Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. SNP Selection and Genotyping
2.3. Statistical, Bioinformatics Analysis
3. Results
3.1. Possible Functionality of the BC-Correlated Locus rs10454142 PPP1R21 (In Silico Data)
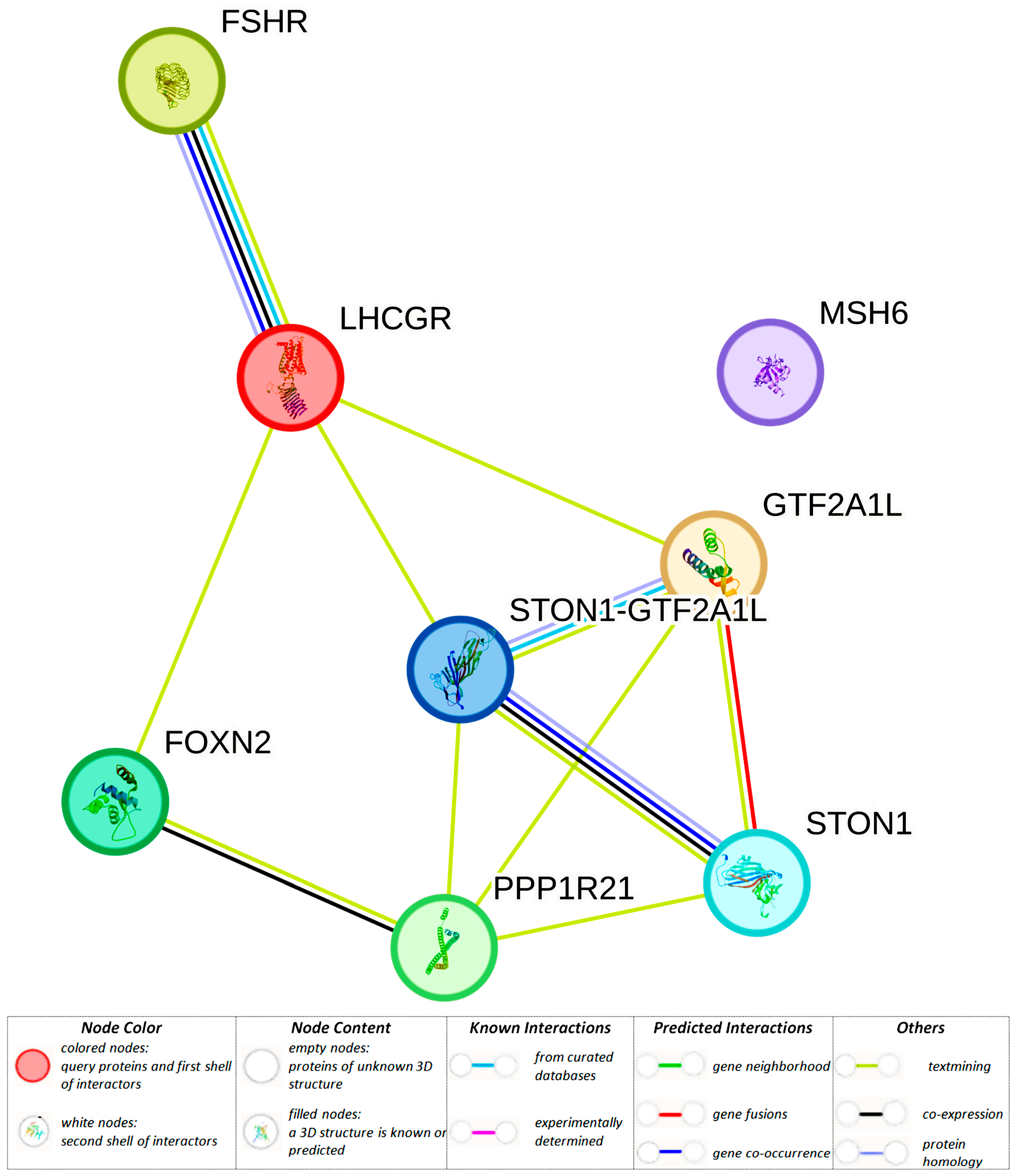
3.1.1. Liver-Specific Regulatory Effects of BC-Causal Loci
3.1.2. Adipose-Specific Regulatory Effects of BC-Causal Loci
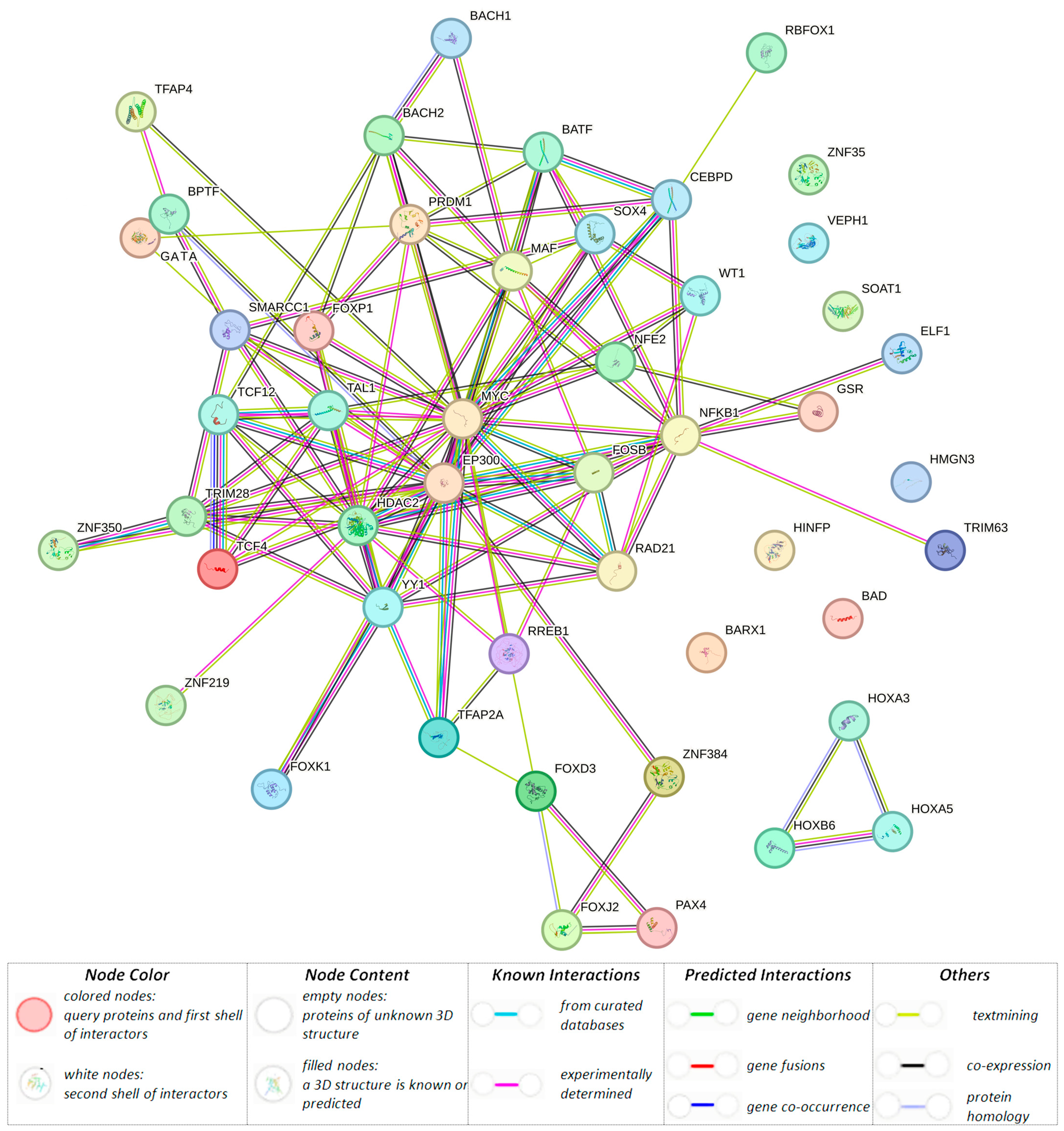
3.1.3. Organism-Significant Regulatory Effects of BC-Causal Loci
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Giaquinto, A.N.; Sung, H.; Miller, K.D.; Kramer, J.L.; Newman, L.A.; Minihan, A.; Jemal, A.; Siegel, R.L. Breast Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 524–541. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 25 August 2023).
