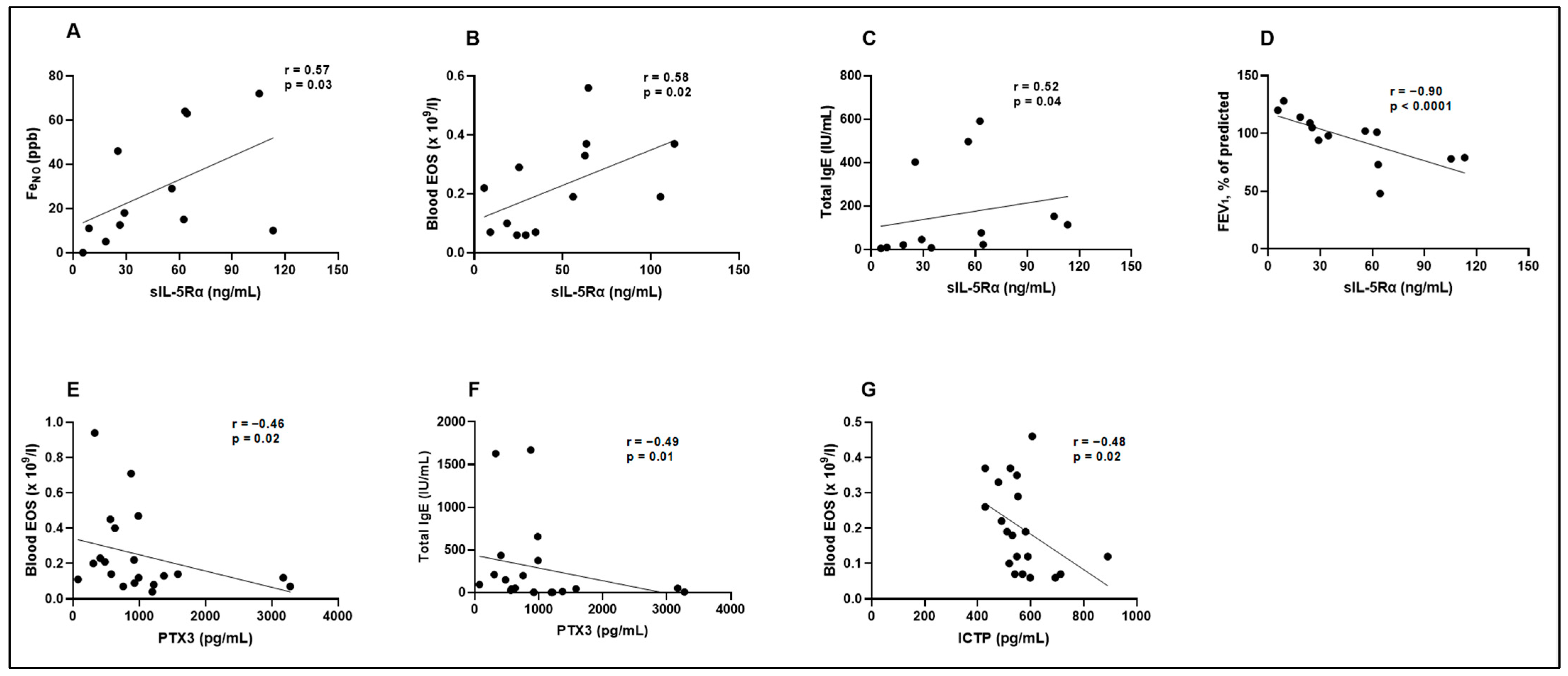
| FeNO (ppb) | Blood EOS (×109/L) | Total IgE (IU/mL) | FEV1, % of Predicted | |
|---|---|---|---|---|
| MET (pg/mL) | r = 0.02 p = 0.47 | r = −0.07 p = 0.39 | r = −0.06 p = 0.41 | r = −0.07 p = 0.40 |
| sIL-5Rα (ng/mL) | r = 0.57 p = 0.03 | r = 0.58 p = 0.02 | r = 0.52 p = 0.04 | r = −0.90 p < 0.0001 |
| PTX3 (pg/mL) | r = −0.32 p = 0.10 | r = −0.46 p = 0.02 | r = −0.49 p = 0.01 | r = 0.23 p = 0.17 |
| TRX1 (ng/mL) | r = 0.25 p = 0.18 | r = 0.19 p = 0.23 | r = 0.35 p = 0.06 | r = −0.23 p = 0.18 |
| ICTP (pg/mL) | r = −0.02 p = 0.46 | r = −0.48 p = 0.02 | r = −0.11 p = 0.32 | r = 0.17 p = 0.24 |
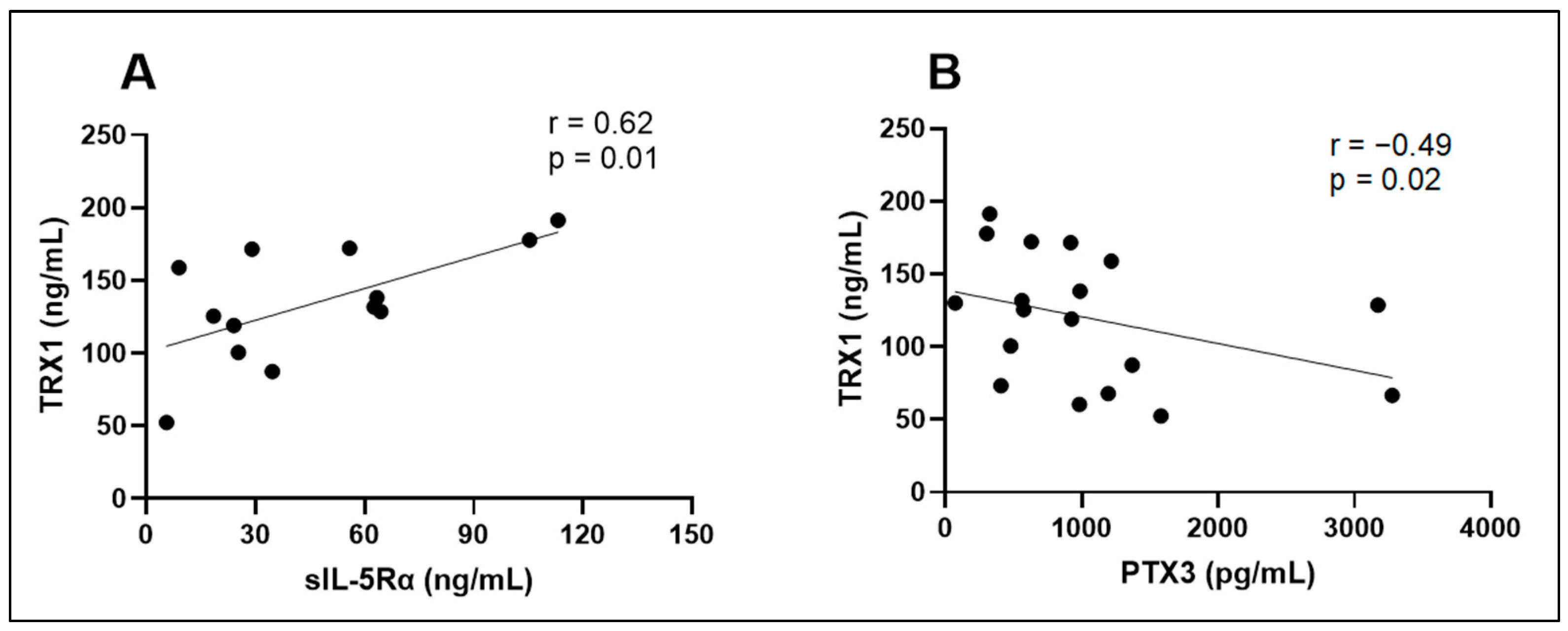
| MET (pg/mL) | sIL-5Rα (ng/mL) | PTX3 (pg/mL) | TRX1 (ng/mL) | ICTP (pg/mL) | |
|---|---|---|---|---|---|
| MET (pg/mL) | – | r = −0.27 p = 0.45 | r = 0.09 p = 0.75 | r = −0.35 p = 0.22 | r = −0.16 p = 0.55 |
| sIL-5Rα (ng/mL) | r = −0.27 p = 0.45 | – | r = −0.38 p = 0.20 | r = 0.62 p = 0.01 | r = −0.06 p = 0.85 |
| PTX3 (pg/mL) | r = 0.09 p = 0.75 | r = −0.38 p = 0.20 | – | r = −0.49 p = 0.02 | r = 0.31 p = 0.19 |
| TRX1 (ng/mL) | r = −0.35 p = 0.22 | r = 0.62 p = 0.01 | r = −0.49 p = 0.02 | – | r = −0.05 p = 0.86 |
