Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins
Abstract
:1. Introduction
2. Results
2.1. Characterization of Donors and Isolation of Antibodies and Their Subfractions
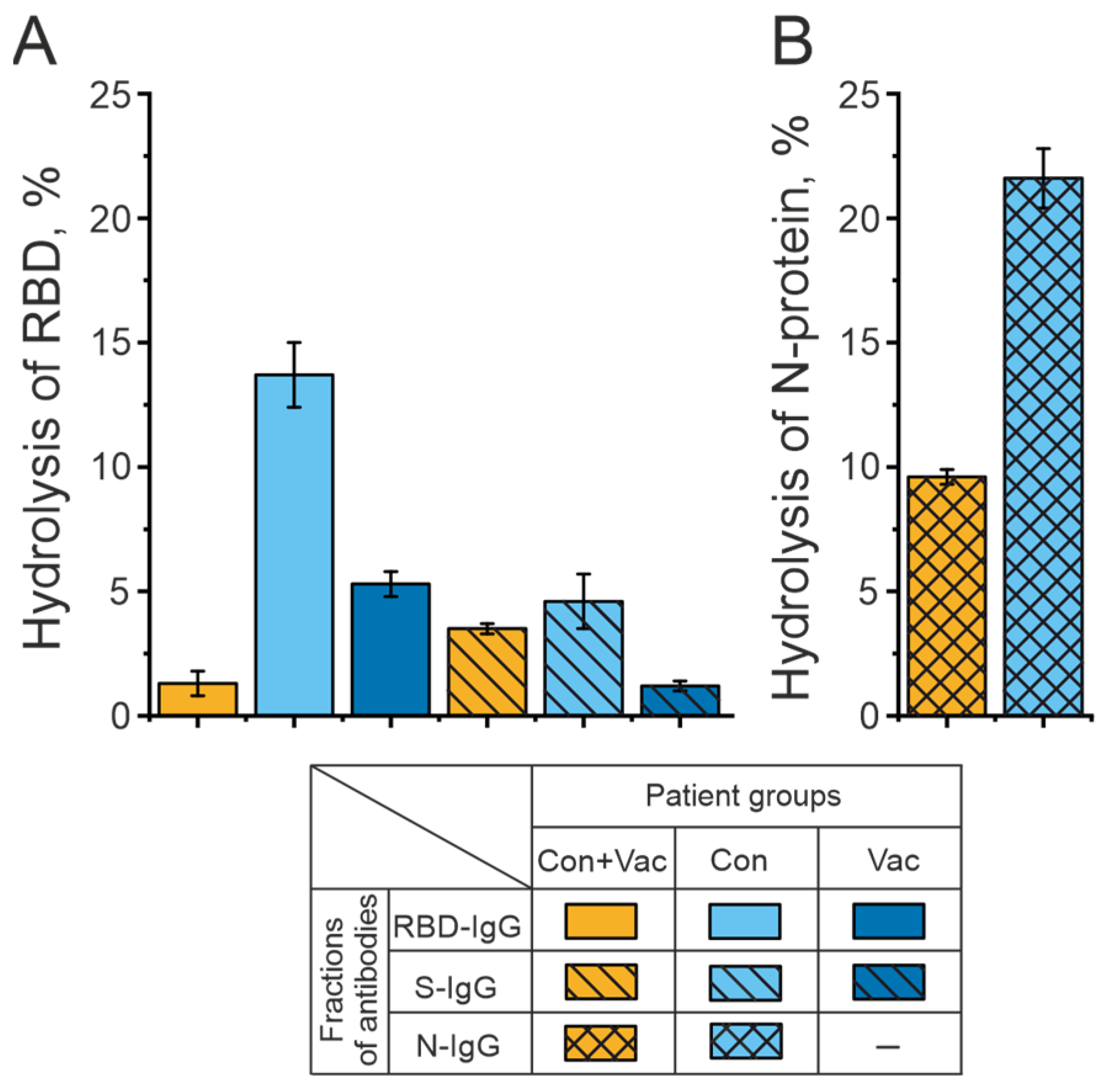
2.2. Analysis of the Catalytic Activity of Antibodies in the Hydrolysis of RBD and N-Protein of the SARS-CoV-2 Coronavirus
2.3. Intrinsic Catalytic Activity of Antibodies
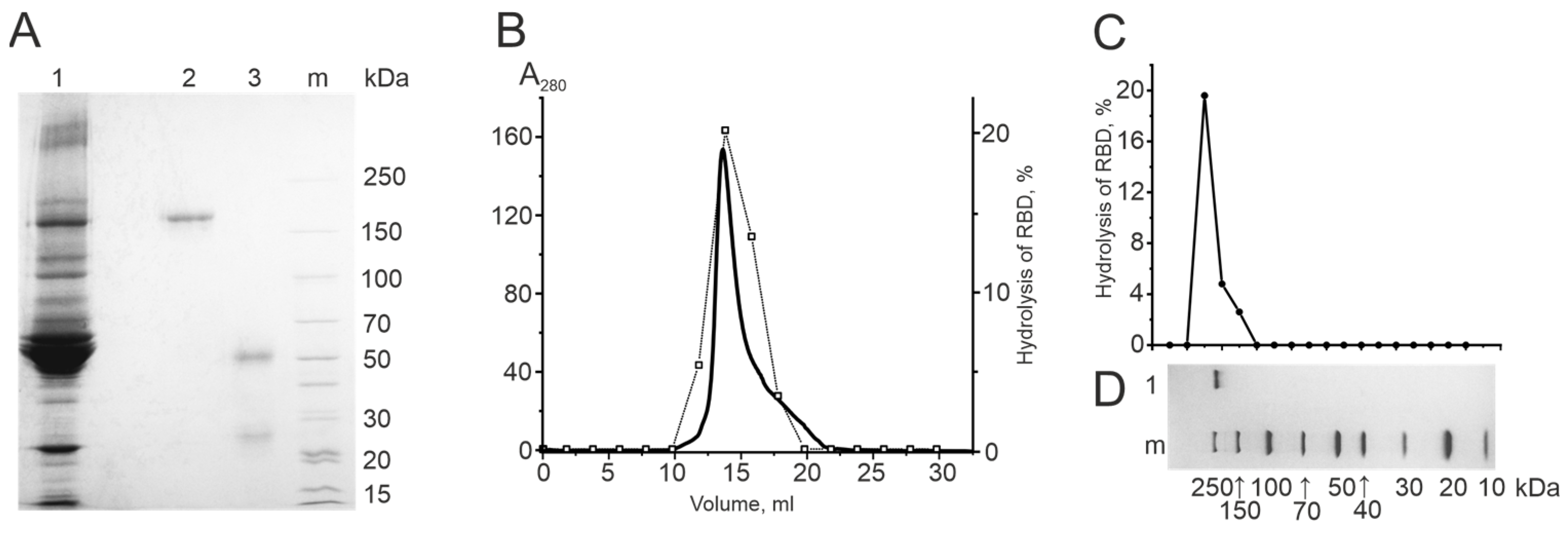
2.3.1. Electrophoretic Homogeneity of IgG Preparations
2.3.2. Gel Filtration of Antibodies under “Acid Shock” Conditions
2.3.3. Determination of the Catalytic Activity of Antibodies in Gel
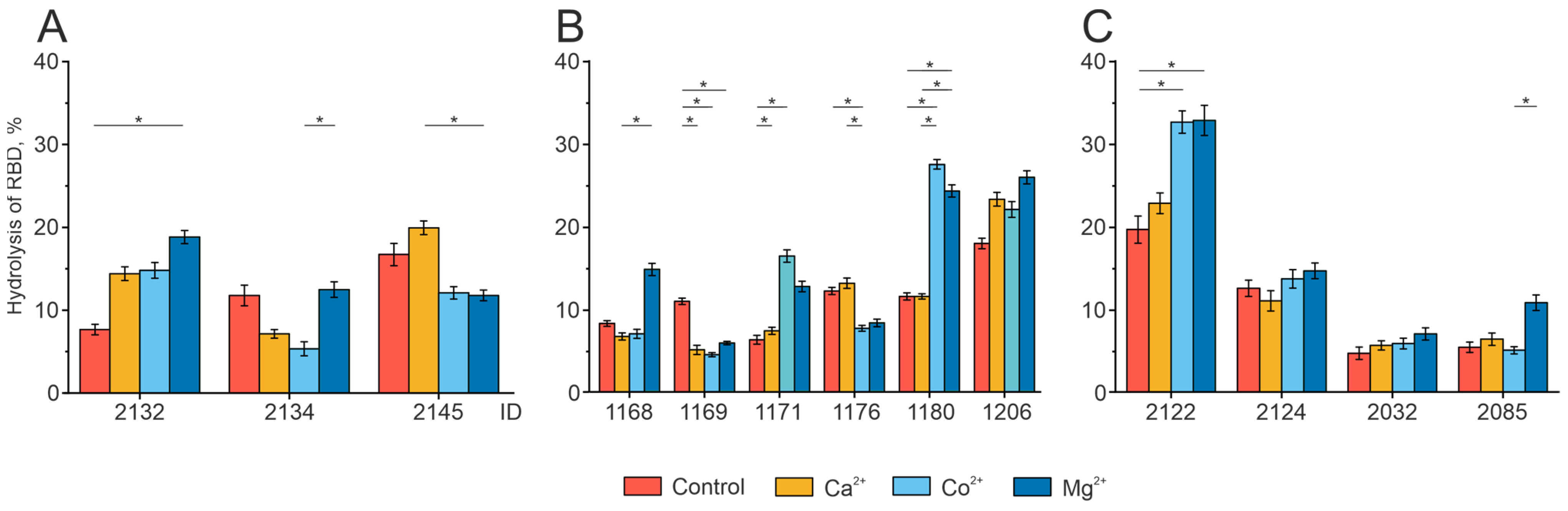
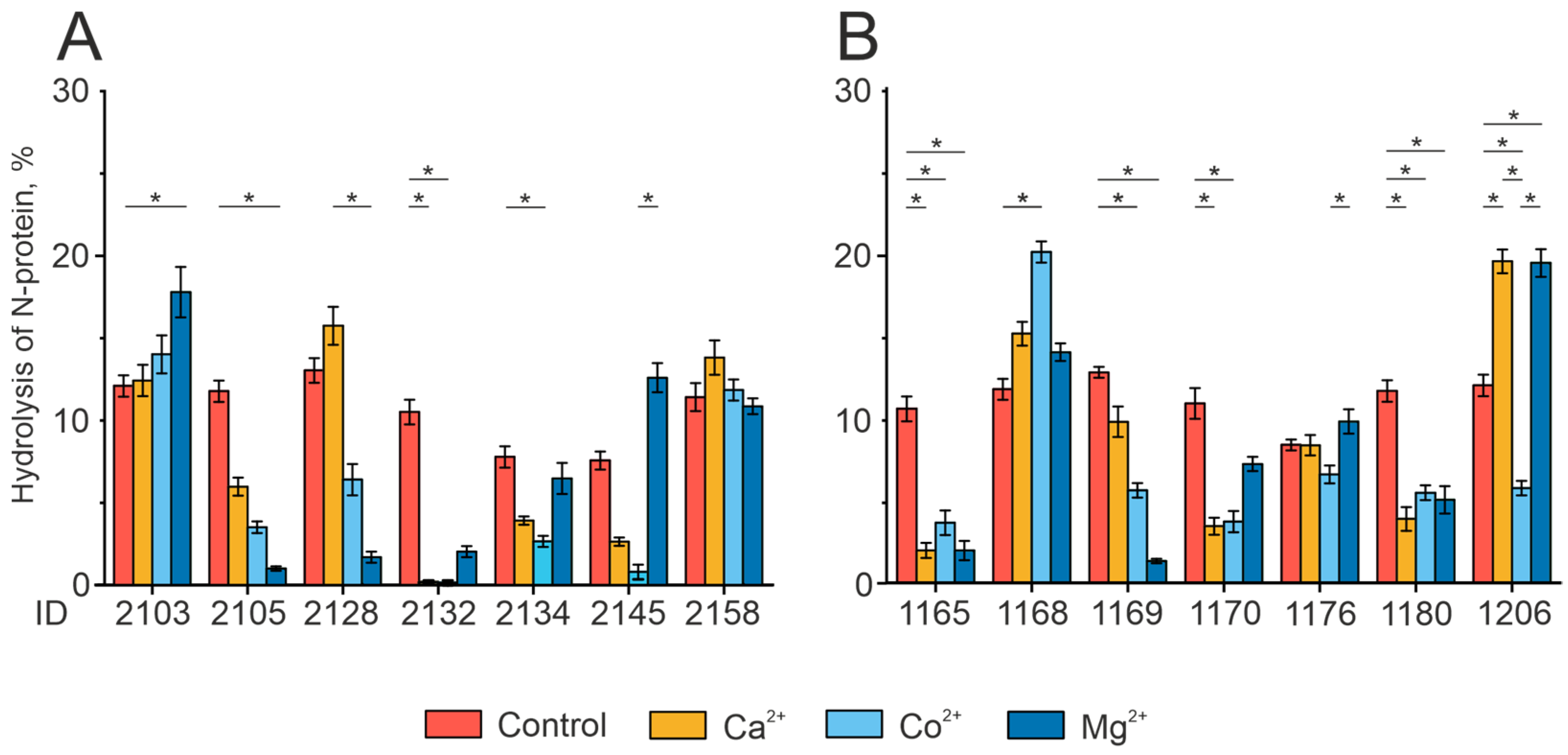
2.4. Analysis of the Individual Peculiarities of the Proteolytic Activity of Antibodies in the Patient Groups
2.5. Effect of Divalent Metal Ions on the Catalytic Activity of IgG Preparations
3. Discussion
4. Materials and Methods
4.1. Donors and Patients
- (Con+Vac group)—COVID-19 convalescents subsequently vaccinated with Sputnik V
- (Con group)—COVID-19 convalescents
- (Vac group)—volunteers vaccinated with Sputnik V
- (Neg group)—a control group of neither infected nor vaccinated donors.
4.2. Antibody Isolation from Blood Plasma
4.3. Gel Filtration under “Acid Shock” Conditions
4.4. Analysis of the Proteolytic Activity of Antibodies in the Gel
4.5. Analysis of Proteolytic IgG Activity
4.6. Investigation of the Effect of Various Metal Ions on Antibody Catalytic Activity
4.7. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lerner, R.A.; Tramontano, A. Antibodies as Enzymes. Trends Biochem. Sci. 1987, 12, 427–430. [Google Scholar] [CrossRef]
- Tanaka, F. Catalytic Antibodies as Designer Proteases and Esterases. Chem. Rev. 2002, 102, 4885–4906. [Google Scholar] [CrossRef]
