The Importance of Sleep in Overcoming Childhood Obesity and Reshaping Epigenetics
Abstract
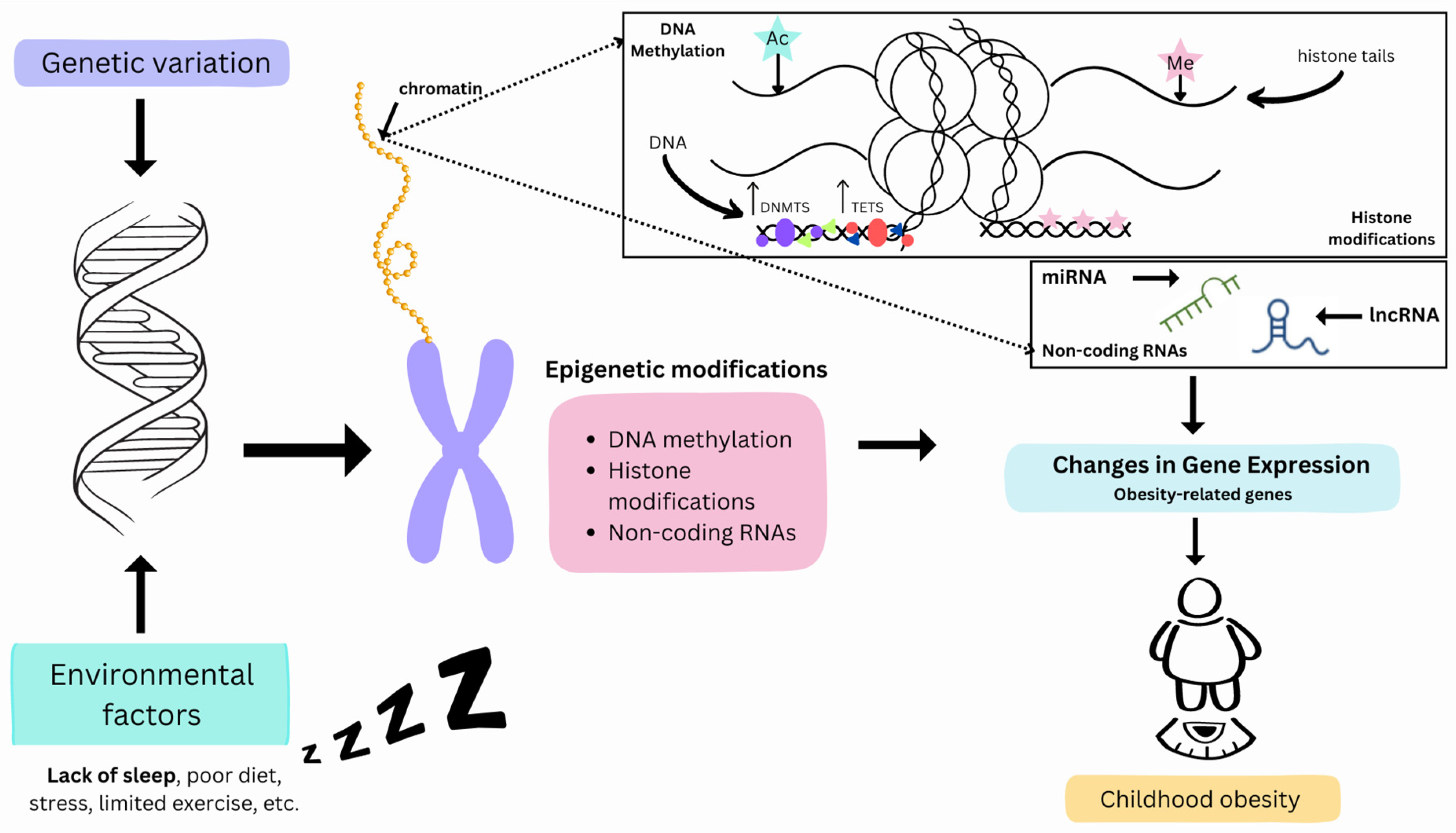
1. Introduction
2. Sleep and Childhood Obesity
3. Sleep and Epigenetics
3.1. DNA Methylation and Sleep
3.2. Histone Modifications and Sleep
3.3. Sleep-Related Non-Coding RNAs
4. Linking Sleep and Childhood Obesity from an Epigenetic Standpoint
5. Transgenerational Epigenetic Changes and Changes in Sleep Patterns
6. Limitations and Future Direction
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Wang, S.; Song, J.; Yang, Y.; Zhang, Y.; Wang, H.; Ma, J. HIF3A DNA methylation is associated with childhood obesity and ALT. PLoS ONE 2015, 10, e0145944. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Obesity and Overweight. World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 5 March 2024).
