Circulating microRNAs as Non-Invasive Biomarkers in Endometriosis Diagnosis—A Systematic Review
Abstract
1. Introduction
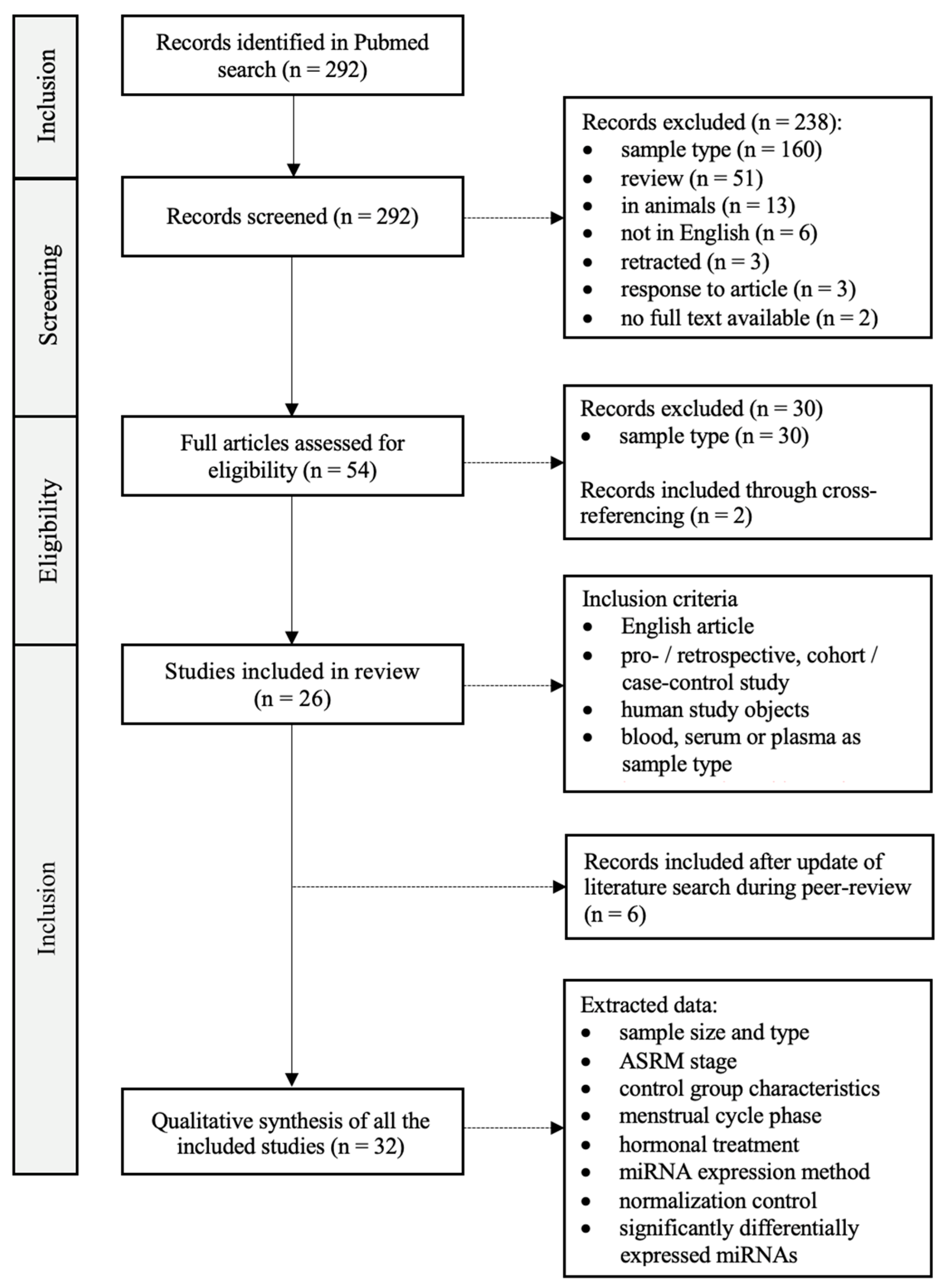
2. Materials and Methods
3. Results
4. Discussion
4.1. Function of the Most Frequently Reported miRNAs
4.2. Lack of Reproducibility
4.2.1. Biological Variability
4.2.2. Pre-Analytical Variability
4.2.3. Analytical Variability
4.3. Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- International Working Group of AAGL, ESGE, ESHRE and WES; Tomassetti, C.; Johnson, N.P.; Petrozza, J.; Abrao, M.S.; Einarsson, J.I.; Horne, A.W.; Lee, T.T.M.; Missmer, S.; Vermeulen, N.; et al. An International Terminology for Endometriosis, 2021. J. Minim. Invasive Gynecol. 2021, 28, 1849–1859. [Google Scholar]
- Foti, P.V.; Farina, R.; Palmucci, S.; Vizzini, I.A.A.; Libertini, N.; Coronella, M.; Spadola, S.; Caltabiano, R.; Iraci, M.; Basile, A.; et al. Endometriosis: Clinical features, MR imaging findings and pathologic correlation. Insights Imaging 2018, 9, 149–172. [Google Scholar] [CrossRef] [PubMed]
- Saunders, P.T.K.; Horne, A.W. Endometriosis: Etiology, pathobiology, and therapeutic prospects. Cell 2021, 184, 2807–2824. [Google Scholar] [CrossRef] [PubMed]
- Sims, O.T.; Gupta, J.; Missmer, S.A.; Aninye, I.O. Stigma and Endometriosis: A Brief Overview and Recommendations to Improve Psychosocial Well-Being and Diagnostic Delay. Int. J. Environ. Res. Public. Health 2021, 18, 8210. [Google Scholar] [CrossRef]
- Koninckx, P.R.; Fernandes, R.; Ussia, A.; Schindler, L.; Wattiez, A.; Al-Suwaidi, S.; Amro, B.; Al-Maamari, B.; Hakim, Z.; Tahlak, M. Pathogenesis Based Diagnosis and Treatment of Endometriosis. Front. Endocrinol. 2021, 12, 745548. [Google Scholar] [CrossRef] [PubMed]
- Kiesel, L.; Sourouni, M. Diagnosis of endometriosis in the 21st century. Climacteric 2019, 22, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.M.; Bokor, A.; Heikinheimo, O.; Horne, A.; Jansen, F.; Kiesel, L.; King, K.; Kvaskoff, M.; Nap, A.; Petersen, K.; et al. ESHRE guideline: Endometriosis. Hum. Reprod. Open 2022, 2022, hoac009. [Google Scholar] [CrossRef]
- Secosan, C.; Balulescu, L.; Brasoveanu, S.; Balint, O.; Pirtea, P.; Dorin, G.; Pirtea, L. Endometriosis in Menopause-Renewed Attention on a Controversial Disease. Diagnostics 2020, 10, 134. [Google Scholar] [CrossRef]
- Della Corte, L.; Di Filippo, C.; Gabrielli, O.; Reppuccia, S.; La Rosa, V.L.; Ragusa, R.; Fichera, M.; Commodari, E.; Bifulco, G.; Giampaolino, P. The Burden of Endometriosis on Women’s Lifespan: A Narrative Overview on Quality of Life and Psychosocial Wellbeing. Int. J. Environ. Res. Public. Health 2020, 17, 4683. [Google Scholar] [CrossRef]
- Gupta, D.; Hull, M.L.; Fraser, I.; Miller, L.; Bossuyt, P.M.; Johnson, N.; Nisenblat, V. Endometrial biomarkers for the non-invasive diagnosis of endometriosis. Cochrane Database Syst. Rev. 2016, 4, CD012165. [Google Scholar] [CrossRef]
- O, D.F.; Flores, I.; Waelkens, E.; D’Hooghe, T. Noninvasive diagnosis of endometriosis: Review of current peripheral blood and endometrial biomarkers. Best. Pract. Res. Clin. Obstet. Gynaecol. 2018, 50, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Rogers, P.A.; Adamson, G.D.; Al-Jefout, M.; Becker, C.M.; D’Hooghe, T.M.; Dunselman, G.A.; Fazleabas, A.; Giudice, L.C.; Horne, A.W.; Hull, M.L.; et al. Research Priorities for Endometriosis. Reprod. Sci. 2017, 24, 202–226. [Google Scholar] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Moreno-Moya, J.M.; Vilella, F.; Simon, C. MicroRNA: Key gene expression regulators. Fertil. Steril. 2014, 101, 1516–1523. [Google Scholar] [CrossRef]