- Malignant Neoplasms in Russia in 2021 (Morbidity and Mortality); Kaprin, A.D., Starinsky, V.V., Shakhzadova, A.O., Eds.; MNIOI im. P.A. Gercena: Moscow, Russia, 2022; 252p, Available online: https://oncology-association.ru/wp-content/uploads/2022/11/zlokachestvennye-novoobrazovaniya-v-rossii-v-2021-g_zabolevaemost-i-smertnost.pdf (accessed on 3 October 2023). (In Russian)
- International Agency for Research on Cancer. Available online: https://gco.iarc.fr (accessed on 25 August 2023).
- Möller, S.; Mucci, L.A.; Harris, J.R.; Scheike, T.; Holst, K.; Halekoh, U.; Adami, H.O.; Czene, K.; Christensen, K.; Holm, N.V.; et al. The Heritability of Breast Cancer among Women in the Nordic Twin Study of Cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 145–150. [Google Scholar] [CrossRef]
- Mucci, L.A.; Hjelmborg, J.B.; Harris, J.R.; Czene, K.; Havelick, D.J.; Scheike, T.; Graff, R.E.; Holst, K.; Möller, S.; Unger, R.H.; et al. Nordic Twin Study of Cancer (NorTwinCan) Collaboration. Familial Risk and Heritability of Cancer among Twins in Nordic Countries. JAMA 2016, 315, 68–76, Erratum in JAMA 2016, 315, 822. [Google Scholar] [CrossRef]
- Shiovitz, S.; Korde, L.A. Genetics of breast cancer: A topic in evolution. Ann. Oncol. 2015, 26, 1291–1299. [Google Scholar] [CrossRef] [PubMed]
- Lilyquist, J.; Ruddy, K.J.; Vachon, C.M.; Couch, F.J. Common Genetic Variation and Breast Cancer Risk-Past, Present, and Future. Cancer Epidemiol. Biomark. Prev. 2018, 27, 380–394. [Google Scholar] [CrossRef]
- Michailidou, K.; Lindström, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemaçon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef]
- Shu, X.; Long, J.; Cai, Q.; Kweon, S.S.; Choi, J.Y.; Kubo, M.; Park, S.K.; Bolla, M.K.; Dennis, J.; Wang, Q.; et al. Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants. Nat. Commun. 2020, 11, 1217. [Google Scholar] [CrossRef]
- Adedokun, B.; Du, Z.; Gao, G.; Ahearn, T.U.; Lunetta, K.L.; Zirpoli, G.; Figueroa, J.; John, E.M.; Bernstein, L.; Zheng, W.; et al. Cross-ancestry GWAS meta-analysis identifies six breast cancer loci in African and European ancestry women. Nat. Commun. 2021, 12, 4198. [Google Scholar] [CrossRef]
- Key, T.J.; Appleby, P.N.; Reeves, G.K.; Travis, R.C.; Brinton, L.A.; Helzlsouer, K.J.; Dorgan, J.F.; Gapstur, S.M.; Gaudet, M.M.; Kaaks, R.; et al. Steroid hormone measurements from different types of assays in relation to body mass index and breast cancer risk in postmenopausal women: Reanalysis of eighteen prospective studies. Steroids 2015, 99, 49–55. [Google Scholar] [CrossRef]
- Tin Tin, S.; Reeves, G.K.; Key, T.J. Endogenous hormones and risk of invasive breast cancer in pre- and post-menopausal women: Findings from the UK Biobank. Br. J. Cancer 2021, 125, 126–134. [Google Scholar] [CrossRef]
- Chen, F.; Wen, W.; Long, J.; Shu, X.; Yang, Y.; Shu, X.O.; Zheng, W. Mendelian randomization analyses of 23 known and suspected risk factors and biomarkers for breast cancer overall and by molecular subtypes. Int. J. Cancer 2022, 151, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Coviello, A.D.; Zhuang, W.V.; Lunetta, K.L.; Bhasin, S.; Ulloor, J.; Zhang, A.; Karasik, D.; Kiel, D.P.; Vasan, R.S.; Murabito, J.M. Circulating testosterone and SHBG concentrations are heritable in women: The Framingham Heart Study. J. Clin. Endocrinol. Metab. 2011, 96, E1491–E1495. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.L. Plasma steroid-binding proteins: Primary gatekeepers of steroid hormone action. J. Endocrinol. 2016, 230, R13–R25. [Google Scholar] [CrossRef] [PubMed]
- Balogh, A.; Karpati, E.; Schneider, A.E.; Hetey, S.; Szilagyi, A.; Juhasz, K.; Laszlo, G.; Hupuczi, P.; Zavodszky, P.; Papp, Z.; et al. Sex hormone-binding globulin provides a novel entry pathway for estradiol and influences subsequent signaling in lymphocytes via membrane receptor. Sci. Rep. 2019, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Sinnott-Armstrong, N.; Naqvi, S.; Rivas, M.; Pritchard, J.K. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. eLife 2021, 10, e58615. [Google Scholar] [CrossRef] [PubMed]
- Varghese, J.S.; Smith, P.L.; Folkerd, E.; Brown, J.; Leyland, J.; Audley, T.; Warren, R.M.; Dowsett, M.; Easton, D.F.; Thompson, D.J. The heritability of mammographic breast density and circulating sex-hormone levels: Two independent breast cancer risk factors. Cancer Epidemiol. Biomark. Prev. 2012, 21, 2167–2175. [Google Scholar] [CrossRef]
- Coviello, A.D.; Haring, R.; Wellons, M.; Vaidya, D.; Lehtimäki, T.; Keildson, S.; Lunetta, K.L.; He, C.; Fornage, M.; Lagou, V.; et al. A genome-wide association meta-analysis of circulating sex hormone—Binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS Genet. 2012, 8, e1002805. [Google Scholar] [CrossRef] [PubMed]
- Dimou, N.L.; Papadimitriou, N.; Gill, D.; Christakoudi, S.; Murphy, N.; Gunter, M.J.; Travis, R.C.; Key, T.J.; Fortner, R.T.; Haycock, P.C. Sex hormone binding globulin and risk of breast cancer: A Mendelian randomization study. Int. J. Epidemiol. 2019, 48, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Renehan, A.G.; Tyson, M.; Egger, M.; Heller, R.F.; Zwahlen, M. Body-mass index and incidence of cancer: A systematic review and meta-analysis of prospective observational studies. Lancet 2008, 371, 569–578. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Douglas, I.; Forbes, H.; dos-Santos-Silva, I.; Leon, D.A.; Smeeth, L. Body-mass index and risk of 22 specific cancers: A population-based cohort study of 5·24 million UK adults. Lancet 2014, 384, 755–765. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, W.; Dai, Z.; Wang, M.; Tian, T.; Liu, X.; Kang, H.; Guan, H.; Zhang, S.; Dai, Z. Association between body mass index and breast cancer risk: Evidence based on a dose-response meta-analysis. Cancer Manag. Res. 2018, 10, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, P.; Au, C.C.; Benito-Martin, A.; Ladumor, H.; Oshchepkova, S.; Moges, R.; Brown, K.A. Estrogens and breast cancer: Mechanisms involved in obesity-related development, growth and progression. J. Steroid Biochem. Mol. Biol. 2019, 189, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Ajabnoor, G.M.A. The Molecular and Genetic Interactions between Obesity and Breast Cancer Risk. Medicina 2023, 59, 1338. [Google Scholar] [CrossRef] [PubMed]
- Trevellin, E.; Bettini, S.; Pilatone, A.; Vettor, R.; Milan, G. Obesity, the Adipose Organ and Cancer in Humans: Association or Causation? Biomedicines 2023, 11, 1319. [Google Scholar] [CrossRef] [PubMed]
- Verkasalo, P.K.; Thomas, H.V.; Appleby, P.N.; Davey, G.K.; Key, T.J. Circulating levels of sex hormones and their relation to risk factors for breast cancer: A cross-sectional study in 1092 pre- and postmenopausal women (United Kingdom). Cancer Causes Control 2001, 12, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Baglietto, L.; English, D.R.; Hopper, J.L.; MacInnis, R.J.; Morris, H.A.; Tilley, W.D.; Krishnan, K.; Giles, G.G. Circulating steroid hormone concentrations in postmenopausal women in relation to body size and composition. Breast Cancer Res. Treat. 2009, 115, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Liedtke, S.; Schmidt, M.E.; Vrieling, A.; Lukanova, A.; Becker, S.; Kaaks, R.; Zaineddin, A.K.; Buck, K.; Benner, A.; Chang-Claude, J.; et al. Postmenopausal sex hormones in relation to body fat distribution. Obesity 2012, 20, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Goudswaard, L.J.; Bell, J.A.; Hughes, D.A.; Corbin, L.J.; Walter, K.; Davey Smith, G.; Soranzo, N.; Danesh, J.; Di Angelantonio, E.; Ouwehand, W.H.; et al. Effects of adiposity on the human plasma proteome: Observational and Mendelian randomisation estimates. Int. J. Obes. 2021, 45, 2221–2229. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. Matrix Metalloproteinase Gene Polymorphisms Are Associated with Breast Cancer in the Caucasian Women of Russia. Int. J. Mol. Sci. 2022, 23, 12638. [Google Scholar] [CrossRef]