- Obri, A.; Serra, D.; Herrero, L.; Mera, P. The role of epigenetics in the development of obesity. Biochem. Pharmacol. 2020, 177, 113973. [Google Scholar] [CrossRef]
- Hochberg, Z. An evolutionary perspective on the obesity epidemic. Trends Endocrinol. Metab. 2018, 29, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. What Is Epigenetics? Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/genomics/disease/epigenetics.htm (accessed on 15 August 2022).
- Milagro, F.I.; Gómez-Abellán, P.; Campión, J.; Martínez, J.A.; Ordovás, J.M.; Garaulet, M. CLOCK, PER2 and BMAL1 DNA methylation: Association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chrono Int. 2012, 29, 1180–1194. [Google Scholar] [CrossRef]
- Huang, H.; Lin, S.; Garcia, B.A.; Zhao, Y. Quantitative proteomic analysis of histone modifications. Chem. Rev. 2015, 115, 2376–2418. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Babu, J.R.; Wang, X.; Geetha, T. Role of macronutrient intake in the epigenetics of obesity. Biochem. Soc. Trans. 2022, 50, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Miska, E.A.; Ferguson-Smith, A.C. Transgenerational inheritance: Models and mechanisms of non-DNA sequence-based inheritance. Science 2016, 354, 59–63. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Laird, P.W. Interplay between the cancer genome and epigenome. Cell 2013, 153, 38–55. [Google Scholar] [CrossRef]
- Zenk, F.; Loeser, E.; Schiavo, R.; Kilpert, F.; Bogdanović, O.; Iovino, N. Germ line–inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 2017, 357, 212–216. [Google Scholar] [CrossRef]
- Ramar, K.; Malhotra, R.K.; Carden, K.A.; Martin, J.L.; Abbasi-Feinberg, F.; Aurora, R.N.; Kapur, V.K.; Olson, E.J.; Rosen, C.L.; Rowley, J.A.; et al. Sleep is essential to health: An American Academy of Sleep Medicine position statement. J. Clin. Sleep Med. 2021, 17, 2115–2119. [Google Scholar] [CrossRef] [PubMed]
- Paruthi, S.; Brooks, L.J.; D’Ambrosio, C.; Hall, W.A.; Kotagal, S.; Lloyd, R.M.; Malow, B.A.; Maski, K.; Nichols, C.; Quan, S.F.; et al. Recommended Amount of Sleep for Pediatric Populations: A Consensus Statement of the American Academy of Sleep Medicine. J. Clin. Sleep Med. 2016, 12, 785–786. [Google Scholar] [CrossRef]
- Campbell, S.S.; Tobler, I. Animal sleep: A review of sleep duration across phylogeny. Neurosci. Biobehav. Rev. 1984, 8, 269–300. [Google Scholar] [CrossRef] [PubMed]
- Joiner, W.J. Unraveling the evolutionary determinants of sleep. Curr. Biol. 2016, 26, R1073–R1087. [Google Scholar] [CrossRef]
- Yin, J.; Jin, X.; Shan, Z.; Li, S.; Huang, H.; Li, P.; Peng, X.; Peng, Z.; Yu, K.; Bao, W.; et al. Relationship of sleep duration with all-cause mortality and cardiovascular events: A systematic review and dose-response meta-analysis of prospective cohort studies. J. Am. Heart Assoc. 2017, 6, e005947. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. (n.d.). FastStats: Sleep in Children. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/sleep/data-research/facts-stats/children-sleep-facts-and-stats.html (accessed on 5 March 2024).