- Hirakawa, T.; Nasu, K.; Abe, W.; Aoyagi, Y.; Okamoto, M.; Kai, K.; Takebayashi, K.; Narahara, H. miR-503, a microRNA epigenetically repressed in endometriosis, induces apoptosis and cell-cycle arrest and inhibits cell proliferation, angiogenesis, and contractility of human ovarian endometriotic stromal cells. Hum. Reprod. 2016, 31, 2587–2597. [Google Scholar] [CrossRef]
- Agrawal, S.; Tapmeier, T.; Rahmioglu, N.; Kirtley, S.; Zondervan, K.; Becker, C. The miRNA Mirage: How Close Are We to Finding a Non-Invasive Diagnostic Biomarker in Endometriosis? A Systematic Review. Int. J. Mol. Sci. 2018, 19, 599. [Google Scholar] [CrossRef] [PubMed]
- Monnaka, V.U.; Hernandes, C.; Heller, D.; Podgaec, S. Overview of miRNAs for the non-invasive diagnosis of endometriosis: Evidence, challenges and strategies. A systematic review. Einstein 2021, 19, eRW5704. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Rev. Esp. Cardiol. 2021, 74, 790–799. [Google Scholar] [CrossRef]
- Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil. Steril. 1997, 67, 817–821. [CrossRef]
- Wang, W.T.; Zhao, Y.N.; Han, B.W.; Hong, S.J.; Chen, Y.Q. Circulating microRNAs identified in a genome-wide serum microRNA expression analysis as noninvasive biomarkers for endometriosis. J. Clin. Endocrinol. Metab. 2013, 98, 281–289. [Google Scholar] [CrossRef]
- Hsu, C.Y.; Hsieh, T.H.; Tsai, C.F.; Tsai, H.P.; Chen, H.S.; Chang, Y.; Chuang, H.Y.; Lee, J.N.; Hsu, Y.L.; Tsai, E.M. miRNA-199a-5p regulates VEGFA in endometrial mesenchymal stem cells and contributes to the pathogenesis of endometriosis. J. Pathol. 2014, 232, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Mutlu, L.; Grechukhina, O.; Taylor, H.S. Circulating microRNAs as potential biomarkers for endometriosis. Fertil. Steril. 2015, 103, 1252–1260.e1. [Google Scholar] [CrossRef] [PubMed]
- Cosar, E.; Mamillapalli, R.; Ersoy, G.S.; Cho, S.; Seifer, B.; Taylor, H.S. Serum microRNAs as diagnostic markers of endometriosis: A comprehensive array-based analysis. Fertil. Steril. 2016, 106, 402–409. [Google Scholar] [CrossRef]
- Wang, L.; Huang, W.; Ren, C.; Zhao, M.; Jiang, X.; Fang, X.; Xia, X. Analysis of Serum microRNA Profile by Solexa Sequencing in Women With Endometriosis. Reprod. Sci. 2016, 23, 1359–1370. [Google Scholar] [CrossRef]
- Nothnick, W.B.; Falcone, T.; Joshi, N.; Fazleabas, A.T.; Graham, A. Serum miR-451a Levels Are Significantly Elevated in Women With Endometriosis and Recapitulated in Baboons (Papio anubis) With Experimentally-Induced Disease. Reprod. Sci. 2017, 24, 1195–1202. [Google Scholar] [CrossRef]
- Maged, A.M.; Deeb, W.S.; El Amir, A.; Zaki, S.S.; El Sawah, H.; Al Mohamady, M.; Metwally, A.A.; Katta, M.A. Diagnostic accuracy of serum miR-122 and miR-199a in women with endometriosis. Int. J. Gynaecol. Obstet. 2018, 141, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Mamillapalli, R.; Taylor, H.S. Increased circulating miR-370-3p regulates steroidogenic factor 1 in endometriosis. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E373–E382. [Google Scholar] [CrossRef]
- Moustafa, S.; Burn, M.; Mamillapalli, R.; Nematian, S.; Flores, V.; Taylor, H.S. Accurate diagnosis of endometriosis using serum microRNAs. Am. J. Obstet. Gynecol. 2020, 223, 557.e1–557.e11. [Google Scholar] [CrossRef]
- Pang, Q.X.; Liu, Z. miR-17-5p mitigates endometriosis by directly regulating VEGFA. J. Biosci. 2020, 45, 78. [Google Scholar] [CrossRef]