- Demakova, N.A. Molecular and genetic characteristics of patients with hyperplasia and endometric polyps. Res. Results Biomed. 2018, 4, 26–39. [Google Scholar] [CrossRef]
- Radzinsky, V.E.; Altuchova, O.B. Molecular-genetic determinants of infertility in genital endometryosis. Res. Results Biomed. 2018, 4, 28–37. [Google Scholar] [CrossRef]
- Golovchenko, I.O. Genetic determinants of sex hormone levels in endometriosis patients. Res. Results Biomed. 2023, 9, 5–21. [Google Scholar] [CrossRef]
- Tikunova, E.; Ovtcharova, V.; Reshetnikov, E.; Dvornyk, V.; Polonikov, A.; Bushueva, O.; Churnosov, M. Genes of tumor necrosis factors and their receptors and the primary open angle glaucoma in the population of Central Russia. Int. J. Ophthalmol. 2017, 10, 1490–1494. [Google Scholar] [CrossRef] [PubMed]
- Moskalenko, M.I.; Milanova, S.N.; Ponomarenko, I.V.; Polonikov, A.V.; Churnosov, M.I. Study of associations of polymorphism of matrix metalloproteinases genes with the development of arterial hypertension in men. Kardiologiia 2019, 59, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Abramova, M.; Churnosova, M.; Efremova, O.; Aristova, I.; Reshetnikov, E.; Polonikov, A.; Churnosov, M.; Ponomarenko, I. Effects of pre-pregnancy over-weight/obesity on the pattern of association of hypertension susceptibility genes with preeclampsia. Life 2022, 12, 2018. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.; Demin, S.; Churnosov, M.; Reshetnikov, E.; Aristova, I.; Churnosova, M.; Ponomarenko, I. The Modifying Effect of Obesity on the Association of Matrix Metalloproteinase Gene Polymorphisms with Breast Cancer Risk. Biomedicines 2022, 10, 2617. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, E.; Churnosova, M.; Reshetnikova, Y.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; et al. Maternal Age at Menarche Genes Determines Fetal Growth Restriction Risk. Int. J. Mol. Sci. 2024, 25, 647. [Google Scholar] [CrossRef]
- Eliseeva, N.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. LOXL1 gene polymorphism candidates for exfoliation glaucoma are also associated with a risk for primary open-angle glaucoma in a Caucasian population from central Russia. Mol. Vis. 2021, 27, 262–269. [Google Scholar] [PubMed]
- Golovchenko, I.; Aizikovich, B.; Golovchenko, O.; Reshetnikov, E.; Churnosova, M.; Aristova, I.; Ponomarenko, I.; Churnosov, M. Sex Hormone Candidate Gene Polymorphisms Are Associated with Endometriosis. Int. J. Mol. Sci. 2022, 23, 13691. [Google Scholar] [CrossRef]
- Novakov, V.; Novakova, O.; Churnosova, M.; Sorokina, I.; Aristova, I.; Polonikov, A.; Reshetnikov, E.; Churnosov, M. Intergenic Interactions of SBNO1, NFAT5 and GLT8D1 Determine the Susceptibility to Knee Osteoarthritis among Europeans of Russia. Life 2023, 13, 405. [Google Scholar] [CrossRef]
- Ponomarenko, I.; Pasenov, K.; Churnosova, M.; Sorokina, I.; Aristova, I.; Churnosov, V.; Ponomarenko, M.; Reshetnikov, E.; Churnosov, M. Sex-Hormone-Binding Globulin Gene Polymorphisms and Breast Cancer Risk in Caucasian Women of Russia. Int. J. Mol. Sci. 2024, 25, 2182. [Google Scholar] [CrossRef] [PubMed]
- Harrison, S.; Davies, N.M.; Howe, L.D.; Hughes, A. Testosterone and socioeconomic position: Mendelian randomization in 306,248 men and women in UK Biobank. Sci. Adv. 2021, 7, eabf8257. [Google Scholar] [CrossRef] [PubMed]
- Fantus, R.J.; Na, R.; Wei, J.; Shi, Z.; Resurreccion, W.K.; Halpern, J.A.; Franco, O.; Hayward, S.W.; Isaacs, W.B.; Zheng, S.L.; et al. Genetic Susceptibility for Low Testosterone in Men and Its Implications in Biology and Screening: Data from the UK Biobank. Eur. Urol. Open Sci. 2021, 29, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Haas, C.B.; Hsu, L.; Lampe, J.W.; Wernli, K.J.; Lindström, S. Cross-ancestry Genome-wide Association Studies of Sex Hormone Concentrations in Pre- and Postmenopausal Women. Endocrinology 2022, 163, bqac020. [Google Scholar] [CrossRef] [PubMed]
- Ruth, K.S.; Day, F.R.; Tyrrell, J.; Thompson, D.J.; Wood, A.R.; Mahajan, A.; Beaumont, R.N.; Wittemans, L.; Martin, S.; Busch, A.S.; et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020, 26, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, C.; Wallaschofski, H.; Lunetta, K.L.; Stolk, L.; Perry, J.R.; Koster, A.; Petersen, A.K.; Eriksson, J.; Lehtimäki, T.; Huhtaniemi, I.T.; et al. Genetic determinants of serum testosterone concentrations in men. PLoS Genet. 2011, 7, e1002313. [Google Scholar] [CrossRef] [PubMed]
- Golovchenko, O.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Polonikov, A.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of ESR1 and PGR and risk of intrauterine growth restriction in population of Central Russia. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 52–57. [Google Scholar] [CrossRef]
- Bushueva, O.; Solodilova, M.; Churnosov, M.; Ivanov, V.; Polonikov, A. The Flavin-Containing Monooxygenase 3 Gene and Essential Hypertension: The Joint Effect of Polymorphism E158K and Cigarette Smoking on Disease Susceptibility. Int. J. Hypertens. 2014, 2014, 712169. [Google Scholar] [CrossRef] [PubMed]
- Churnosov, M.; Abramova, M.; Reshetnikov, E.; Lyashenko, I.V.; Efremova, O.; Churnosova, M.; Ponomarenko, I. Polymorphisms of hypertension susceptibility genes as a risk factors of preeclampsia in the Caucasian population of central Russia. Placenta 2022, 129, 51–61. [Google Scholar] [CrossRef]
- Ivanova, T.; Churnosova, M.; Abramova, M.; Plotnikov, D.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Sex-Specific Features of the Correlation between GWAS-Noticeable Polymorphisms and Hypertension in Europeans of Russia. Int. J. Mol. Sci. 2023, 24, 7799. [Google Scholar] [CrossRef]
- Ivanova, T.A. Sex-specific features of interlocus interactions determining susceptibility to hypertension. Res. Results Biomed. 2024, 10, 53–68. [Google Scholar]
- Che, R.; Jack, J.R.; Motsinger-Reif, A.A.; Brown, C.C. An adaptive permutation approach for genome-wide association study: Evaluation and recommendations for use. BioData Min. 2014, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikova, Y.; Churnosova, M.; Stepanov, V.; Bocharova, A.; Serebrova, V.; Trifonova, E.; Ponomarenko, I.; Sorokina, I.; Efremova, O.; Orlova, V.; et al. Maternal Age at Menarche Gene Polymorphisms Are Associated with Offspring Birth Weight. Life 2023, 13, 1525. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, E.; Ponomarenko, I.; Golovchenko, O.; Sorokina, I.; Batlutskaya, I.; Yakunchenko, T.; Dvornyk, V.; Polonikov, A.; Churnosov, M. The VNTR polymorphism of the endothelial nitric oxide synthase gene and blood pressure in women at the end of pregnancy. Taiwan. J. Obstet. Gynecol. 2019, 58, 390–395. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, T.; Churnosova, M.; Abramova, M.; Ponomarenko, I.; Reshetnikov, E.; Aristova, I.; Sorokina, I.; Churnosov, M. Risk Effects of rs1799945 Polymorphism of the HFE Gene and Intergenic Interactions of GWAS-Significant Loci for Arterial Hypertension in the Caucasian Population of Central Russia. Int. J. Mol. Sci. 2023, 24, 8309. [Google Scholar] [CrossRef] [PubMed]
- Gauderman, W.; Morrison, J. QUANTO 1.1: A Computer Program for Power and Sample Size Calculations Genetic–Epidemiology Studies. 2006. Available online: http://hydra.usc.edu/gxe (accessed on 18 June 2023).