- Suzuki, H. Recent Advances in Abzyme Studies1. J. Biochem. 1994, 115, 623–628. [Google Scholar] [CrossRef]
- Paul, S.; Volle, D.J.; Beach, C.M.; Johnson, D.R.; Powell, M.J.; Massey, R.J. Catalytic Hydrolysis of Vasoactive Intestinal Peptide by Human Autoantibody. Science 1989, 244, 1158–1162. [Google Scholar] [CrossRef] [PubMed]
- Bowen, A.; Wear, M.; Casadevall, A. Antibody-Mediated Catalysis in Infection and Immunity. Infect. Immun. 2017, 85. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xia, Y. Anti-Double Stranded DNA Antibodies: Origin, Pathogenicity, and Targeted Therapies. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Shuster, A.M.; Gololobov, G.V.; Kvashuk, O.A.; Bogomolova, A.E.; Smirnov, I.V.; Gabibov, A.G. DNA Hydrolyzing Autoantibodies. Science 1992, 256, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-H.; Chang, C.-J.; Chuang, Y.-H.; Hsu, H.-Y.; Chen, P.P.; Chiang, B.-L. Identification of Anti-Prothrombin Antibodies in the Anti-Phospholipid Syndrome That Display the Prothrombinase Activity. Rheumatology 2010, 49, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Planque, S.A.; Nishiyama, Y.; Hanson, C.V.; Massey, R.J. Nature and Nurture of Catalytic Antibodies. In Naturally Occurring Antibodies; Springer: Berlin/Heidelberg, Germany, 2012; pp. 56–75. [Google Scholar]
- Planque, S.; Nishiyama, Y.; Taguchi, H.; Salas, M.; Hanson, C.; Paul, S. Catalytic Antibodies to HIV: Physiological Role and Potential Clinical Utility. Autoimmun. Rev. 2008, 7, 473–479. [Google Scholar] [CrossRef]
- Li, L.; Paul, S.; Tyutyulkova, S.; Kazatchkine, M.D.; Kaveri, S. Catalytic Activity of Anti-Thyroglobulin Antibodies. J. Immunol. 1995, 154, 3328–3332. [Google Scholar] [CrossRef]