- Misir, S.; Hepokur, C.; Oksasoglu, B.; Yildiz, C.; Yanik, A.; Aliyazicioglu, Y. Circulating serum miR-200c and miR-34a-5p as diagnostic biomarkers for endometriosis. J. Gynecol. Obstet. Hum. Reprod. 2021, 50, 102092. [Google Scholar] [CrossRef]
- He, S.; Li, J.; Ma, D.; Liu, Z.; Lv, N. MicroRNA-148a targets ADAMTS5 to inhibit proliferation of endometriosis cells. Pak. J. Pharm. Sci. 2022, 35, 335–341. [Google Scholar] [PubMed]
- Neuhausser, W.M.; Faure-Kumar, E.; Mahurkar-Joshi, S.; Iliopoulos, D.; Sakkas, D. Identification of miR-34-3p as a candidate follicular phase serum marker for endometriosis: A pilot study. F S Sci. 2022, 3, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Kumari, P.; Sharma, I.; Saha, S.C.; Srinivasan, R.; Bhardwaj, P. Role of serum microRNAs as biomarkers for endometriosis, endometrioid carcinoma of ovary & endometrioid endometrial cancer. Indian. J. Med. Res. 2022, 156, 516–523. [Google Scholar] [PubMed]
- Lin, C.; Zeng, S.; Li, M. miR-424-5p combined with miR-17-5p has high diagnostic efficacy for endometriosis. Arch. Gynecol. Obstet. 2023, 307, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tao, Y.; Jin, O.; Lai, J.; Yang, X. MiR-17-5p promoter methylation regulated by DNA methyltransferase 3 beta (DNMT3B) expedites endometriosis via the Kruppel-like factor 12 (KLF12)/Wnt/beta-catenin axis. J. Reprod. Immunol. 2023, 158, 103974. [Google Scholar] [CrossRef] [PubMed]
- Suryawanshi, S.; Vlad, A.M.; Lin, H.M.; Mantia-Smaldone, G.; Laskey, R.; Lee, M.; Lin, Y.; Donnellan, N.; Klein-Patel, M.; Lee, T.; et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin. Cancer Res. 2013, 19, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.Z.; Yang, Y.; Lang, J.; Sun, P.; Leng, J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum. Reprod. 2013, 28, 322–330. [Google Scholar] [CrossRef]
- Rekker, K.; Saare, M.; Roost, A.M.; Kaart, T.; Soritsa, D.; Karro, H.; Soritsa, A.; Simon, C.; Salumets, A.; Peters, M. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil. Steril. 2015, 104, 938–946.e2. [Google Scholar] [CrossRef]
- Bashti, O.; Noruzinia, M.; Garshasbi, M.; Abtahi, M. miR-31 and miR-145 as Potential Non-Invasive Regulatory Biomarkers in Patients with Endometriosis. Cell J. 2018, 20, 293. [Google Scholar]
- Wang, F.; Wang, H.; Jin, D.; Zhang, Y. Serum miR-17, IL-4, and IL-6 levels for diagnosis of endometriosis. Medicine 2018, 97, e10853. [Google Scholar] [CrossRef]
- Pateisky, P.; Pils, D.; Szabo, L.; Kuessel, L.; Husslein, H.; Schmitz, A.; Wenzl, R.; Yotova, I. hsa-miRNA-154-5p expression in plasma of endometriosis patients is a potential diagnostic marker for the disease. Reprod. Biomed. Online 2018, 37, 449–466. [Google Scholar] [CrossRef]
- Nisenblat, V.; Sharkey, D.J.; Wang, Z.; Evans, S.F.; Healey, M.; Ohlsson Teague, E.M.C.; Print, C.G.; Robertson, S.A.; Hull, M.L. Plasma miRNAs Display Limited Potential as Diagnostic Tools for Endometriosis. J. Clin. Endocrinol. Metab. 2019, 104, 1999–2022. [Google Scholar] [CrossRef]
- Vanhie, A.; O, D.; Peterse, D.; Beckers, A.; Cuellar, A.; Fassbender, A.; Meuleman, C.; Mestdagh, P.; D’Hooghe, T. Plasma miRNAs as biomarkers for endometriosis. Hum. Reprod. 2019, 34, 1650–1660. [Google Scholar] [CrossRef]
- Hossein Razi, M.; Eftekhar, M.; Ghasemi, N.; Hasan Sheikhha, M.; Dehghani Firoozabadi, A. Expression levels of circulatory mir-185-5p, vascular endothelial growth factor, and platelet-derived growth factor target genes in endometriosis. Int. J. Reprod. Biomed. 2020, 18, 347–358. [Google Scholar] [CrossRef]