- Polonikov, A.; Kharchenko, A.; Bykanova, M.; Sirotina, S.; Ponomarenko, I.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Bushueva, O.; Churnosov, M.; et al. Polymorphisms of CYP2C8, CYP2C9 and CYP2C19 and risk of coronary heart disease in Russian population. Gene 2017, 627, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Sirotina, S.; Ponomarenko, I.; Kharchenko, A.; Bykanova, M.; Bocharova, A.; Vagaytseva, K.; Stepanov, V.; Churnosov, M.; Solodilova, M.; Polonikov, A. A Novel Polymorphism in the Promoter of the CYP4A11 Gene Is Associated with Susceptibility to Coronary Artery Disease. Dis. Markers 2018, 2018, 5812802. [Google Scholar] [CrossRef] [PubMed]
- Novakov, V.; Novakova, O.; Churnosova, M.; Aristova, I.; Ponomarenko, M.; Reshetnikova, Y.; Churnosov, V.; Sorokina, I.; Ponomarenko, I.; Efremova, O.; et al. Polymorphism rs143384 GDF5 reduces the risk of knee osteoarthritis development in obese individuals and increases the disease risk in non-obese population. Arthroplasty 2024, 6, 12. [Google Scholar] [CrossRef]
- Polonikov, A.; Rymarova, L.; Klyosova, E.; Volkova, A.; Azarova, I.; Bushueva, O.; Bykanova, M.; Bocharova, I.; Zhabin, S.; Churnosov, M.; et al. Matrix metalloproteinases as target genes for gene regulatory networks driving molecular and cellular pathways related to a multistep pathogenesis of cerebrovascular disease. J. Cell. Biochem. 2019, 120, 16467–16482. [Google Scholar] [CrossRef]
- Minyaylo, O.; Ponomarenko, I.; Reshetnikov, E.; Dvornyk, V.; Churnosov, M. Functionally significant polymorphisms of the MMP-9 gene are associated with peptic ulcer disease in the Caucasian population of Central Russia. Sci. Rep. 2021, 11, 13515. [Google Scholar] [CrossRef]
- Zheng, Z.; Huang, D.; Wang, J.; Zhao, K.; Zhou, Y.; Guo, Z.; Zhai, S.; Xu, H.; Cui, H.; Yao, H.; et al. QTLbase: An Integrative Resource for Quantitative Trait Loci across Multiple Human 846 Molecular Phenotypes. Nucleic Acids Res. 2020, 48, D983–D991. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; He, Y.; Liu, H.; Zhai, H.; Huang, D.; Yi, X.; Dong, X.; Wang, Z.; Zhao, K.; Zhou, Y.; et al. regBase: Whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants. Nucleic Acids Res. 2019, 47, e134. [Google Scholar] [CrossRef] [PubMed]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 36, 1318–1330. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.L. Diverse roles for sex hormone-binding globulin in reproduction. Biol. Reprod. 2011, 85, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Fortunati, N.; Catalano, M.G.; Boccuzzi, G.; Frairia, R. Sex Hormone-Binding Globulin (SHBG), estradiol and breast cancer. Mol. Cell. Endocrinol. 2010, 316, 86–92. [Google Scholar] [CrossRef]
- He, X.Y.; Liao, Y.D.; Yu, S.; Zhang, Y.; Wang, R. Sex hormone binding globulin and risk of breast cancer in postmenopausal women: A meta-analysis of prospective studies. Horm. Metab. Res. 2015, 47, 485–490. [Google Scholar] [CrossRef]
- Arthur, R.S.; Xue, X.; Rohan, T.E. Prediagnostic Circulating Levels of Sex Steroid Hormones and SHBG in Relation to Risk of Ductal Carcinoma In Situ of the Breast among UK Women. Cancer Epidemiol. Biomark. Prev. 2020, 29, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.N.; Zuber, V.; Tsilidis, K.K. Identifying and ranking causal biochemical biomarkers for breast cancer: A Mendelian randomisation study. BMC Med. 2022, 20, 457. [Google Scholar] [CrossRef] [PubMed]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GTF2A1L (accessed on 25 August 2023).
- Wang, J.; Zhao, S.; He, W.; Wei, Y.; Zhang, Y.; Pegg, H.; Shore, P.; Roberts, S.G.E.; Deng, W. A transcription factor IIA-binding site differentially regulates RNA polymerase II-mediated transcription in a promoter context-dependent manner. J. Biol. Chem. 2017, 292, 11873–11885. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Yang, X.; Gao, L.; Xu, Y.; Yi, C. Differences in potential key genes and pathways between primary and radiation-associated angiosarcoma of the breast. Transl. Oncol. 2022, 19, 101385. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.C.; Freitas, M.; Coutinho, L.; Falcon, T.; Matte, U. Machine Learning Supports Long Noncoding RNAs as Expression Markers for Endometrial Carcinoma. Biomed Res. Int. 2020, 2020, 3968279. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, C.; Wu, Y.; He, W.; Gou, X. Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in bladder urothelial carcinoma. Cancer Cell Int. 2019, 19, 327. [Google Scholar] [CrossRef]
- Park, A.K.; Lee, J.Y.; Cheong, H.; Ramaswamy, V.; Park, S.H.; Kool, M.; Phi, J.H.; Choi, S.A.; Cavalli, F.; Taylor, M.D.; et al. Subgroup-specific prognostic signaling and metabolic pathways in pediatric medulloblastoma. BMC Cancer 2019, 19, 571. [Google Scholar] [CrossRef]
- Guo, Y.; Warren Andersen, S.; Shu, X.O.; Michailidou, K.; Bolla, M.K.; Wang, Q.; Garcia-Closas, M.; Milne, R.L.; Schmidt, M.K.; Chang-Claude, J.; et al. Genetically Predicted Body Mass Index and Breast Cancer Risk: Mendelian Randomization Analyses of Data from 145,000 Women of European Descent. PLoS Med. 2016, 13, e1002105. [Google Scholar] [CrossRef] [PubMed]
- Ooi, B.N.S.; Loh, H.; Ho, P.J.; Milne, R.L.; Giles, G.; Gao, C.; Kraft, P.; John, E.M.; Swerdlow, A.; Brenner, H.; et al. The genetic interplay between body mass index, breast size and breast cancer risk: A Mendelian randomization analysis. Int. J. Epidemiol. 2019, 48, 781–794. [Google Scholar] [CrossRef]
- Glassman, I.; Le, N.; Asif, A.; Goulding, A.; Alcantara, C.A.; Vu, A.; Chorbajian, A.; Mirhosseini, M.; Singh, M.; Venketaraman, V. The Role of Obesity in Breast Cancer Pathogenesis. Cells 2023, 12, 2061. [Google Scholar] [CrossRef]
- Campbell, K.L.; Foster-Schubert, K.E.; Alfano, C.M.; Wang, C.C.; Wang, C.Y.; Duggan, C.R.; Mason, C.; Imayama, I.; Kong, A.; Xiao, L.; et al. Reduced-calorie dietary weight loss, exercise, and sex hormones in postmenopausal women: Randomized controlled trial. J. Clin. Oncol. 2012, 30, 2314–2326. [Google Scholar] [CrossRef] [PubMed]
- van Gemert, W.A.; Schuit, A.J.; van der Palen, J.; May, A.M.; Iestra, J.A.; Wittink, H.; Peeters, P.H.; Monninkhof, E.M. Effect of weight loss, with or without exercise, on body composition and sex hormones in postmenopausal women: The SHAPE-2 trial. Breast Cancer Res. 2015, 17, 120. [Google Scholar] [CrossRef]
- de Roon, M.; May, A.M.; McTiernan, A.; Scholten, R.J.P.M.; Peeters, P.H.M.; Friedenreich, C.M.; Monninkhof, E.M. Effect of exercise and/or reduced calorie dietary interventions on breast cancer-related endogenous sex hormones in healthy postmenopausal women. Breast Cancer Res. 2018, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.W.; Hang, D.; Kværner, A.S.; Giovannucci, E.; Song, M. Associations between body shape across the life course and adulthood concentrations of sex hormones in men and pre- and postmenopausal women: A multicohort study. Br. J. Nutr. 2022, 127, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Zeleniuch-Jacquotte, A.; Shore, R.E.; Koenig, K.L.; Akhmedkhanov, A.; Afanasyeva, Y.; Kato, I.; Kim, M.Y.; Rinaldi, S.; Kaaks, R.; Toniolo, P. Postmenopausal levels of oestrogen, androgen, and SHBG and breast cancer: Long-term results of a prospective study. Br. J. Cancer 2004, 90, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.E.; Swain, C.T.V.; Brown, K.A.; Dixon-Suen, S.C.; Boing, L.; van Roekel, E.H.; Moore, M.M.; Gaunt, T.R.; Milne, R.L.; English, D.R.; et al. Linking Physical Activity to Breast Cancer via Sex Steroid Hormones, Part 2: The Effect of Sex Steroid Hormones on Breast Cancer Risk. Cancer Epidemiol. Biomark. Prev. 2022, 31, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Caldon, C.E. Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front. Oncol. 2014, 4, 106. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.F.; Nisula, B.C.; Rodbard, D. Transport of steroid hormones: Binding of 21 endogenous steroids to both testosterone-binding globulin and corticosteroid-binding globulin in human plasma. J. Clin. Endocrinol. Metab. 1981, 53, 58–68. [Google Scholar] [CrossRef] [PubMed]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=STON1 (accessed on 25 August 2023).