- Liu, Y.; Wheaton, A.; Chapman, D.; Cunningham, T.; Lu, H.; Croft, J. Prevalence of healthy sleep duration among adults—United States, 2014. Morb. Mortal. Wkly. Rep. 2016, 65, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Dashti, H.S.; Cade, E.B.; Stutaite, G.; Saxena, R.; Redline, S.; Karlson, E.W. Sleep health, diseases, and pain syndromes: Findings from an electronic health record biobank. Sleep 2020, 44, zsaa189. [Google Scholar] [CrossRef]
- Cappuccio, F.P.; Taggart, F.M.; Kandala, N.-B.; Currie, A.; Peile, E.; Stranges, S.; Miller, M.A. Meta-analysis of short sleep duration and obesity in children and adults. Sleep 2008, 31, 619–626. [Google Scholar] [CrossRef]
- Ogilvie, R.P.; Patel, S.R. The epidemiology of sleep and obesity. Sleep Health 2017, 3, 383–388. [Google Scholar] [CrossRef]
- Patel, S.R.; Hu, F.B. Short sleep duration and weight gain: A systematic review. Obesity 2008, 16, 643–653. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, Q.; Zou, Z.; Li, H.; Zhang, X. Short sleep duration and obesity among children: A systematic review and meta-analysis of prospective studies. Obes. Res. Clin. Pract. 2017, 11, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Cappuccio, F.P.; D’Elia, L.; Strazzullo, P.; Miller, M.A. Quantity and quality of sleep and incidence of type 2 diabetes: A systematic review and meta-analysis. Diabetes Care 2010, 33, 414–420. [Google Scholar] [CrossRef]
- Daghlas, I.; Dashti, H.S.; Lane, J.; Aragam, K.G.; Rutter, M.K.; Saxena, R.; Vetter, C. Sleep Duration and myocardial infarction. J. Am. Coll. Cardiol. 2019, 74, 1304–1314. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lane, J.M.; Jones, S.E.; Dashti, H.S.; Ollila, H.M.; Wood, A.R.; van Hees, V.T.; Brumpton, B.; Winsvold, B.S.; Kantojärvi, K.; et al. Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat. Commun. 2019, 10, 3503. [Google Scholar] [CrossRef]
- Barnes, C.M.; Drake, C.L. Prioritizing sleep health: Public health policy recommendations. Perspect. Psychol. Sci. 2015, 10, 733–737. [Google Scholar] [CrossRef]
- Luyster, F.S.; Strollo, P.J.; Zee, P.C.; Walsh, J.K. Sleep: A health imperative. Sleep 2012, 35, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Deacon-Crouch, M.; Begg, S.; Skinner, T. Is sleep duration associated with overweight/obesity in indigenous Australian adults? BMC Public Health 2020, 20, 1229. [Google Scholar] [CrossRef] [PubMed]
- Sa, J.; Choe, S.; Cho, B.Y.; Chaput, J.P.; Kim, G.; Park, C.H.; Chung, J.; Choi, Y.; Nelson, B.; Kim, Y. Relationship between sleep and obesity among U.S. and South Korean college students. BMC Public Health 2020, 20, 96. [Google Scholar] [CrossRef]
- Dashti, H.S.; Ordovás, J.M. Genetics of Sleep and Insights into Its Relationship with Obesity. Annu. Rev. Nutr. 2021, 41, 223–252. [Google Scholar] [CrossRef] [PubMed]
- Stranges, S.; Cappuccio, F.P.; Kandala, N.B.; Miller, M.A.; Taggart, F.M.; Kumari, M.; Ferrie, J.E.; Shipley, M.J.; Brunner, E.J.; Marmot, M.G. Cross-sectional versus prospective associations of sleep duration with changes in relative weight and body fat distribution: The Whitehall II Study. Am. J. Epidemiol. 2008, 167, 321–329. [Google Scholar] [CrossRef]