- Thiagarajan, P.; Dannenbring, R.; Matsuura, K.; Tramontano, A.; Gololobov, G.; Paul, S. Monoclonal Antibody Light Chain with Prothrombinase Activity. Biochemistry 2000, 39, 6459–6465. [Google Scholar] [CrossRef]
- Lacroix-Desmazes, S.; Moreau, A.; Sooryanarayana; Bonnemain, C.; Stieltjes, N.; Pashov, A.; Sultan, Y.; Hoebeke, J.; Kazatchkine, M.D.; Kaveri, S.V. Catalytic Activity of Antibodies against Factor VIII in Patients with Hemophilia A. Nat. Med. 1999, 5, 1044–1047. [Google Scholar] [CrossRef] [PubMed]
- Lacroix-Desmazes, S.; Wootla, B.; Delignat, S.; Dasgupta, S.; Nagaraja, V.; Kazatchkine, M.D.; Kaveri, S.V. Pathophysiology of Catalytic Antibodies. Immunol. Lett. 2006, 103, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, N.A.; Durova, O.M.; Vorobiev, I.I.; Belogurov, A.A.; Kurkova, I.N.; Petrenko, A.G.; Telegin, G.B.; Suchkov, S.V.; Kiselev, S.L.; Lagarkova, M.A.; et al. Autoantibodies to Myelin Basic Protein Catalyze Site-Specific Degradation of Their Antigen. Proc. Natl. Acad. Sci. USA 2006, 103, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Bezuglova, A.M.; Konenkova, L.P.; Doronin, B.M.; Buneva, V.N.; Nevinsky, G.A. Affinity and Catalytic Heterogeneity and Metal-Dependence of Polyclonal Myelin Basic Protein-Hydrolyzing IgGs from Sera of Patients with Systemic Lupus Erythematosus. J. Mol. Recognit. 2011, 24, 960–974. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, H.; Planque, S.; Nishiyama, Y.; Symersky, J.; Boivin, S.; Szabo, P.; Friedland, R.P.; Ramsland, P.A.; Edmundson, A.B.; Weksler, M.E.; et al. Autoantibody-Catalyzed Hydrolysis of Amyloid β Peptide. J. Biol. Chem. 2008, 283, 4714–4722. [Google Scholar] [CrossRef] [PubMed]