- Feutlinske, F.; Browarski, M.; Ku, M.C.; Trnka, P.; Waiczies, S.; Niendorf, T.; Stallcup, W.B.; Glass, R.; Krause, E.; Maritzen, T. Stonin1 mediates endocytosis of the proteoglycan NG2 and regulates focal adhesion dynamics and cell motility. Nat. Commun. 2015, 6, 8535. [Google Scholar] [CrossRef]
- Zheng, A.; Bai, J.; Ha, Y.; Yu, Y.; Fan, Y.; Liang, M.; Lu, Y.; Shen, Z.; Luo, B.; Jie, W. Integrated analysis of the relation to tumor immune microenvironment and predicted value of Stonin1 gene for immune checkpoint blockage and targeted treatment in kidney renal clear cell carcinoma. BMC Cancer 2023, 23, 135. [Google Scholar] [CrossRef]
- Shanle, E.K.; Zhao, Z.; Hawse, J.; Wisinski, K.; Keles, S.; Yuan, M.; Xu, W. Research resource: Global identification of estrogen receptor β target genes in triple negative breast cancer cells. Mol. Endocrinol. 2013, 27, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Fan, J.; Wang, S.; Wang, Z.; Wang, C.; Zuo, Z.; Chow, M.S.; Shi, L.; Wen, Z.; Huang, Y. Transcriptional profiling of Chinese medicinal formula Si-Wu-Tang on breast cancer cells reveals phytoestrogenic activity. BMC Complement. Altern. Med. 2013, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Horvath, A.; Pakala, S.B.; Mudvari, P.; Reddy, S.D.; Ohshiro, K.; Casimiro, S.; Pires, R.; Fuqua, S.A.; Toi, M.; Costa, L.; et al. Novel insights into breast cancer genetic variance through RNA sequencing. Sci. Rep. 2013, 3, 2256. [Google Scholar] [CrossRef] [PubMed]
- Idichi, T.; Seki, N.; Kurahara, H.; Fukuhisa, H.; Toda, H.; Shimonosono, M.; Okato, A.; Arai, T.; Kita, Y.; Mataki, Y.; et al. Molecular pathogenesis of pancreatic ductal adenocarcinoma: Impact of passenger strand of pre-miR-148a on gene regulation. Cancer Sci. 2018, 109, 2013–2026. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Luo, W.; Zhou, P.; Cheng, Y.; Qian, L. Bioinformatics Analysis and Functional Verification of ADAMTS9-AS1/AS2 in Lung Adenocarcinoma. Front. Oncol. 2021, 11, 681777. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Z.; Zhang, Y.; Zhao, C.; Wang, H.; Lin, J.; Liu, C.; Wang, X.; Wang, H. Genomic Variations and Immune-Related Features of TMB, PD-L1 Expression and CD8+ T Cell Infiltration in Chinese Pulmonary Sarcomatoid Carcinoma. Int. J. Gen. Med. 2022, 15, 4209–4220. [Google Scholar] [CrossRef]
- Nazarian, A.; Arbeev, K.G.; Yashkin, A.P.; Kulminski, A.M. Genome-wide analysis of genetic predisposition to common polygenic cancers. J. Appl. Genet. 2022, 63, 315–325. [Google Scholar] [CrossRef]
- Slattery, M.L.; Pellatt, A.J.; Lee, F.Y.; Herrick, J.S.; Samowitz, W.S.; Stevens, J.R.; Wolff, R.K.; Mullany, L.E. Infrequently expressed miRNAs influence survival after diagnosis with colorectal cancer. Oncotarget 2017, 8, 83845–83859. [Google Scholar] [CrossRef]
- Zheng, H.; Liu, H.; Li, H.; Dou, W.; Wang, J.; Zhang, J.; Liu, T.; Wu, Y.; Liu, Y.; Wang, X. Characterization of stem cell landscape and identification of stemness-relevant prognostic gene signature to aid immunotherapy in colorectal cancer. Stem Cell Res. Ther. 2022, 13, 244. [Google Scholar] [CrossRef]
- Cao, Y.; Deng, S.; Yan, L.; Gu, J.; Mao, F.; Xue, Y.; Qin, L.; Jiang, Z.; Cai, W.; Zheng, C.; et al. The Prognostic Significance of RIMKLB and Related Immune Infiltrates in Colorectal Cancers. Front. Genet. 2022, 13, 818994. [Google Scholar] [CrossRef]
- Wu, P.; Xiang, T.; Wang, J.; Lv, R.; Ma, S.; Yuan, L.; Wu, G.; Che, X. Identification of immunization-related new prognostic biomarkers for papillary renal cell carcinoma by integrated bioinformatics analysis. BMC Med. Genom. 2021, 14, 241. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, D.; Zhang, C.; Zhang, Z.; Chen, X.; Lian, J.; Liu, J.; Wang, G.; Yuan, W.; Sun, Z.; et al. Identification of liver metastasis-associated genes in human colon carcinoma by mRNA profiling. Chin. J. Cancer Res. 2018, 30, 633–646. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Lin, K.; Huang, Y.; Lu, Y.; Chen, W.Q.; Zhang, X.R.; He, B.S.; Pan, Y.Q.; Wang, S.K.; Fan, W.X. Identification of critically carcinogenesis-related genes in basal cell carcinoma. OncoTargets Ther. 2018, 11, 6957–6967. [Google Scholar] [CrossRef]
- Li, Y.; Zu, X.; Hu, X.; Zhao, C.; Mo, M.; Fan, B. Competing endogenous RNA network analysis reveals pivotal ceRNAs in bladder urothelial carcinoma. Transl. Androl. Urol. 2021, 10, 797–808. [Google Scholar] [CrossRef]
- Zhang, Y.; Hua, S.; Jiang, Q.; Xie, Z.; Wu, L.; Wang, X.; Shi, F.; Dong, S.; Jiang, J. Identification of Feature Genes of a Novel Neural Network Model for Bladder Cancer. Front. Genet. 2022, 13, 912171. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Song, Z.; Wang, Z.; Chen, G. Protective Prognostic Biomarkers Negatively Correlated with Macrophage M2 Infiltration in Low-Grade Glioma. J. Oncol. 2022, 2022, 3623591. [Google Scholar] [CrossRef] [PubMed]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=STON1-GTF2A1L (accessed on 25 August 2023).