- Mindell, J.A.; Owens, J.A. A Clinical Guide to Pediatric Sleep: Diagnosis and Management of Sleep Problems; Lippincott Williams and Wilkins: Philadelphia, PA, USA, 2015. [Google Scholar]
- Liu, J.; Feng, R.; Ji, X.; Cui, N.; Raine, A.; Mednick, S.C. Midday napping in children: Associations between nap frequency and duration across cognitive, positive psychological well-being, behavioral, and metabolic health outcomes. Sleep 2019, 42, zsz126. [Google Scholar] [CrossRef] [PubMed]
- Honaker, S.M.; Meltzer, L.J. Sleep in pediatric primary care: A review of the literature. Sleep Med. Rev. 2016, 25, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Ranum, B.M.; Wichstrøm, L.; Pallesen, S.; Steinsbekk, S. Prevalence and stability of insufficient sleep measured by actigraphy: A prospective community study. Pediatr. Res. 2020, 88, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Chattu, V.K.; Sakhamuri, S.M.; Kumar, R.; Spence, D.W.; BaHammam, A.S.; Pandi-Perumal, S.R. Insufficient sleep syndrome: Is it time to classify it as a major noncommunicable disease? Sleep Sci. 2018, 11, 56. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. (n.d.-a). About Sleep. Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/sleep/about/?CDC_AAref_Val=https%3A%2F%2Fwww.cdc.gov%2Fsleep%2Fabout_sleep%2Fhow_much_sleep.html (accessed on 15 May 2024).
- Laposky, A.D.; Van Cauter, E.; Diez-Roux, A.V. Reducing health disparities: The role of sleep deficiency and sleep disorders. Sleep Med. 2016, 18, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Skjåkødegård, H.F.; Danielsen, Y.S.; Frisk, B.; Hystad, S.W.; Roelants, M.; Pallesen, S.; Conlon, R.P.K.; Wilfley, D.E.; Juliusson, P.B. Beyond sleep duration: Sleep timing as a risk factor for childhood obesity. Pediatr. Obes. 2021, 16, e12698. [Google Scholar] [CrossRef]
- Patel, S.R.; Hayes, A.L.; Blackwell, T.; Evans, D.S.; Ancoli-Israel, S.; Wing, Y.K.; Stone, K.L. The association between sleep patterns and obesity in older adults. Int. J. Obes. 2014, 38, 1159–1164. [Google Scholar] [CrossRef]
- Miller, A.L.; Kaciroti, N.; LeBourgeois, M.K.; Chen, Y.P.; Sturza, J.; Lumeng, J.C. Sleep timing moderates the concurrent sleep duration–body mass index association in low-income preschool-age children. Acad. Pediatr. 2014, 14, 207–213. [Google Scholar] [CrossRef]
- Golley, R.K.; Maher, A.C.; Matricciani, L.; Olds, T.S. Sleep duration or bedtime? Exploring the association between sleep timing behaviour, diet and BMI in children and adolescents. Int. J. Obes. 2013, 37, 546–551. [Google Scholar] [CrossRef]
- Jarrin, D.C.; McGrath, J.J.; Drake, C.L. Beyond sleep duration: Distinct sleep dimensions are associated with obesity in children and adolescents. Int. J. Obes. 2013, 37, 552–558. [Google Scholar] [CrossRef]
- Adamo, K.B.; Wilson, S.; Belanger, K.; Chaput, J.-P. Later bedtime is associated with greater daily energy intake and screen time in obese adolescents independent of sleep duration. J. Sleep Disord. Ther. 2013, 2, 126–130. [Google Scholar] [CrossRef]