- Loeffler, D.A. Antibody-Mediated Clearance of Brain Amyloid-β: Mechanisms of Action, Effects of Natural and Monoclonal Anti-Aβ Antibodies, and Downstream Effects. J. Alzheimer’s Dis. Rep. 2023, 7, 873–899. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Taguchi, H.; Hara, M.; Planque, S.A.; Mitsuda, Y.; Paul, S. Metal-Dependent Amyloid β-Degrading Catalytic Antibody Construct. J. Biotechnol. 2014, 180, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Planque, S.; Nishiyama, Y. Immunological Origin and Functional Properties of Catalytic Autoantibodies to Amyloid β Peptide. J. Clin. Immunol. 2010, 30, 43–49. [Google Scholar] [CrossRef]
- Odintsova, E.S.; Baranova, S.V.; Dmitrenok, P.S.; Rasskazov, V.A.; Calmels, C.; Parissi, V.; Andreola, M.-L.; Buneva, V.N.; Zakharova, O.D.; Nevinsky, G.A. Antibodies to HIV Integrase Catalyze Site-Specific Degradation of Their Antigen. Int. Immunol. 2011, 23, 601–612. [Google Scholar] [CrossRef]
- Paul, S.; Karle, S.; Planque, S.; Taguchi, H.; Salas, M.; Nishiyama, Y.; Handy, B.; Hunter, R.; Edmundson, A.; Hanson, C. Naturally Occurring Proteolytic Antibodies. J. Biol. Chem. 2004, 279, 39611–39619. [Google Scholar] [CrossRef] [PubMed]
- Planque, S.; Mitsuda, Y.; Taguchi, H.; Salas, M.; Morris, M.-K.; Nishiyama, Y.; Kyle, R.; Okhuysen, P.; Escobar, M.; Hunter, R.; et al. Characterization of Gp120 Hydrolysis by IgA Antibodies from Humans without HIV Infection. AIDS Res. Hum. Retroviruses 2007, 23, 1541–1554. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Planque, S.; Zhou, Y.-X.; Taguchi, H.; Bhatia, G.; Karle, S.; Hanson, C.; Nishiyama, Y. Specific HIV Gp120-Cleaving Antibodies Induced by Covalently Reactive Analog of Gp120. J. Biol. Chem. 2003, 278, 20429–20435. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Karle, S.; Planque, S.; Taguchi, H.; Paul, S. Antibodies to the Superantigenic Site of HIV-1 Gp120: Hydrolytic and Binding Activities of the Light Chain Subunit. Mol. Immunol. 2007, 44, 2707–2718. [Google Scholar] [CrossRef] [PubMed]