- Zhang, X.; Wei, X.; Bai, G.; Huang, X.; Hu, S.; Mao, H.; Liu, P. Identification of Three Potential Prognostic Genes in Platinum-Resistant Ovarian Cancer via Integrated Bioinformatics Analysis. Cancer Manag. Res. 2021, 13, 8629–8646. [Google Scholar] [CrossRef]
- Yang, H.; Qu, X.; Huang, J.; Zhang, F.; Fang, Z.; Zhao, B.; Wang, Y. E2F4 may be a core transcription factor in the lncRNA-TF regulatory network in cervical cancer. J. Clin. Lab. Anal. 2022, 36, e24322. [Google Scholar] [CrossRef]
- Ebert, K.; Haffner, I.; Zwingenberger, G.; Keller, S.; Raimúndez, E.; Geffers, R.; Wirtz, R.; Barbaria, E.; Hollerieth, V.; Arnold, R.; et al. Combining gene expression analysis of gastric cancer cell lines and tumor specimens to identify biomarkers for anti-HER therapies—The role of HAS2, SHB and HBEGF. BMC Cancer 2022, 22, 254. [Google Scholar] [CrossRef]
- He, Y.; Zhang, H.; Zhang, Y.; Wang, P.; Zhu, K.; Ba, Y. Comprehensive Characterization of Transforming Growth Factor Beta Receptor 1 in Stomach Adenocarcinoma Identifies a Prognostic Signature for Predicting Clinical Outcomes and Immune Infiltrates. Int. J. Gen. Med. 2022, 15, 3375–3391. [Google Scholar] [CrossRef]
- Tini, G.; Varma, V.; Lombardo, R.; Nolen, G.T.; Lefebvre, G.; Descombes, P.; Métairon, S.; Priami, C.; Kaput, J.; Scott-Boyer, M.P. DNA methylation during human adipogenesis and the impact of fructose. Genes Nutr. 2020, 15, 21. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.H.; Wei, Y.; Liu, R.; Lin, X.R.; Luo, J.Q.; Zhang, Q.J.; Lin, S.R.; Geng, L.; Ye, S.K.; Shi, Y.; et al. Three-Dimensional Genome Interactions Identify Potential Adipocyte Metabolism-Associated Gene STON1 and Immune-Correlated Gene FSHR at the rs13405728 Locus in Polycystic Ovary Syndrome. Front. Endocrinol. 2021, 12, 686054. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, G.; Vastrad, B.; Tengli, A.; Vastrad, C.; Kotturshetti, I. Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules. BMC Endocr. Disord. 2021, 21, 80. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Zhu, H.; Zhang, J.; Yang, X.; Zhao, L. Genome-wide screening for circRNAs in epicardial adipose tissue of heart failure patients with preserved ejection fraction. Am. J. Transl. Res. 2023, 15, 4610–4619. [Google Scholar] [PubMed]
- Yengo, L.; Vedantam, S.; Marouli, E.; Sidorenko, J.; Bartell, E.; Sakaue, S.; Graff, M.; Eliasen, A.U.; Jiang, Y.; Raghavan, S.; et al. A saturated map of common genetic variants associated with human height. Nature 2022, 610, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Tachmazidou, I.; Süveges, D.; Min, J.L.; Ritchie, G.R.S.; Steinberg, J.; Walter, K.; Iotchkova, V.; Schwartzentruber, J.; Huang, J.; Memari, Y.; et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am. J. Hum. Genet. 2017, 100, 865–884. [Google Scholar] [CrossRef] [PubMed]
- Lotta, L.A.; Wittemans, L.B.L.; Zuber, V.; Stewart, I.D.; Sharp, S.J.; Luan, J.; Day, F.R.; Li, C.; Bowker, N.; Cai, L.; et al. Association of Genetic Variants Related to Gluteofemoral vs Abdominal Fat Distribution with Type 2 Diabetes, Coronary Disease, and Cardiovascular Risk Factors. JAMA 2018, 320, 2553–2563. [Google Scholar] [CrossRef]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef]
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef]
- Christakoudi, S.; Evangelou, E.; Riboli, E.; Tsilidis, K.K. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci. Rep. 2021, 11, 10688. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Guo, Y.; Shi, H.; Liu, C.L.; Panganiban, R.A.; Chung, W.; O’Connor, L.J.; Himes, B.E.; Gazal, S.; Hasegawa, K.; et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 2022, 145, 537–549, Erratum in J. Allergy Clin. Immunol. 2022, 149, 1486–1489. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Wang, M.; Klarqvist, M.D.R.; Smith, K.; Shin, J.; Dashti, H.; Diamant, N.; Choi, S.H.; Jurgens, S.J.; Ellinor, P.T.; et al. Inherited basis of visceral, abdominal subcutaneous and gluteofemoral fat depots. Nat. Commun. 2022, 13, 3771. [Google Scholar] [CrossRef] [PubMed]
- Haiman, C.A.; Han, Y.; Feng, Y.; Xia, L.; Hsu, C.; Sheng, X.; Pooler, L.C.; Patel, Y.; Kolonel, L.N.; Carter, E.; et al. Genome-wide testing of putative functional exonic variants in relationship with breast and prostate cancer risk in a multiethnic population. PLoS Genet. 2013, 9, e1003419. [Google Scholar] [CrossRef]
- Akiyama, M.; Okada, Y.; Kanai, M.; Takahashi, A.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017, 49, 1458–1467. [Google Scholar] [CrossRef] [PubMed]
- Koskeridis, F.; Evangelou, E.; Said, S.; Boyle, J.J.; Elliott, P.; Dehghan, A.; Tzoulaki, I. Pleiotropic genetic architecture and novel loci for C-reactive protein levels. Nat. Commun. 2022, 13, 6939. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Huffman, J.E.; Huang, Y.; Do Valle, Í.; Assimes, T.L.; Raghavan, S.; Voight, B.F.; Liu, C.; Barabási, A.L.; Huang, R.D.L.; et al. Genomics and phenomics of body mass index reveals a complex disease network. Nat. Commun. 2022, 13, 7973. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.G.; Sanderson, E.; Elsworth, B.; Tilling, K.; Davey Smith, G. Use of genetic variation to separate the effects of early and later life adiposity on disease risk: Mendelian randomisation study. BMJ 2020, 369, m1203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wei, X.T.; Niu, J.J.; Lin, Z.X.; Xu, Q.; Ni, J.J.; Zhang, W.L.; Han, B.X.; Yan, S.S.; Feng, G.J.; et al. Joint Genome-Wide Association Analyses Identified 49 Novel Loci For Age at Natural Menopause. J. Clin. Endocrinol. Metab. 2021, 106, 2574–2591. [Google Scholar] [CrossRef]
- Ripatti, P.; Rämö, J.T.; Mars, N.J.; Fu, Y.; Lin, J.; Söderlund, S.; Benner, C.; Surakka, I.; Kiiskinen, T.; Havulinna, A.S.; et al. Polygenic Hyperlipidemias and Coronary Artery Disease Risk. Circ. Genom. Precis. Med. 2020, 13, e002725. [Google Scholar] [CrossRef]
- Richardson, T.G.; Sanderson, E.; Palmer, T.M.; Ala-Korpela, M.; Ference, B.A.; Davey Smith, G.; Holmes, M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable Mendelian randomisation analysis. PLoS Med. 2020, 17, e1003062. [Google Scholar] [CrossRef] [PubMed]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194, Erratum in Nat Genet. 2021, 53, 1622. [Google Scholar] [CrossRef]