- Olds, T.S.; Maher, C.A.; Matricciani, L. Sleep duration or bedtime? Exploring the relationship between sleep habits and weight status and activity patterns. Sleep 2011, 34, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.L.; Lumeng, J.C.; LeBourgeois, M.K. Sleep patterns and obesity in childhood. Curr. Opin. Endocrinol. Diabetes 2015, 22, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, B.; Taveras, E.; Allender, S.; Strugnell, C. Sleep and obesity among children: A systematic review of multiple sleep dimensions. Pediatr. Obes. 2020, 15, e12619. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Lin, L.; Austin, D.; Young, T.; Mignot, E. Short sleep duration is associated with reduced leptin, elevated ghrelin, and increased body mass index. PLOS Med. 2004, 1, e62. [Google Scholar] [CrossRef] [PubMed]
- Boeke, C.E.; Storfer-Isser, A.; Redline, S.; Taveras, E.M. Childhood sleep duration and quality in relation to leptin concentration in two cohort studies. Sleep 2013, 37, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Hart, C.N.; Carskadon, M.A.; Considine, R.V.; Fava, J.L.; Lawton, J.; Raynor, H.A.; Jelalian, E.; Owens, J.; Wing, R. Changes in children’s sleep duration on food intake, weight, and leptin. Pediatrics 2013, 132, e1473–e1480. [Google Scholar] [CrossRef] [PubMed]
- St-Onge, M.-P. The Role of sleep duration in the regulation of energy balance: Effects on energy intakes and expenditure. J. Clin. Sleep Med. 2013, 9, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Chaput, J.P.; St-Onge, M.P. Increased food intake by insufficient sleep in humans: Are we jumping the gun on the hormonal explanation? Front. Endocrinol. 2014, 5, 116. [Google Scholar] [CrossRef]
- Gonnissen, H.K.; Hulshof, T.; Westerterp-Plantenga, M.S. Chronobiology, endocrinology, and energy- and food-reward homeostasis. Obes. Rev. 2013, 14, 405–416. [Google Scholar] [CrossRef]
- Garaulet, M.; Gómez-Abellán, P. Timing of food intake and obesity: A novel association. Physiol. Behav. 2014, 134, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Magee, C.A.; Caputi, P.; Iverson, D.C. The longitudinal relationship between sleep duration and body mass index in children: A growth mixture modeling approach. J. Dev. Behav. Pediatr. 2013, 34, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Figueroa, R.; Brink, H.V.; Vorland, C.J.; Auckburally, S.; Johnson, L.; Garay, J.; Brown, T.; Simon, S.; Ells, L. The efficacy of sleep lifestyle interventions for the management of overweight or obesity in children: A systematic review and meta-analysis. BMC Public Health 2024, 24, 321. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M. An Overview of Epigenetics in Obesity: The Role of Lifestyle and Therapeutic Interventions. Int. J. Mol. Sci. 2022, 23, 1341. [Google Scholar] [CrossRef] [PubMed]
- Gaine, M.E.; Chatterjee, S.; Abel, T. Sleep Deprivation and the Epigenome. Front. Neural Circuits 2018, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef] [PubMed]
- Nan, X.; Ng, H.H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a, histone deacetylase complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef]
- Espada, J.; Esteller, M. Mouse models in epigenetics: Insights in development and disease. Briefings Funct. Genom. 2013, 12, 279–287. [Google Scholar] [CrossRef]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Kaiser, S.; Jurkowski, T.P.; Kellner, S.; Schneider, D.; Jeltsch, A.; Helm, M. The RNA methyltransferase Dnmt2 methylates DNA in the structural context of a tRNA. RNA Biol. 2016, 14, 1241–1251. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, M. Distribution of 5-hydroxymethylcytosine in different human tissues. J. Nucleic Acids 2011, 2011, 870726. [Google Scholar] [CrossRef] [PubMed]