| ICTP (pg/mL) | r = −0.16 p = 0.55 | r = −0.06 p = 0.85 | r = 0.31 p = 0.19 | r = −0.05 p = 0.86 | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palacionyte, J.; Januskevicius, A.; Vasyle, E.; Rimkunas, A.; Bajoriuniene, I.; Vitkauskiene, A.; Miliauskas, S.; Malakauskas, K. Novel Serum Biomarkers for Patients with Allergic Asthma Phenotype. Biomedicines 2024, 12, 232. https://doi.org/10.3390/biomedicines12010232
Palacionyte J, Januskevicius A, Vasyle E, Rimkunas A, Bajoriuniene I, Vitkauskiene A, Miliauskas S, Malakauskas K. Novel Serum Biomarkers for Patients with Allergic Asthma Phenotype. Biomedicines. 2024; 12(1):232. https://doi.org/10.3390/biomedicines12010232
Chicago/Turabian StylePalacionyte, Jolita, Andrius Januskevicius, Egle Vasyle, Airidas Rimkunas, Ieva Bajoriuniene, Astra Vitkauskiene, Skaidrius Miliauskas, and Kestutis Malakauskas. 2024. "Novel Serum Biomarkers for Patients with Allergic Asthma Phenotype" Biomedicines 12, no. 1: 232. https://doi.org/10.3390/biomedicines12010232
APA StylePalacionyte, J., Januskevicius, A., Vasyle, E., Rimkunas, A., Bajoriuniene, I., Vitkauskiene, A., Miliauskas, S., & Malakauskas, K. (2024). Novel Serum Biomarkers for Patients with Allergic Asthma Phenotype. Biomedicines, 12(1), 232. https://doi.org/10.3390/biomedicines12010232