- Papari, E.; Noruzinia, M.; Kashani, L.; Foster, W.G. Identification of candidate microRNA markers of endometriosis with the use of next-generation sequencing and quantitative real-time polymerase chain reaction. Fertil. Steril. 2020, 113, 1232–1241. [Google Scholar] [CrossRef]
- Gu, C.L.; Zhang, Z.; Fan, W.S.; Li, L.A.; Ye, M.X.; Zhang, Q.; Zhang, N.N.; Li, Z.; Meng, Y.G. Identification of MicroRNAs as Potential Biomarkers in Ovarian Endometriosis. Reprod. Sci. 2020, 27, 1715–1723. [Google Scholar] [CrossRef]
- Zafari, N.; Tarafdari, A.M.; Izadi, P.; Noruzinia, M.; Yekaninejad, M.S.; Bahramy, A.; Mohebalian, A. A Panel of Plasma miRNAs 199b-3p, 224-5p and Let-7d-3p as Non-Invasive Diagnostic Biomarkers for Endometriosis. Reprod. Sci. 2021, 28, 991–999. [Google Scholar] [CrossRef]
- Bahramy, A.; Zafari, N.; Izadi, P.; Soleymani, F.; Kavousi, S.; Noruzinia, M. The Role of miRNAs 340-5p, 92a-3p, and 381-3p in Patients with Endometriosis: A Plasma and Mesenchymal Stem-Like Cell Study. Biomed. Res. Int. 2021, 2021, 5298006. [Google Scholar] [CrossRef]
- Bendifallah, S.; Dabi, Y.; Suisse, S.; Jornea, L.; Bouteiller, D.; Touboul, C.; Puchar, A.; Darai, E. MicroRNome analysis generates a blood-based signature for endometriosis. Sci. Rep. 2022, 12, 4051. [Google Scholar] [CrossRef]
- Tahermanesh, K.; Hakimpour, S.; Govahi, A.; Rokhgireh, S.; Mehdizadeh, M.; Minaeian, S.; Barati, M.; Chaichian, S.; Kashi, A.M.; Nassiri, S.; et al. Evaluation of expression of biomarkers of PLAGL1 (ZAC1), microRNA, and their non-coding RNAs in patients with endometriosis. J. Gynecol. Obstet. Hum. Reprod. 2023, 52, 102568. [Google Scholar] [CrossRef]
- Walasik, I.; Klicka, K.; Grzywa, T.M.; Szymusik, I.; Wlodarski, P.; Wielgos, M.; Pietrzak, B.; Ludwin, A. Circulating miR-3613-5p but not miR-125b-5p, miR-199a-3p, and miR-451a are biomarkers of endometriosis. Reprod. Biol. 2023, 23, 100796. [Google Scholar] [CrossRef]
- Grechukhina, O.; Petracco, R.; Popkhadze, S.; Massasa, E.; Paranjape, T.; Chan, E.; Flores, I.; Weidhaas, J.B.; Taylor, H.S. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol. Med. 2012, 4, 206–217. [Google Scholar] [CrossRef]
- Sahin, C.; Mamillapalli, R.; Yi, K.W.; Taylor, H.S. microRNA Let-7b: A Novel treatment for endometriosis. J. Cell Mol. Med. 2018, 22, 5346–5353. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Gu, L.; Di, W. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKbeta/NF-kappaB pathway and reduced interleukin-8 expression. Mol. Hum. Reprod. 2012, 18, 136–145. [Google Scholar] [CrossRef]
- D’Hooghe, T.M.; Grechukhina, O.; Cho, S.; Fassbender, A.; O, D.; Peterse, D.; Weidhaas, J.; Taylor, H.S. Lack of an Association between a Polymorphism in the KRAS 3′ Untranslated Region (rs61764370) and Endometriosis in a Large European Case-Control Study. Gynecol. Obstet. Invest. 2019, 84, 575–582. [Google Scholar] [CrossRef]
- Dabi, Y.; Suisse, S.; Jornea, L.; Bouteiller, D.; Touboul, C.; Puchar, A.; Darai, E.; Bendifallah, S. Clues for Improving the Pathophysiology Knowledge for Endometriosis Using Serum Micro-RNA Expression. Diagnostics 2022, 12, 175. [Google Scholar] [CrossRef]
- Vitonis, A.F.; Vincent, K.; Rahmioglu, N.; Fassbender, A.; Buck Louis, G.M.; Hummelshoj, L.; Giudice, L.C.; Stratton, P.; Adamson, G.D.; Becker, C.M.; et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: II. Clinical and covariate phenotype data collection in endometriosis research. Fertil. Steril. 2014, 102, 1223–1232. [Google Scholar] [CrossRef]