- Khare, T.; Pai, S.; Koncevicius, K.; Pal, M.; Kriukiene, E.; Liutkeviciute, Z.; Irimia, M.; Jia, P.; Ptak, C.; Xia, M.; et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exonintron boundary. Nat. Struct. Mol. Biol. 2012, 19, 1037–1043. [Google Scholar] [CrossRef]
- Wong, C.C.; Parsons, M.J.; Lester, K.J.; Burrage, J.; Eley, T.C.; Mill, J.; Dempster, E.L.; Gregory, A.M. Epigenome-wide DNA methylation analysis of monozygotic twins discordant for diurnal preference. Twin Res. Hum. Genet. 2015, 18, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Zhu, Y.; Eliot, M.N.; Knopik, V.S.; McGeary, J.E.; Carskadon, M.A.; Hart, A.C. Combining human epigenetics and sleep studies in caenorhabditis elegans: A cross-species approach for finding conserved genes regulating sleep. Sleep 2017, 40, zsx063. [Google Scholar] [CrossRef]
- Qureshi, I.A.; Mehler, M.F. Epigenetics of Sleep and Chronobiology. Curr. Neurol. Neurosci. Rep. 2014, 14, 432. [Google Scholar] [CrossRef] [PubMed]
- Cedernaes, J.; Osler, M.E.; Voisin, S.; Broman, J.-E.; Vogel, H.; Dickson, S.L.; Zierath, J.R.; Schiöth, H.B.; Benedict, C. Acute sleep loss induces tissue-specific epigenetic and transcriptional alterations to circadian clock genes in men. J. Clin. Endocrinol. Metab. 2015, 100, E1255–E1261. [Google Scholar] [CrossRef]
- Mews, P.; Donahue, G.; Drake, A.M.; Luczak, V.; Abel, T.; Berger, S.L. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 2017, 546, 381–386. [Google Scholar] [CrossRef]
- Skuladottir, G.V.; Nilsson, E.K.; Mwinyi, J.; Schiöth, H.B. One-night sleep deprivation induces changes in the DNA methylation and serum activity indices of stearoyl-CoA desaturase in young healthy men. Lipids Health Dis. 2016, 15, 137. [Google Scholar] [CrossRef] [PubMed]
- Massart, R.; Freyburger, M.; Suderman, M.; Paquet, J.; El Helou, J.; Belanger-Nelson, E.; Rachalski, A.; Koumar, O.C.; Carrier, J.; Szyf, M.; et al. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl. Psychiatry 2014, 4, e347. [Google Scholar] [CrossRef] [PubMed]
- Möller-Levet, C.S.; Archer, S.N.; Bucca, G.; Laing, E.E.; Slak, A.; Kabiljo, R.; Lo, J.C.; Santhi, N.; von Schantz, M.; Smith, C.P.; et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl. Acad. Sci. USA 2013, 110, E1132–E1141. [Google Scholar] [CrossRef]
- Doyon, Y.; Cayrou, C.; Ullah, M.; Landry, A.-J.; Côté, V.; Selleck, W.; Lane, W.S.; Tan, S.; Yang, X.-J.; Côté, J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 2006, 21, 51–64. [Google Scholar] [CrossRef] [PubMed]
- O’callaghan, E.K.; Roig, M.N.B.; Mongrain, V. Cell adhesion molecules and sleep. Neurosci. Res. 2017, 116, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, L.; Abel, T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 2013, 38, 62–76. [Google Scholar] [CrossRef]
- Hernandez, P.J.; Abel, T. The role of protein synthesis in memory consolidation: Progress amid decades of debate. Neurobiol. Learn. Mem. 2008, 89, 293–311. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, L.L.; Wimmer, M.E.; Poplawski, S.G.; Tudor, J.C.; Kenworthy, C.A.; Liu, S.; Mizuno, K.; Garcia, B.A.; Zhang, N.R.; Giese, K.P.; et al. Memory acquisition and retrieval impact different epigenetic processes that regulate gene expression. BMC Genom. 2015, 16, S5. [Google Scholar] [CrossRef] [PubMed]