- Hifumi, E.; Mitsuda, Y.; Ohara, K.; Uda, T. Targeted Destruction of the HIV-1 Coat Protein Gp41 by a Catalytic Antibody Light Chain. J. Immunol. Methods 2002, 269, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Sedykh, S.E.; Sedykh, T.A.; Nevinsky, G.A. Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Recognize and Hydrolyze Oligopeptides Corresponding to the S-Protein of SARS-CoV-2. Vaccines 2023, 11, 1494. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.M.; Sedykh, S.E.; Dmitrenok, P.S.; Nevinsky, G.A. Identification of Antibody-Mediated Hydrolysis Sites of Oligopeptides Corresponding to the SARS-CoV-2 S-Protein by MALDI-TOF Mass Spectrometry. Int. J. Mol. Sci. 2023, 24, 14342. [Google Scholar] [CrossRef]
- McConnell, S.A.; Sachithanandham, J.; Mudrak, N.J.; Zhu, X.; Farhang, P.A.; Cordero, R.J.B.; Wear, M.P.; Shapiro, J.R.; Park, H.-S.; Klein, S.L.; et al. Spike-Protein Proteolytic Antibodies in COVID-19 Convalescent Plasma Contribute to SARS-CoV-2 Neutralization. Cell Chem. Biol. 2023, 30, 726–738. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, V.A.; Dolzhikova, I.V.; Shchetinin, A.M.; Odintsova, A.S.; Siniavin, A.E.; Nikiforova, M.A.; Pochtovyi, A.A.; Shidlovskaya, E.V.; Kuznetsova, N.A.; Burgasova, O.A.; et al. Neutralizing Activity of Sera from Sputnik V-Vaccinated People against Variants of Concern (VOC: B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.617.3) and Moscow Endemic SARS-CoV-2 Variants. Vaccines 2021, 9, 779. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Sedykh, S.E.; Ermakov, E.A.; Matveev, A.L.; Odegova, E.I.; Sedykh, T.A.; Shcherbakov, D.N.; Merkuleva, I.A.; Volosnikova, E.A.; Nesmeyanova, V.S.; et al. Natural IgG against S-Protein and RBD of SARS-CoV-2 Do Not Bind and Hydrolyze DNA and Are Not Autoimmune. Int. J. Mol. Sci. 2022, 23, 13681. [Google Scholar] [CrossRef]