- Rahmioglu, N.; Fassbender, A.; Vitonis, A.F.; Tworoger, S.S.; Hummelshoj, L.; D’Hooghe, T.M.; Adamson, G.D.; Giudice, L.C.; Becker, C.M.; Zondervan, K.T.; et al. World Endometriosis Research Foundation Endometriosis Phenome and Biobanking Harmonization Project: III. Fluid biospecimen collection, processing, and storage in endometriosis research. Fertil. Steril. 2014, 102, 1233–1243. [Google Scholar] [CrossRef]
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE 2013, 8, e64795. [Google Scholar] [CrossRef]
- Zhelankin, A.V.; Iulmetova, L.N.; Sharova, E.I. The Impact of the Anticoagulant Type in Blood Collection Tubes on Circulating Extracellular Plasma MicroRNA Profiles Revealed by Small RNA Sequencing. Int. J. Mol. Sci. 2022, 23, 340. [Google Scholar] [CrossRef]
- Mussbacher, M.; Krammer, T.L.; Heber, S.; Schrottmaier, W.C.; Zeibig, S.; Holthoff, H.P.; Pereyra, D.; Starlinger, P.; Hackl, M.; Assinger, A. Impact of Anticoagulation and Sample Processing on the Quantification of Human Blood-Derived microRNA Signatures. Cells 2020, 9, 1915. [Google Scholar] [CrossRef]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef]
- Appierto, V.; Callari, M.; Cavadini, E.; Morelli, D.; Daidone, M.G.; Tiberio, P. A lipemia-independent NanoDrop((R))-based score to identify hemolysis in plasma and serum samples. Bioanalysis 2014, 6, 1215–1226. [Google Scholar] [CrossRef]
- Kupec, T.; Bleilevens, A.; Iborra, S.; Najjari, L.; Wittenborn, J.; Maurer, J.; Stickeler, E. Stability of circulating microRNAs in serum. PLoS ONE 2022, 17, e0268958. [Google Scholar] [CrossRef]
- Matias-Garcia, P.R.; Wilson, R.; Mussack, V.; Reischl, E.; Waldenberger, M.; Gieger, C.; Anton, G.; Peters, A.; Kuehn-Steven, A. Impact of long-term storage and freeze-thawing on eight circulating microRNAs in plasma samples. PLoS ONE 2020, 15, e0227648. [Google Scholar] [CrossRef]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Kloten, V.; Neumann, M.H.D.; Di Pasquale, F.; Sprenger-Haussels, M.; Shaffer, J.M.; Schlumpberger, M.; Herdean, A.; Betsou, F.; Ammerlaan, W.; Af Hallstrom, T.; et al. Multicenter Evaluation of Circulating Plasma MicroRNA Extraction Technologies for the Development of Clinically Feasible Reverse Transcription Quantitative PCR and Next-Generation Sequencing Analytical Work Flows. Clin. Chem. 2019, 65, 1132–1140. [Google Scholar] [CrossRef]
- Setti, G.; Pezzi, M.E.; Viani, M.V.; Pertinhez, T.A.; Cassi, D.; Magnoni, C.; Bellini, P.; Musolino, A.; Vescovi, P.; Meleti, M. Salivary MicroRNA for Diagnosis of Cancer and Systemic Diseases: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 907. [Google Scholar] [CrossRef]
- Kang, J.W.; Eun, Y.G.; Lee, Y.C. Diagnostic Value of Salivary miRNA in Head and Neck Squamous Cell Cancer: Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2021, 22, 7026. [Google Scholar] [CrossRef]
- Bendifallah, S.; Dabi, Y.; Suisse, S.; Delbos, L.; Spiers, A.; Poilblanc, M.; Golfier, F.; Jornea, L.; Bouteiller, D.; Fernandez, H.; et al. Validation of a Salivary miRNA Signature of Endometriosis—Interim Data. NEJM Evidence 2023, 2, EVIDoa2200282. [Google Scholar] [CrossRef]
- Feng, X.; Liu, Y.; Wan, N. Plasma microRNA detection standardization test. J. Clin. Lab. Anal. 2020, 34, e23058. [Google Scholar] [CrossRef] [PubMed]
- Mari-Alexandre, J.; Sanchez-Izquierdo, D.; Gilabert-Estelles, J.; Barcelo-Molina, M.; Braza-Boils, A.; Sandoval, J. miRNAs Regulation and Its Role as Biomarkers in Endometriosis. Int. J. Mol. Sci. 2016, 17, 93. [Google Scholar] [CrossRef] [PubMed]
- Faraldi, M.; Gomarasca, M.; Banfi, G.; Lombardi, G. Free Circulating miRNAs Measurement in Clinical Settings: The Still Unsolved Issue of the Normalization. Adv. Clin. Chem. 2018, 87, 113–139. [Google Scholar] [PubMed]