- Doi, M.; Hirayama, J.; Sassone-Corsi, P. Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell 2006, 125, 497–508. [Google Scholar] [CrossRef]
- Hirayama, J.; Sahar, S.; Grimaldi, B.; Tamaru, T.; Takamatsu, K.; Nakahata, Y.; Sassone-Corsi, P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 2007, 450, 1086–1090. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Lee, C.; Wade, P.A.; Reppert, S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 2003, 421, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Seo, S.B.; Westgate, E.J.; Rudic, R.D.; Smyth, E.M.; Chakravarti, D.; FitzGerald, G.A.; McNamara, P. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2004, 279, 7091–7097. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, L.; Qin, C. Long non-coding RNAs in brain development, synaptic biology, and Alzheimer’s disease. Brain Res. Bull. 2017, 132, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, J.; Pandey, G.K.; Kanduri, C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim. Biophys. Acta BBA Gen. Subj. 2009, 1790, 936–947. [Google Scholar] [CrossRef]
- Powell, W.T.; Coulson, R.L.; Crary, F.K.; Wong, S.S.; Ach, R.A.; Tsang, P.; Yamada, N.A.; Yasui, D.H.; LaSalle, J.M. A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet. 2013, 22, 4318–4328. [Google Scholar] [CrossRef]
- Sammallahti, S.; Koopman-Verhoeff, M.E.; Binter, A.-C.; Mulder, R.H.; Cabré-Riera, A.; Kvist, T.; Malmberg, A.L.K.; Pesce, G.; Plancoulaine, S.; Heiss, J.A.; et al. Longitudinal associations of DNA methylation and sleep in children: A meta-analysis. Clin. Epigenetics 2022, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Leader, B.A.; Koritala, B.S.C.; Moore, C.A.; Grigg Dean, E.H.; Kottyan, L.C.; Smith, D.F. Epigenetics of obstructive sleep apnea syndrome: A systematic review. JCSM J. Clin. Sleep Med. 2021, 17, 2533–2541. [Google Scholar] [CrossRef]
- Lahtinen, A.; Puttonen, S.; Vanttola, P.; Viitasalo, K.; Sulkava, S.; Pervjakova, N.; Joensuu, A.; Salo, P.; Toivola, A.; Härmä, M.; et al. A distinctive DNA methylation pattern in insufficient sleep. Sci. Rep. 2019, 9, 1193. [Google Scholar] [CrossRef] [PubMed]
- Champagne, F.A. Epigenetic mechanisms and the transgenerational effects of maternal care. Front. Neuroendocrinol. 2008, 29, 386–397. [Google Scholar] [CrossRef]
- Chang, J.J.; Pien, G.W.; Duntley, S.P.; Macones, G.A. Sleep deprivation during pregnancy and maternal and fetal outcomes: Is there a relationship? Sleep Med. Rev. 2010, 14, 107–114. [Google Scholar] [CrossRef]
- Gazerani, P. Epigenetics of Sleep Disruption. OBM Neurobiol. 2020, 4, 1–25. [Google Scholar] [CrossRef]
- Davenport, M.H.; Meah, V.L.; Ruchat, S.M.; Davies, G.A.; Skow, R.J.; Barrowman, N.; Adamo, K.B.; Poitras, V.J.; Gray, C.E.; Garcia, A.J.; et al. Impact ofprenatal exercise on neonatal and childhood outcomes: A systematic review and meta-analysis. Br. J. Sports Med. 2018, 52, 1386–1396. [Google Scholar] [CrossRef] [PubMed]