- Grant, O.C.; Montgomery, D.; Ito, K.; Woods, R.J. Analysis of the SARS-CoV-2 Spike Protein Glycan Shield Reveals Implications for Immune Recognition. Sci. Rep. 2020, 10, 14991. [Google Scholar] [CrossRef] [PubMed]
- Legostaeva, G.A.; Polosukhina, D.I.; Bezuglova, A.M.; Doronin, B.M.; Buneva, V.N.; Nevinsky, G.A. Affinity and Catalytic Heterogeneity of Polyclonal Myelin Basic Protein-Hydrolyzing IgGs from Sera of Patients with Multiple Sclerosis. J. Cell. Mol. Med. 2009, 14, 699–709. [Google Scholar] [CrossRef]
- Odintsova, E.S.; Baranova, S.V.; Dmitrenok, P.S.; Calmels, C.; Parissi, V.; Andreola, M.-L.; Buneva, V.N.; Nevinsky, G.A. Anti-Integrase Abzymes from the Sera of HIV-Infected Patients Specifically Hydrolyze Integrase but Nonspecifically Cleave Short Oligopeptides. J. Mol. Recognit. 2012, 25, 193–207. [Google Scholar] [CrossRef] [PubMed]
- McLain, L.; Dimmock, N.J. Single- and Multi-Hit Kinetics of Immunoglobulin G Neutralization of Human Immunodeficiency Virus Type 1 by Monoclonal Antibodies. J. Gen. Virol. 1994, 75, 1457–1460. [Google Scholar] [CrossRef] [PubMed]
- Magnus, C. Virus Neutralisation: New Insights from Kinetic Neutralisation Curves. PLoS Comput. Biol. 2013, 9, e1002900. [Google Scholar] [CrossRef]
- Li, C.-J.; Chang, S.-C. SARS-CoV-2 Spike S2-Specific Neutralizing Antibodies. Emerg. Microbes Infect. 2023, 12. [Google Scholar] [CrossRef]
- Gruell, H.; Vanshylla, K.; Weber, T.; Barnes, C.O.; Kreer, C.; Klein, F. Antibody-Mediated Neutralization of SARS-CoV-2. Immunity 2022, 55, 925–944. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Han, X.; Yan, J. Structure-Based Neutralizing Mechanisms for SARS-CoV-2 Antibodies. Emerg. Microbes Infect. 2022, 11, 2412–2422. [Google Scholar] [CrossRef] [PubMed]