- Selvaraj, M.S.; Paruchuri, K.; Haidermota, S.; Bernardo, R.; Rich, S.S.; Peloso, G.M.; Natarajan, P. Genome-wide discovery for diabetes-dependent triglycerides-associated loci. PLoS ONE 2022, 17, e0275934. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.E.; Clarke, S.L.; Wu, K.H.; Kanoni, S.; Zajac, G.J.M.; Ramdas, S.; Surakka, I.; Ntalla, I.; Vedantam, S.; Winkler, T.W.; et al. The power of genetic diversity in genome-wide association studies of lipids. Nature 2023, 600, 675–679, Erratum in Nature 2023, 618, E19–E20. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.; Zhao, H.; He, L.; Shi, Y.; Qin, Y.; Shi, Y.; Li, Z.; You, L.; Zhao, J.; Liu, J.; et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat. Genet. 2011, 43, 55–59. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, H.; Shi, Y.; Cao, Y.; Yang, D.; Li, Z.; Zhang, B.; Liang, X.; Li, T.; Chen, J.; et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat. Genet. 2012, 44, 1020–1025. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.R.; Brower, M.A.; Xu, N.; Cui, J.; Mengesha, E.; Chen, Y.D.; Taylor, K.D.; Azziz, R.; Goodarzi, M.O. Systems Genetics Reveals the Functional Context of PCOS Loci and Identifies Genetic and Molecular Mechanisms of Disease Heterogeneity. PLoS Genet. 2015, 11, e1005455. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Shu, X.O.; Cai, Q.; Jin, F.; Cheng, J.R.; Cai, H.; Gao, Y.T.; Zheng, W. Association of breast cancer risk with a common functional polymorphism (Asp327Asn) in the sex hormone-binding globulin gene. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Beeghly-Fadiel, A.; Lu, W.; Cai, Q.; Xiang, Y.B.; Zheng, Y.; Long, J.; Ye, C.; Gu, K.; Shu, X.O.; et al. Evaluation of functional genetic variants for breast cancer risk: Results from the Shanghai breast cancer study. Am. J. Epidemiol. 2011, 173, 1159–1170. [Google Scholar] [CrossRef]
- Collaborative Group on Hormonal Factors in Breast Cancer. Type and timing of menopausal hormone therapy and breast cancer risk: Individual participant meta-analysis of the worldwide epidemiological evidence. Lancet 2019, 394, 1159–1168. [Google Scholar] [CrossRef]
- McTiernan, A.; Friedenreich, C.M.; Katzmarzyk, P.T.; Powell, K.E.; Macko, R.; Buchner, D.; Pescatello, L.S.; Bloodgood, B.; Tennant, B.; Vaux-Bjerke, A.; et al. 2018 Physical Activity Guidelines Advisory Committee*. Physical Activity in Cancer Prevention and Survival: A Systematic Review. Med. Sci. Sports Exerc. 2019, 51, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Łukasiewicz, S.; Czeczelewski, M.; Forma, A.; Baj, J.; Sitarz, R.; Stanisławek, A. Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review. Cancers 2021, 13, 4287. [Google Scholar] [CrossRef] [PubMed]
- Fiolet, T.; Srour, B.; Sellem, L.; Kesse-Guyot, E.; Allès, B.; Méjean, C.; Deschasaux, M.; Fassier, P.; Latino-Martel, P.; Beslay, M.; et al. Consumption of ultra-processed foods and cancer risk: Results from NutriNet-Santé prospective cohort. BMJ 2018, 360, k322. [Google Scholar] [CrossRef] [PubMed]
- Castelló, A.; Pollán, M.; Buijsse, B.; Ruiz, A.; Casas, A.; Baena-Cañada, J.M.; Lope, V.; Antolín, S.; Ramos, M.; Muñoz, M.; et al. GEICAM researchers. Spanish Mediterranean diet and other dietary patterns and breast cancer risk: Case-control EpiGEICAM study. Br. J. Cancer 2014, 111, 1454–1462. [Google Scholar] [CrossRef]
| Parameters | BMI ≥ 30 | BMI < 30 | ||||
|---|---|---|---|---|---|---|
| BC Patients ± SD/%(n) | Controls ± SD/%(n) | p | BC Patients ± SD/%(n) | Controls ± SD/%(n) | p | |
| N | 119 | 253 | - | 239 | 887 | - |
| Age, years (min–max) | 58.97 ± 10.67 (33–84) | 58.01 ± 10.01 (31–80) | 0.22 | 53.58 ± 13.12 (28–82) | 52.77 ± 12.27 (29–80) | 0.14 |
| <50 years | 26.89 (32) | 27.67 (70) | 0.98 | 37.24 (89) | 40.70 (361) | 0.37 |
| ≥50 years | 73.11 (87) | 72.73 (183) | 62.76 (150) | 59.30 (526) | ||
| BMI, kg/m2 | 34.95 ± 4.76 | 33.12 ± 4.04 | 0.001 | 27.55 ± 2.85 | 26.13 ± 2.59 | 0.0004 |
| Age at menarche, years | 12.11 ± 1.02 | 12.30 ± 1.04 | 0.71 | 12.57 ± 1.05 | 12.78 ± 1.08 | 0.46 |
| Age at menopause, years | 48.58 ± 4.13 | 48.21 ± 4.01 | 0.65 | 48.08 ± 4.07 | 47.79 ± 4.02 | 0.36 |
| Menstruation status | ||||||
| Premenopausal | 24.37 (29) | 27.27 (69) | 0.64 | 35.56 (85) | 39.01 (346) | 0.37 |
| Postmenopausal | 75.63 (90) | 72.73 (184) | 64.44 (154) | 60.99 (541) | ||
| Smoker (yes) | 20.17 (24) | 16.60 (42) | 0.49 | 23.01 (55) | 19.73 (175) | 0.31 |
| Biochemical parameters | ||||||
| Fasting blood glucose (mmol/L) | 8.76 ± 0.89 | 8.08 ± 0.75 | <0.001 | 6.17 ± 0.75 | 5.19 ± 0.69 | <0.001 |
| TC (mmol/L) | 6.34 ± 1.10 | 5.87 ± 1.02 | <0.001 | 5.26 ± 1.01 | 4.76 ± 0.91 | <0.001 |
| HDL-C (mmol/L) | 1.13 ± 0.45 | 1.26 ± 0.36 | <0.001 | 1.40 ± 0.40 | 1.49 ± 0.42 | <0.001 |
| LDL-C (mmol/L) | 4.31 ± 0.95 | 4.00 ± 0.86 | <0.001 | 3.39 ± 0.79 | 3.07 ± 0.72 | <0.001 |
| TG (mmol/L) | 1.98 ± 1.03 | 1.72 ± 1.00 | <0.001 | 1.38 ± 0.64 | 1.21 ± 0.52 | <0.001 |
| Clinicopathological parameters of BC patients | ||||||
| Stage of the cancer | T0–T2—79%, T3–T4—21% | T0–T2—72%, T3–T4—28% | ||||
| Lymph node involvement (N) | negative—50%, positive—50% | negative—46%, positive—54% | ||||
| Estrogen receptor (ER) | negative—31%, positive—69% | negative—36%, positive—64% | ||||
| Progesterone receptor (PR) | negative—38%, positive—62% | negative—43%, positive—57% | ||||
| Human epidermal growth factor receptor 2 (HER2) | negative—60%, positive—40% | negative—66%, positive—34% | ||||
| Tumor histological type | ductal—95%, lobular—5% | ductal—94%, lobular—6% | ||||
| Tumor histological grade (G) | G1/G2—70%, G3—30% | G1/G2—67%, G3—33% | ||||
| Progression | absent—68%, present—32% | absent—65%, present—35% | ||||
| Metastasis | absent—80%, present—20% | absent—77%, present—23% | ||||
| Death | absent—76%, present—24% | absent—83%, present—17% | ||||
| Chr | SNP | Gene | Minor Allele | n | Allelic Model | Additive Model | Dominant Model | Recessive Model | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | OR | 95%CI | p | |||||||||
| L95 | U95 | L95 | U95 | L95 | U95 | L95 | U95 | |||||||||||||
| Females with BMI < 30 | ||||||||||||||||||||
| 1 | rs17496332 | PRMT6 | G | 1065 | 0.97 | 0.79 | 1.21 | 0.809 | 0.97 | 0.75 | 1.27 | 0.847 | 0.87 | 0.60 | 1.25 | 0.447 | 1.19 | 0.72 | 1.98 | 0.497 |
| 2 | rs780093 | GCKR | T | 1087 | 1.00 | 0.81 | 1.23 | 0.987 | 0.87 | 0.67 | 1.14 | 0.314 | 0.85 | 0.58 | 1.25 | 0.407 | 0.81 | 0.49 | 1.34 | 0.415 |
| 2 | rs10454142 | PPP1R21 | C | 1072 | 1.02 | 0.82 | 1.28 | 0.837 | 1.15 | 0.87 | 1.53 | 0.331 | 1.13 | 0.78 | 1.65 | 0.508 | 1.38 | 0.75 | 2.53 | 0.301 |
| 7 | rs3779195 | BAIAP2L1 | A | 1073 | 1.05 | 0.81 | 1.36 | 0.726 | 1.12 | 0.80 | 1.56 | 0.522 | 1.18 | 0.80 | 1.75 | 0.405 | 0.89 | 0.32 | 2.51 | 0.827 |
| 8 | rs440837 | ZBTB10 | G | 1053 | 1.00 | 0.79 | 1.27 | 0.988 | 0.98 | 0.73 | 1.32 | 0.911 | 0.82 | 0.57 | 1.02 | 0.312 | 1.71 | 0.90 | 3.27 | 0.102 |
| 10 | rs7910927 | JMJD1C | T | 1087 | 0.89 | 0.73 | 1.10 | 0.276 | 0.90 | 0.70 | 1.17 | 0.446 | 0.84 | 0.56 | 1.27 | 0.415 | 0.91 | 0.59 | 1.40 | 0.668 |