- Faraldi, M.; Gomarasca, M.; Sansoni, V.; Perego, S.; Banfi, G.; Lombardi, G. Normalization strategies differently affect circulating miRNA profile associated with the training status. Sci. Rep. 2019, 9, 1584. [Google Scholar] [CrossRef]
| No | Author | Sample Size | Sample Type | ASRM Stage * | Control Group Characteristics | Methods | Normalization Control | |
|---|---|---|---|---|---|---|---|---|
| 1 | Wang et al., 2013 [20] | Cases Controls | 60 25 | Serum | I–IV | NLP ILP: severe DM, PM, IF | 1. Microarray 2. RT-qPCR | U6 snRNA |
| 2 | Hsu et al., 2014 [21] | Cases Controls | 40 25 | Serum | II–IV | NLP ILP: OBGP | 1. Microarray 2. RT-qPCR | 18s RNA |
| 3 | Cho et al., 2015 [22] | Cases Controls | 24 24 | Serum | III–IV | NLP ILP: PM, PP, SE, IF, OBGP | RT-qPCR | U6 snRNA |
| 4 | Cosar et al., 2016 [23] | Cases Controls | 24 24 | Serum | III–IV | NLP ILP: PM, PP, IF, SE | 1. Microarray 2. RT-qPCR | U6 snRNA |
| 5 | Wang et al., 2016 [24] | Cases Controls | 30 20 | Serum | I–II | NLP ILP: PM, IF | 1. Solexa sequencing 2. RT-qPCR | Cel-miR-39 |
| 6 | Nothnick et al., 2017 [25] | Cases Controls | 41 40 | Serum | I–IV | n = 20: NLP ILP: PP, pain with bleeding n = 20: (SR)HW | RT-qPCR | U6 snRNA |
| 7 | Maged et al., 2018 [26] | Cases Controls | 45 35 | Serum | I–IV | NLP ILP: PP, IF, benign neoplasms | RT-qPCR | U6 snRNA |
| 8 | Hu et al., 2019 [27] | Cases Controls | 20 26 | Serum | III–IV | NLP or NLT ILP or ILT: PM, PP, IF, SE | RT-qPCR | U6 snRNA |
| 9 | Moustafa et al., 2020 [28] | Cases Controls | 41 59 | Serum | I–IV | NLP ILP: OBGP | RT-qPCR | U6 snRNA |
| 10 | Pang et al., 2020 [29] | Cases Controls | 20 30 | Serum | NS | Uterine fibroids, endometriosis free (NFS) | NS | NS |
| 11 | Misir et al., 2021 [30] | Cases Controls | 71 65 | Serum | I–IV | NLP or NLT | RT-qPCR | U6 snRNA |
| 12 | He et al., 2022 [31] | Cases Controls | 23 20 | Serum | I–IV | Tubal obstruction with NLP | RT-qPCR | U6 snRNA |
| 13 | Neuhausser et al., 2022 [32] | 1. DC Cases Controls 2. VC Cases Controls | 21 24 27 24 | Serum | NS | Asymptomatic egg donors with normal pelvic US | 1. NanoString nCounter 2. RT-qPCR | U6 snRNA |
| 14 | Kumari et al., 2022 [33] | Cases Controls | 10 10 | Serum | NS | NLP or NLT ILP: PP, SE, PM, IF, OBGP | RT-qPCR | miR-39 |
| 15 | Lin et al., 2023 [34] | Cases Controls | 80 80 | Serum | I-IV | HW: normal gynecological examination and US | RT-qPCR | U6 snRNA |
| 16 | Yang et al., 2023 [35] | Cases Controls | 17 13 | Serum | NS | HT for uterine fibroids or CIN grade II-III | RT-qPCR | U6 snRNA |
| 17 | Suryawanshi et al., 2013 [36] | Cases Controls | 33 20 | Plasma | NS | SRHW | 1. RT-qPCR 2. RT-qPCR | miR-132 |
| 18 | Jia et al., 2013 [37] | Cases Controls | 23 23 | Plasma | III–IV | NLP ILP: PM, PP, IF, UL | 1. Microarray 2. RT-qPCR | miR-16 |
| 19 | Rekker et al., 2015 [38] | Cases Controls | 61 65 | Plasma | I–IV | n = 35: NLP ILP: SE, DM, IF, PP, polycystic ovaries n = 30: SRHW | RT-qPCR | miR-30e-5p, miR-99a-5p |
| 20 | Bashti et al., 2018 [39] | Cases Controls | 55 23 | Plasma | I–IV | NLP ILP: prolapsed uterus, ovarian cyst, urinary incontinence | RT-qPCR | miR-103-3p |
| 21 | Wang et al., 2018 [40] | Cases Controls | 80 60 | Plasma | I–IV | NLP ILP: SE, PM, PP, IF, UL | RT-qPCR | Beta-actin |
| 22 | Pateisky et al., 2018 [41] | Cases Controls | 51 41 | Plasma | I–IV | NLP ILP: SE, PP, IF, adnexal cysts, uterine fibroids | q-PCR-array based microarrays | miR-199a |