- Kamaeva, D.A.; Smirnova, L.P.; Vasilieva, S.N.; Kazantseva, D.V.; Vasilieva, A.R.; Ivanova, S.A. Catalytic Antibodies in Bipolar Disorder: Serum IgGs Hydrolyze Myelin Basic Protein. Int. J. Mol. Sci. 2022, 23, 7397. [Google Scholar] [CrossRef]
- Zavialova, M.; Kamaeva, D.; Kazieva, L.; Skvortsov, V.S.; Smirnova, L. Some Structural Features of the Peptide Profile of Myelin Basic Protein-Hydrolyzing Antibodies in Schizophrenic Patients. PeerJ 2023, 11, e15584. [Google Scholar] [CrossRef]
- WANG, J.; HAN, Y.; WILKINSON, M.F. An Active Immunization Approach to Generate Protective Catalytic Antibodies. Biochem. J. 2001, 360, 151. [Google Scholar] [CrossRef]
- Lacroix-Desmazes, S.; Bayry, J.; Kaveri, S.V.; Hayon-Sonsino, D.; Thorenoor, N.; Charpentier, J.; Luyt, C.-E.; Mira, J.-P.; Nagaraja, V.; Kazatchkine, M.D.; et al. High Levels of Catalytic Antibodies Correlate with Favorable Outcome in Sepsis. Proc. Natl. Acad. Sci. USA 2005, 102, 4109–4113. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Li, L.; Kalaga, R.; Wilkins-Stevens, P.; Stevens, F.J.; Solomon, A. Natural Catalytic Antibodies: Peptide-Hydrolyzing Activities of Bence Jones Proteins and VL Fragment. J. Biol. Chem. 1995, 270, 15257–15261. [Google Scholar] [CrossRef]
- Nevinsky, G.A.; Buneva, V.N. Natural Catalytic Antibodies in Norm, Autoimmune, Viral, and Bacterial Diseases. Sci. World J. 2010, 10, 1203–1233. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Said, S.I.; Thompson, A.B.; Volle, D.J.; Agrawal, D.K.; Foda, H.; de la Rocha, S. Characterization of Autoantibodies to Vasoactive Intestinal Peptide in Asthma. J. Neuroimmunol. 1989, 23, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Castanares-Zapatero, D.; Chalon, P.; Kohn, L.; Dauvrin, M.; Detollenaere, J.; Maertens de Noordhout, C.; Primus-de Jong, C.; Cleemput, I.; Van den Heede, K. Pathophysiology and Mechanism of Long COVID: A Comprehensive Review. Ann. Med. 2022, 54, 1473–1487. [Google Scholar] [CrossRef]