- Rogozińska, E.; Marlin, N.; Jackson, L.; Rayanagoudar, G.; E Ruifrok, A.; Dodds, J.; Molyneaux, E.; van Poppel, M.N.; Poston, L.; A Vinter, C.; et al. Effects of antenatal diet and physical activity on maternal and fetal outcomes: Individual patient data meta-analysis and health economic evaluation. Health Technol. Assess. 2017, 21, 1–158. [Google Scholar] [CrossRef] [PubMed]
- Mindell, J.A.; Cook, R.A.; Nikolovski, J. Sleep patterns and sleep disturbances across pregnancy. Sleep Med. 2015, 16, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, Z.M.; Chaput, J.-P.; Gruslin, A.; Adamo, K.B. The potential value of sleep hygiene for a healthy pregnancy: A brief review. ISRN Fam. Med. 2014, 2014, 928293. [Google Scholar] [CrossRef]
- Kawai, T.; Yamada, T.; Abe, K.; Okamura, K.; Kamura, H.; Akaishi, R.; Minakami, H.; Nakabayashi, K.; Hata, K. Increased epigenetic alterations at the promoters of transcriptional regulators following inadequate maternal gestational weight gain. Sci. Rep. 2015, 5, 14224. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.E.; Ross, K.M.; Horvath, S.; Okun, M.; Hobel, C.; Rentscher, K.E.; Coussons-Read, M.; Schetter, C.D. Postpartum sleep loss and accelerated epigenetic aging. Sleep Health 2021, 7, 362–367. [Google Scholar] [CrossRef]
- Harskamp-van Ginkel, M.W.; Ierodiakonou, D.; Margetaki, K.; Vafeiadi, M.; Karachaliou, M.; Kogevinas, M.; Vrijkotte, T.G.M.; Chatzi, L. Gestational sleep deprivation is associated with higher offspring body mass index and blood pressure. Sleep 2020, 43, zsaa110. [Google Scholar] [CrossRef] [PubMed]
- Briollais, L.; Rustand, D.; Allard, C.; Wu, Y.; Xu, J.; Rajan, S.G.; Hivert, M.-F.; Doyon, M.; Bouchard, L.; McGowan, P.O.; et al. DNA methylation mediates the association between breastfeeding and early-life growth trajectories. Clin. Epigenet. 2021, 13, 231. [Google Scholar] [CrossRef]
- Jafar, N.K.A.; Tham, E.K.; Pang, W.W.; Fok, D.; Chua, M.C.; Teoh, O.H.; Goh, D.Y.; Shek, L.P.; Yap, F.; Tan, K.H.; et al. Association between breastfeeding and sleep patterns in infants and preschool children. Am. J. Clin. Nutr. 2021, 114, 1986–1996. [Google Scholar] [CrossRef]
- Meng, M.; Jiang, Y.; Lin, J.; Zhang, J.; Wang, G.; Zhu, Q.; Lin, Q.; Jiang, F. The mediating effect of DNA methylation in the association between maternal sleep during pregnancy and offspring adiposity status: A prospective cohort study. Clin. Epigenet. 2022, 14, 66. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richter, E.; Patel, P.; Babu, J.R.; Wang, X.; Geetha, T. The Importance of Sleep in Overcoming Childhood Obesity and Reshaping Epigenetics. Biomedicines 2024, 12, 1334. https://doi.org/10.3390/biomedicines12061334
Richter E, Patel P, Babu JR, Wang X, Geetha T. The Importance of Sleep in Overcoming Childhood Obesity and Reshaping Epigenetics. Biomedicines. 2024; 12(6):1334. https://doi.org/10.3390/biomedicines12061334
Chicago/Turabian StyleRichter, Erika, Priyadarshni Patel, Jeganathan Ramesh Babu, Xu Wang, and Thangiah Geetha. 2024. "The Importance of Sleep in Overcoming Childhood Obesity and Reshaping Epigenetics" Biomedicines 12, no. 6: 1334. https://doi.org/10.3390/biomedicines12061334
APA StyleRichter, E., Patel, P., Babu, J. R., Wang, X., & Geetha, T. (2024). The Importance of Sleep in Overcoming Childhood Obesity and Reshaping Epigenetics. Biomedicines, 12(6), 1334. https://doi.org/10.3390/biomedicines12061334