| 12 | rs4149056 | SLCO1B1 | C | 1041 | 0.88 | 0.69 | 1.14 | 0.336 | 1.03 | 0.75 | 1.42 | 0.838 | 1.08 | 0.73 | 1.58 | 0.709 | 0.88 | 0.36 | 2.14 | 0.779 |
| 15 | rs8023580 | NR2F2 | C | 1085 | 0.87 | 0.69 | 1.10 | 0.246 | 0.96 | 0.72 | 1.27 | 0.760 | 1.07 | 0.74 | 1.55 | 0.707 | 0.60 | 0.29 | 1.24 | 0.169 |
| 17 | rs12150660 | SHBG | T | 1090 | 0.96 | 0.76 | 1.21 | 0.712 | 0.83 | 0.62 | 1.12 | 0.231 | 0.83 | 0.57 | 1.20 | 0.326 | 0.67 | 0.31 | 1.44 | 0.307 |
| Females with BMI ≥ 30 | ||||||||||||||||||||
| 1 | rs17496332 | PRMT6 | G | 357 | 0.86 | 0.62 | 1.19 | 0.362 | 1.02 | 0.69 | 1.50 | 0.919 | 0.93 | 0.55 | 1.58 | 0.787 | 1.27 | 0.59 | 2.76 | 0.542 |
| 2 | rs780093 | GCKR | T | 358 | 0.86 | 0.63 | 1.19 | 0.371 | 0.91 | 0.62 | 1.34 | 0.635 | 1.03 | 0.60 | 1.77 | 0.920 | 0.65 | 0.29 | 1.44 | 0.289 |
| 2 | rs10454142 | PPP1R21 | C | 352 | 1.52 | 1.10 | 2.11 | 0.012 | 1.73 | 1.15 | 2.62 | 0.009 | 1.95 | 1.13 | 3.37 | 0.016 | 2.06 | 0.87 | 4.89 | 0.099 |
| 7 | rs3779195 | BAIAP2L1 | A | 348 | 1.15 | 0.75 | 1.76 | 0.514 | 0.98 | 0.60 | 1.61 | 0.949 | 1.09 | 0.61 | 1.94 | 0.768 | 0.43 | 0.08 | 2.45 | 0.345 |
| 8 | rs440837 | ZBTB10 | G | 355 | 0.79 | 0.54 | 1.17 | 0.235 | 0.76 | 0.48 | 1.19 | 0.230 | 0.72 | 0.42 | 1.23 | 0.230 | 0.70 | 0.19 | 2.57 | 0.594 |
| 10 | rs7910927 | JMJD1C | T | 359 | 1.04 | 0.76 | 1.42 | 0.801 | 1.03 | 0.70 | 1.51 | 0.888 | 0.98 | 0.54 | 1.78 | 0.949 | 1.10 | 0.58 | 2.09 | 0.761 |
| 12 | rs4149056 | SLCO1B1 | C | 344 | 0.97 | 0.66 | 1.42 | 0.877 | 0.95 | 0.59 | 1.53 | 0.831 | 0.88 | 0.51 | 1.52 | 0.646 | 1.54 | 0.36 | 6.54 | 0.557 |
| 15 | rs8023580 | NR2F2 | C | 355 | 0.98 | 0.69 | 1.40 | 0.930 | 0.82 | 0.53 | 1.27 | 0.375 | 0.82 | 0.49 | 1.39 | 0.470 | 0.63 | 0.19 | 2.11 | 0.453 |
| 17 | rs12150660 | SHBG | T | 362 | 1.11 | 0.77 | 1.58 | 0.583 | 1.16 | 0.77 | 1.76 | 0.480 | 1.10 | 0.65 | 1.85 | 0.728 | 1.72 | 0.63 | 4.69 | 0.288 |
| SNP (Position hg38) (r2, LD) | Haploreg and GTE-Portal Data | GTE-Portal Data | Haploreg Data | |||||
|---|---|---|---|---|---|---|---|---|
| Liver | Adipocyte-Cultured Cells | Visceral Adipose | Subcutaneous Adipose | Transcription Factors | ||||
| Mesenchymal Stem-Cell-Derived Adipocyte-Cultured Cells | Adipose Nuclei | eQTL | sQTL | eQTL | sQTL | |||
| rs17855177 (48375113) (r2 = 0.81, LD = 0.99) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L, PPP1R21 | GTF2A1L, STON1, PPP1R21 | |||
| rs78597273 (48380665) (r2 = 0.81, LD = 0.99) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L, PPP1R21 | GTF2A1L, STON1, PPP1R21 | MIZF | ||
| rs11689645 (48381420) (r2 = 0.81, LD = 0.99) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L | GTF2A1L, STON1, PPP1R21 | AP-1, AP-2, BAF155, BATF, GR, Myc, BCL, Bach1, Bach2, GATA, HMGN3, KAP1, Maf, NF-E2, STAT, PRDM1, TCF4, p300 | ||
| rs111960813 (48404376) (r2 = 0.80, LD = 0.93) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L | GTF2A1L, STON1, PPP1R21 | ELF1, Myc, ZBRK1 | ||
| rs56391806 (48404838) (r2 = 0.85, LD = 0.98) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L, PPP1R21 | GTF2A1L, STON1, PPP1R21 | Fox, Hoxb6 | ||
| rs55744465 (48405316) (r2 = 0.85, LD = 0.98) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L | GTF2A1L, STON1, PPP1R21 | Hoxa5 | ||
| rs201414717 (48419259) (r2 = 1.00, LD = 1.00) | * H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | H3K4me1_Enh H3K9ac_Pro | H3K4me1_Enh H3K27ac_Enh H3K9ac_Pro | * | * | * | * | AP-4, CACD, WT1, YY1, TAL1, TCF12, Rad21, LBP-1, ZNF219 |
| rs10454142 (48419260) | GTF2A1L H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | H3K4me1_Enh H3K9ac_Pro | H3K4me1_Enh H3K27ac_Enh H3K9ac_Pro | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1, PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L | GTF2A1L, STON1, PPP1R21 | NF-kappaB |
| rs10454143 (48419261) (r2 = 1.00, LD = 1.00) | GTF2A1L H3K4me1_Enh H3K4me3_Pro H3K27ac_Enh H3K9ac_Pro | H3K4me1_Enh H3K9ac_Pro | H3K4me1_Enh H3K27ac_Enh H3K9ac_Pro | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1,PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L | GTF2A1L, STON1, PPP1R21 | Barx1, CEBPD, Hoxa3 |
| rs13399936 (48426987) (r2 = 0.87, LD = 0.96) | GTF2A1L | GTF2A1L, STON1-GTF2A1L, RP11-460M2.1 | GTF2A1L, STON1,PPP1R21, STON1-GTF2A1L | GTF2A1L, STON1-GTF2A1L | GTF2A1L, STON1, PPP1R21 | |||
| rs4638844 (48427445) (r2 = 0.81, LD = 0.94) | * | * | * | * | * | CIZ, FAC1, Foxa, Foxd3, Foxj2, Foxk1 Foxo, Foxp1, HDAC2, Irf, Pax-4, Sox, RREB-1, Zfp105, p300 | ||
| SNP (Position hg38) (r2, LD) | Score | Phred Score | Potential Role |
|---|---|---|---|
| rs17855177 (48375113) (r2 = 0.81, LD = 0.99) | 0.9983 | 22.4912 | Likely cancer driver |
| rs78597273 (48380665) (r2 = 0.81, LD = 0.99) | 0.8876 | 8.1965 | Likely cancer driver |
| rs11689645 (48381420) (r2 = 0.81, LD = 0.99) | 0.4049 | 4.4773 | - |
| rs111960813 (48404376) (r2 = 0.80, LD = 0.93) | 0.3714 | 4.2968 | - |
| rs56391806 (48404838) (r2 = 0.85, LD = 0.98) | 0.8828 | 8.1117 | Likely cancer driver |
| rs55744465 (48405316) (r2 = 0.85, LD = 0.98) | 0.7368 | 6.4687 | Likely cancer driver |
| rs10454142 (48419260) | 0.7992 | 7.0231 | Likely cancer driver |
| rs10454143 (48419261) (r2 = 1.00, LD = 1.00) | 0.7429 | 6.5174 | Likely cancer driver |
| rs13399936 (48426987) (r2 = 0.87, LD = 0.96) | 0.9669 | 10.8931 | Likely cancer driver |
| rs4638844 (48427445) (r2 = 0.81, LD = 0.94) | 0.9989 | 25.0004 | Likely cancer driver |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ponomarenko, I.; Pasenov, K.; Churnosova, M.; Sorokina, I.; Aristova, I.; Churnosov, V.; Ponomarenko, M.; Reshetnikova, Y.; Reshetnikov, E.; Churnosov, M. Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer. Biomedicines 2024, 12, 818. https://doi.org/10.3390/biomedicines12040818
Ponomarenko I, Pasenov K, Churnosova M, Sorokina I, Aristova I, Churnosov V, Ponomarenko M, Reshetnikova Y, Reshetnikov E, Churnosov M. Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer. Biomedicines. 2024; 12(4):818. https://doi.org/10.3390/biomedicines12040818
Chicago/Turabian StylePonomarenko, Irina, Konstantin Pasenov, Maria Churnosova, Inna Sorokina, Inna Aristova, Vladimir Churnosov, Marina Ponomarenko, Yuliya Reshetnikova, Evgeny Reshetnikov, and Mikhail Churnosov. 2024. "Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer" Biomedicines 12, no. 4: 818. https://doi.org/10.3390/biomedicines12040818
APA StylePonomarenko, I., Pasenov, K., Churnosova, M., Sorokina, I., Aristova, I., Churnosov, V., Ponomarenko, M., Reshetnikova, Y., Reshetnikov, E., & Churnosov, M. (2024). Obesity-Dependent Association of the rs10454142 PPP1R21 with Breast Cancer. Biomedicines, 12(4), 818. https://doi.org/10.3390/biomedicines12040818