| 23 | Nisenblat et al., 2019 [42] | 1.Cases Controls 2.a) Cases Controls 2.b) Cases Controls 3.Cases Controls 4.Cases Controls | 8 8 8 8 10 10 51 27 80 39 | Plasma | I–IV | 1. Asymptomatic (SR)HW 2.a) Asymptomatic (SR)HW 2.b) NLP ILP: PP and/or IF 3. NLP ILP: DM, DP, chronic PP, IF 4. NLP ILP: DM, DP, chronic PP | 1. Microarray 2. RT-qPCR | miR-30b |
| 24 | Vanhie et al., 2019 [43] | 1. DC Cases Controls 2. VC Cases Controls | 82 38 60 30 | Plasma | I–IV | NLP ILP: surgical treatment of endometriosis diagnosed on imaging or diagnostic laparoscopy, PP with SE, IF | 1. NGS 2. RT-qPCR 3. RT-qPCR | hsa-miR-423-3p, hsa-miR-28-3p |
| 25 | Hossein Razi et al., 2019 [44] | Cases Controls | 25 25 | Plasma | III–IV | NLP ILP: PP, ovarian cyst | 1. NGS 2. RT-qPCR | SNORD47 |
| 26 | Papari et al., 2020 [45] | Cases Controls | 25 28 | Plasma | III–IV | NLP ILP: PM, PM, IF, UL | RT-qPCR | hsa-miR-148b-3p, hsa-miR-30e-5p |
| 27 | Gu et al., 2020 [46] | Cases Controls | 19 21 | Plasma | Ovarian endometriosis, NS | HW (NFS) | RT-qPCR | NS |
| 28 | Zafari et al., 2021 [47] | Cases Controls | 25 25 | Plasma | III-IV | NLP ILP: PP, IF, DM | RT-qPCR | miR-16 |
| 29 | Bahramy et al., 2021 [48] | Cases Controls | 30 30 | Plasma | III–IV | NLP ILP: PP, IF, DM | RT-qPCR | miR-16 |
| 30 | Bendifallah et al., 2022 [49] | Cases Controls | 153 47 | Plasma | I–IV | Negative MRI and NLP ILP: OBGP, SE | NGS | NS |
| 31 | Tahermanesh et al., 2023 [50] | Cases Controls | 30 30 | Plasma | III-IV | HW with NLP | RT-qPCR | U6 snRNA |
| 32 | Walasik et al., 2023 [51] | Cases Controls | 24 25 | Plasma | III-IV | PM, OBGP, HW, NLP | RT-qPCR | miR-39 |
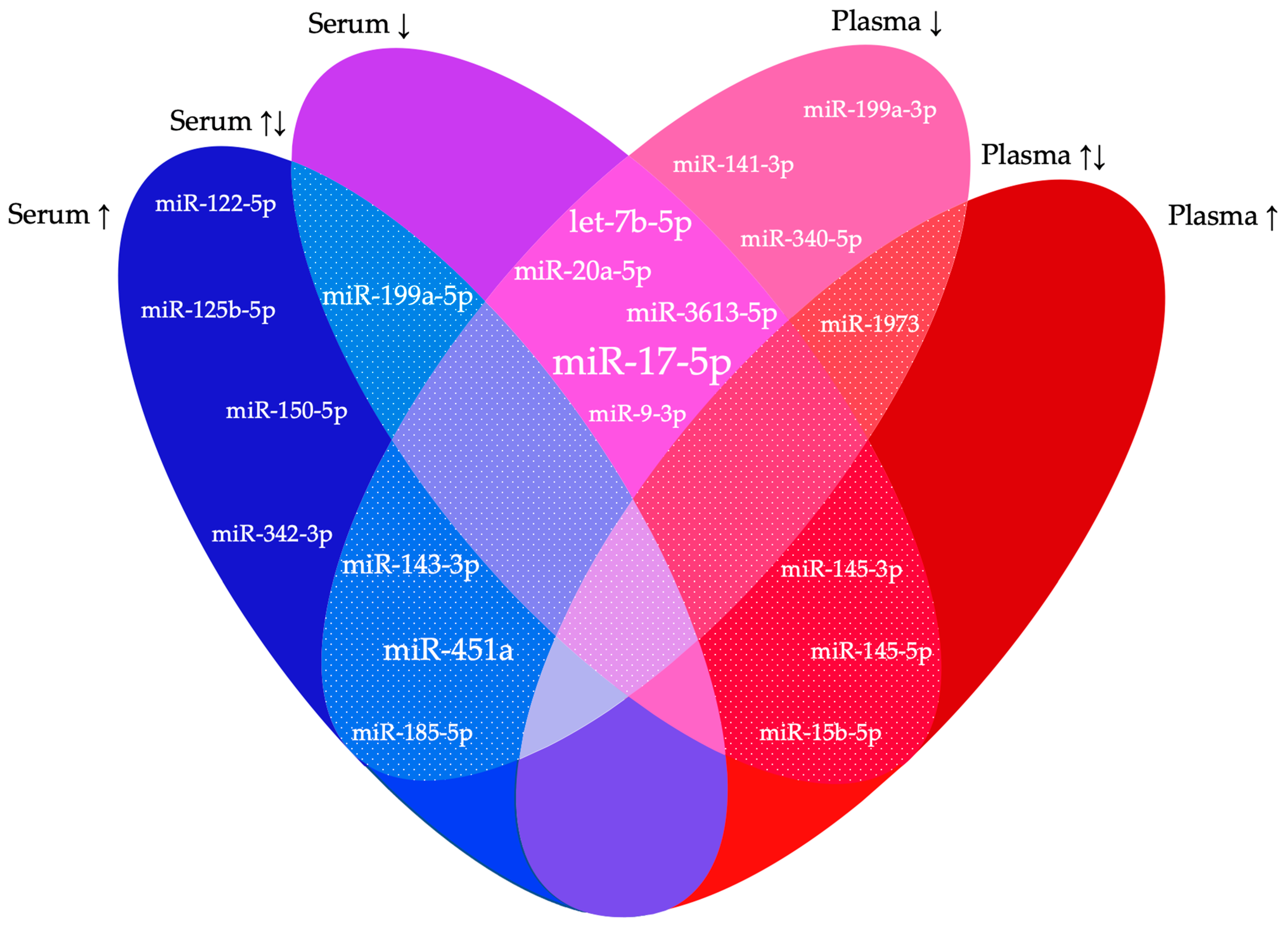
| Serum | |||
|---|---|---|---|
| No | Author | Dysregulated miRNAs | |
| 1 | Wang et al., 2013 [20] | ↑ | hsa-miR-122-5p, hsa-miR-199a-5p |
| ↓ | hsa-miR-9-3p, hsa-miR-141-5p, hsa-miR-145-3p, hsa-miR-542-3p | ||
| 2 | Hsu et al., 2014 [21] | ↓ | hsa-miR-199a-5p |
| 3 | Cho et al., 2015 [22] | ↓ | hsa-let-7b-5p, hsa-mir-135a-5p |
| 4 | Cosar et al., 2016 [23] | ↑ | hsa-miR-18a-5p, hsa-miR-125b-5p, hsa-miR-143-3p, hsa-miR-150-5p, hsa-miR-342-3p, hsa-miR-451a, hsa-miR-500a-3p |
| ↓ | hsa-miR-3613-5p, hsa-miR-6755-3p | ||
| 5 | Wang et al., 2016 [24] | ↑ | hsa-miR-185-5p, hsa-miR-424-3p |
| ↓ | hsa-miR-15b-5p, hsa-miR-20a-5p, hsa-miR-30c-5p, hsa-miR-99b-5p, hsa-miR-127-3p | ||
| 6 | Nothnick et al., 2017 [25] | ↑ | hsa-miR-451a |
| 7 | Maged et al., 2018 [26] | ↑ | hsa-miR-122-5p, hsa-miR-199a-5p |
| 8 | Hu et al., 2019 [27] | ↓ | hsa-miR-370-3p |
| 9 | Moustafa et al., 2020 [28] | ↑ | hsa-miR-125b-5p, hsa-miR-150-5p, hsa-miR-342-3p, hsa-miR-451a |
| ↓ | hsa-let-7b-5p, hsa-miR-3613-5p | ||
| 10 | Pang et al., 2020 [29] | ↓ | hsa-miR-17-5p |
| 11 | Misir et al., 2021 [30] | ↑ | hsa-miR-200c |
| ↓ | hsa-miR-34a-5p | ||
| 12 | He et al., 2022 [31] | ↓ | hsa-mir-148a |
| 13 | Neuhausser et al., 2022 [32] | DC ↑ | hsa-miR-10a-5p, hsa-miR-182-3p, hsa-miR-210-5p, hsa-miR-219a-5p, hsa-miR-363-3p, hsa-miR-478a-3p, hsa-miR-495-3p, hsa-miR-513b-5p, hsa-miR-518c-3p, hsa-miR-626, hsa-miR-802, hsa-miR-942-5p, hsa-miR-1228-3p, hsa-miR-1249-3p, hsa-miR-1266-5p, hsa-miR-1306-5p, hsa-miR-4443 and hsa-miR-4516 |
| DC ↓ | hsa-miR-34c-3p | ||
| VC ↓ | hsa-miR-34c-3p | ||
| 14 | Kumari et al., 2022 [33] | ↑ | hsa-miR-99b-5p, hsa-miR-125a-5p, hsa-miR-143-3p, hsa-miR-145-5p |
| ↓ | hsa-miR-16 | ||
| 15 | Lin et al., 2023 [34] | ↓ | hsa-miR-17-5p, hsa-miR-424-5p |
| 16 | Yang et al., 2023 [35] | ↓ | hsa-miR-17-5p |
| Plasma | |||
| 17 | Suryawanshi et al., 2013 [36] | ↑ | hsa-miR-15b-5p, hsa-miR-16-5p, hsa-miR-191-5p, hsa-miR-195-5p, hsa-miR-362-5p, hsa-miR-1973, hsa-miR-1974, hsa-miR-1978, hsa-miR-1979, hsa-miR-4284 |