- Bezuglova, A.M.; Konenkova, L.P.; Buneva, V.N.; Nevinsky, G.A. IgGs Containing Light Chains of the λ- and κ- Type and of All Subclasses (IgG1–IgG4) from the Sera of Patients with Systemic Lupus Erythematosus Hydrolyze Myelin Basic Protein. Int. Immunol. 2012, 24, 759–770. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timofeeva, A.M.; Shayakhmetova, L.S.; Nikitin, A.O.; Sedykh, T.A.; Matveev, A.L.; Shanshin, D.V.; Volosnikova, E.A.; Merkuleva, I.A.; Shcherbakov, D.N.; Tikunova, N.V.; et al. Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins. Biomedicines 2024, 12, 1007. https://doi.org/10.3390/biomedicines12051007
Timofeeva AM, Shayakhmetova LS, Nikitin AO, Sedykh TA, Matveev AL, Shanshin DV, Volosnikova EA, Merkuleva IA, Shcherbakov DN, Tikunova NV, et al. Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins. Biomedicines. 2024; 12(5):1007. https://doi.org/10.3390/biomedicines12051007
Chicago/Turabian StyleTimofeeva, Anna M., Liliya Sh. Shayakhmetova, Artem O. Nikitin, Tatyana A. Sedykh, Andrey L. Matveev, Daniil V. Shanshin, Ekaterina A. Volosnikova, Iuliia A. Merkuleva, Dmitriy N. Shcherbakov, Nina V. Tikunova, and et al. 2024. "Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins" Biomedicines 12, no. 5: 1007. https://doi.org/10.3390/biomedicines12051007
APA StyleTimofeeva, A. M., Shayakhmetova, L. S., Nikitin, A. O., Sedykh, T. A., Matveev, A. L., Shanshin, D. V., Volosnikova, E. A., Merkuleva, I. A., Shcherbakov, D. N., Tikunova, N. V., Sedykh, S. E., & Nevinsky, G. A. (2024). Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins. Biomedicines, 12(5), 1007. https://doi.org/10.3390/biomedicines12051007