| 18 | Jia et al., 2013 [37] | ↓ | hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR-22-5p |
| 19 | Rekker et al., 2015 [38] | ↓ | hsa-mir-141-3p, hsa-mir-200a-3p |
| 20 | Bashti et al., 2018 [39] | ↑ | hsa-miR-145-5p |
| ↓ | hsa-miR-31-5p | ||
| 21 | Wang et al., 2018 [40] | ↓ | hsa-miR-17-5p |
| 22 | Pateisky et al., 2018 [41] | ↑ | hsa-miR-33a-5p |
| ↓ | hsa-miR-154-5p, hsa-miR-196b-5p, hsa-miR-378a-3p | ||
| 23 | Nisenblat et al., 2019 [42] | DC ↑ | hsa-miR-145-3p |
| DC ↓ | hsa-miR-9-3p, hsa-miR-135b-5p, hsa-miR-139-3p, hsa-miR-141-3p, hsa-miR-155-5p, hsa-miR-574-3p, hsa-miR-923 | ||
| VC ↑ | No differentially expressed miRNAs | ||
| VC ↓ | hsa-miR-139-3p, hsa-miR-155-5p, hsa-miR-574-3p | ||
| 24 | Vanhie et al., 2019 [43] | No differentially expressed miRNAs | |
| 25 | Hossein Razi et al., 2019 [44] | ↓ | hsa-miR-185-5p |
| 26 | Papari et al., 2020 [45] | ↓ | hsa-let-7b-5p, hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR-21-5p, hsa-miR-103a-3p, hsa-miR-143-3p, hsa-miR-199a-3p, hsa-miR-340-5p |
| 27 | Gu et al., 2020 [46] | ↓ | hsa-let-7a-5p, hsa-let-7b-5p, hsa-let-7d-5p, hsa-let-7f-5p, hsa-let-7g-5p, hsa-let-7i-5p, hsa-miR-199a-3p, hsa-miR-320a, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, hsa-miR-328-3p, hsa-miR-131-3p, hsa-miR-320e |
| 28 | Zafari et al., 2021 [47] | ↑ | hsa-miR-199b-3p |
| ↓ | hsa-let-7d-3p, hsa-miR-224-5p | ||
| 29 | Bahramy et al., 2021 [48] | ↓ | hsa-miR-92a-3p, hsa-miR-340-5p, hsa-miR-381-3p |
| 30 | Bendifallah et al., 2022 [49] | ↑ | hsa-miR-29b-1-5p, hsa-miR-3122, hsa-miR-4536-3p, hsa-miR-4715-5p, hsa-miR-6502-5p |
| ↓ | hsa-miR-203a-5p, hsa-miR-208a-5p, hsa-miR-208a-3p, hsa-miR-216b-3p, hsa-miR-504-3p, hsa-miR-514b-5p, hsa-miR-573, hsa-miR-889-5p, hsa-miR-1180-5p, hsa-miR-1253, hsa-miR-1910-5p, hsa-miR-1973, hsa-miR-3064-3p, hsa-miR-3137, hsa-miR-3168, hsa-miR-3185, hsa-miR-3622a-3p, hsa-miR-3923, hsa-miR-4674, hsa-miR-4703-5p, hsa-miR-4725-5p, hsa-miR-4740-5p, hsa-miR-4749-5p, hsa-miR-4750-3p, hsa-miR-4764-5p, hsa-miR-5004-3p, hsa-miR-6075, hsa-miR-6811-3p, hsa-miR-6824-3p, hsa-miR-6875-3p, hsa-miR-6788-3p, hsa-miR-6799-3p, hsa-miR-7108-3p, hsa-miR-7109-5p, hsa-miR-7150, hsa-miR-7152-5p | ||
| 31 | Tahermanesh et al., 2023 [50] | ↑ | hsa-miR-490-3p, hsa-miR-1271-5p |
| 32 | Walasik et al., 2023 [51] | ↓ | hsa-miR-451a, hsa-miR-3613-5p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanhie, A.; Caron, E.; Vermeersch, E.; O, D.; Tomassetti, C.; Meuleman, C.; Mestdagh, P.; D’Hooghe, T.M. Circulating microRNAs as Non-Invasive Biomarkers in Endometriosis Diagnosis—A Systematic Review. Biomedicines 2024, 12, 888. https://doi.org/10.3390/biomedicines12040888
Vanhie A, Caron E, Vermeersch E, O D, Tomassetti C, Meuleman C, Mestdagh P, D’Hooghe TM. Circulating microRNAs as Non-Invasive Biomarkers in Endometriosis Diagnosis—A Systematic Review. Biomedicines. 2024; 12(4):888. https://doi.org/10.3390/biomedicines12040888
Chicago/Turabian StyleVanhie, Arne, Ellen Caron, Eveline Vermeersch, Dorien O, Carla Tomassetti, Christel Meuleman, Pieter Mestdagh, and Thomas M. D’Hooghe. 2024. "Circulating microRNAs as Non-Invasive Biomarkers in Endometriosis Diagnosis—A Systematic Review" Biomedicines 12, no. 4: 888. https://doi.org/10.3390/biomedicines12040888
APA StyleVanhie, A., Caron, E., Vermeersch, E., O, D., Tomassetti, C., Meuleman, C., Mestdagh, P., & D’Hooghe, T. M. (2024). Circulating microRNAs as Non-Invasive Biomarkers in Endometriosis Diagnosis—A Systematic Review. Biomedicines, 12(4), 888. https://doi.org/10.3390/biomedicines12040888




