Predictive Value and Diagnostic Potential of IL-10, IL-17A, IL1-β, IL-6, CXCL, and MCP for Severe COVID-19 and COVID-19 Mortality
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Inclusion and Exclusion Criteria
2.3. Biochemical Analysis
2.4. Statistical Analysis
3. Results
4. Discussion
4.1. Literature Findings
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sawicka, B.; Aslan, I.; Della Corte, V.; Periasamy, A.; Krishnamurthy, S.K.; Mohammed, A.; Tolba Said, M.M.; Saravanan, P.; Del Gaudio, G.; Adom, D.; et al. The coronavirus global pandemic and its impacts on society. In Coronavirus Drug Discovery: SARS-CoV-2 (COVID-19) Prevention, Diagnosis, and Treatment. Volume 1 in Drug Discovery Update; Elsevier: Amsterdam, The Netherlands, 2022; pp. 267–311. [Google Scholar] [CrossRef] [PubMed Central]
- Del Re, D.; Palla, L.; Meridiani, P.; Soffi, L.; Loiudice, M.T.; Antinozzi, M.; Cattaruzza, M.S. The spread in time and space of COVID-19 pandemic waves: The Italian experience from mortality data analyses. Front. Public Health 2024, 12, 1324033. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Padilla-Bórquez, D.L.; Matuz-Flores, M.G.; Hernández-Bello, J.; Sánchez-Zuno, G.A.; García-Arellano, S.; Oregon-Romero, E.; Herrera-Godina, M.G.; González-Estevez, G.; Adan-Bante, N.P.; Rosas-Rodríguez, J.A.; et al. Seroprevalence of IgM/IgG and Neutralizing Antibodies against SARS-CoV-2 in Unvaccinated Young Adults from Mexico Who Reported Not Having Had a Previous COVID-19 Infection. Can. J. Infect. Dis. Med. Microbiol. 2024, 2024, 8871439. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Çelik, I.; Öztürk, R. From asymptomatic to critical illness: Decoding various clinical stages of COVID-19. Turk. J. Med. Sci. 2021, 51, 3284–3300. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Marks, M.; O’Hara, G.; Houlihan, C.; Bell, L.; Heightman, M.; Hart, N. Severe Acute Respiratory Syndrome Coronavirus 2. In Encyclopedia of Respiratory Medicine, 2nd ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 231–242. [Google Scholar] [CrossRef] [PubMed Central]
- Maveddat, A.; Mallah, H.; Rao, S.; Ali, K.; Sherali, S.; Nugent, K. Severe Acute Respiratory Distress Syndrome Secondary to Coronavirus 2 (SARS-CoV-2). Int. J. Occup. Environ. Med. 2020, 11, 157–178. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Singhal, T. A Review of Coronavirus Disease-2019 (COVID-19). Indian J. Pediatr. 2020, 87, 281–286. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krynytska, I.; Marushchak, M.; Birchenko, I.; Dovgalyuk, A.; Tokarskyy, O. COVID-19-associated acute respiratory distress syndrome versus classical acute respiratory distress syndrome (a narrative review). Iran. J. Microbiol. 2021, 13, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Empson, S.; Rogers, A.J.; Wilson, J.G. COVID-19 Acute Respiratory Distress Syndrome: One Pathogen, Multiple Phenotypes. Crit. Care Clin. 2022, 38, 505–519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, R.; Liu, Y.; Zhang, B.; Wu, C.; Zhou, J.; Zhang, Y.; Yang, W.; Li, Z.; Shi, S. Coagulopathy is associated with multiple organ damage and prognosis of COVID-19. EXCLI J. 2021, 20, 174–191. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, T.; Zuo, Z.; Kang, S.; Jiang, L.; Luo, X.; Xia, Z.; Liu, J.; Xiao, X.; Ye, M.; Deng, M. Multi-organ Dysfunction in Patients with COVID-19: A Systematic Review and Meta-analysis. Aging Dis. 2020, 11, 874–894. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kabir, M.A.; Ahmed, R.; Iqbal, S.M.A.; Chowdhury, R.; Paulmurugan, R.; Demirci, U.; Asghar, W. Diagnosis for COVID-19: Current status and future prospects. Expert Rev. Mol. Diagn. 2021, 21, 269–288. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yüce, M.; Filiztekin, E.; Özkaya, K.G. COVID-19 diagnosis -A review of current methods. Biosens. Bioelectron. 2021, 172, 112752. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nojiri, S.; Irie, Y.; Kanamori, R.; Naito, T.; Nishizaki, Y. Mortality Prediction of COVID-19 in Hospitalized Patients Using the 2020 Diagnosis Procedure Combination Administrative Database of Japan. Intern Med. 2023, 62, 201–213. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Villegas, M.; Gonzalez-Agirre, A.; Gutiérrez-Fandiño, A.; Armengol-Estapé, J.; Carrino, C.P.; Pérez-Fernández, D.; Soares, F.; Serrano, P.; Pedrera, M.; García, N.; et al. Predicting the evolution of COVID-19 mortality risk: A Recurrent Neural Network approach. Comput. Methods Programs Biomed. 2023, 3, 100089. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rai, N.; Kaushik, N.; Kumar, D.; Raj, C.; Ali, A. Mortality prediction of COVID-19 patients using soft voting classifier. Int. J. Cogn. Comput. Eng. 2022, 3, 172–179. [Google Scholar] [CrossRef] [PubMed Central]
- Moulaei, K.; Ghasemian, F.; Bahaadinbeigy, K.; Ershad Sarbi, R.; Mohamadi Taghiabad, Z. Predicting Mortality of COVID-19 Patients based on Data Mining Techniques. J. Biomed. Phys. Eng. 2021, 11, 653–662. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Onuk, S.; Sipahioğlu, H.; Karahan, S.; Yeşiltepe, A.; Kuzugüden, S.; Karabulut, A.; Beştepe Dursun, Z.; Akın, A. Cytokine Levels and Severity of Illness Scoring Systems to Predict Mortality in COVID-19 Infection. Healthcare 2023, 11, 387. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, H.; Pan, H.; Li, R.; He, K.; Zhang, H.; Liu, L. Increased Circulating Cytokines Have a Role in COVID-19 Severity and Death with a More Pronounced Effect in Males: A Systematic Review and Meta-Analysis. Front. Pharmacol. 2022, 13, 802228. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lucijanić, M.; Piskač Živković, N.; Režić, T.; Durlen, I.; Stojić, J.; Jurin, I.; Šakota, S.; Filipović, D.; Kurjaković, I.; Jordan, A.; et al. The performance of the WHO COVID-19 severity classification, COVID-GRAM, VACO Index, 4C Mortality, and CURB-65 prognostic scores in hospitalized COVID-19 patients: Data on 4014 patients from a tertiary center registry. Croat. Med. J. 2023, 64, 13–20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Leulseged, T.W.; Hassen, I.S.; Ayele, B.T.; Tsegay, Y.G.; Abebe, D.S.; Edo, M.G.; Maru, E.H.; Zewde, W.C.; Naylor, L.K.; Semane, D.F.; et al. Laboratory biomarkers of COVID-19 disease severity and outcome: Findings from a developing country. PLoS ONE 2021, 16, e0246087. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kumari, S.; Nayak, S.; Tripathy, S.; Bhuniya, S.; Mangaraj, M.; Ramadass, B.; Sahu, S.; Bandyopadhyay, D.; Dash, P.; Saharia, G.K. Analysis of Biochemical and Inflammatory Markers for Predicting COVID-19 Severity: Insights from a Tertiary Healthcare Institution of Eastern India. Cureus 2023, 15, e33893. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hariyanto, T.I.; Japar, K.V.; Kwenandar, F.; Damay, V.; Siregar, J.I.; Lugito, N.P.H.; Tjiang, M.M.; Kurniawan, A. Inflammatory and hematologic markers as predictors of severe outcomes in COVID-19 infection: A systematic review and meta-analysis. Am. J. Emerg. Med. 2021, 41, 110–119. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, J.J.; Cao, Y.Y.; Tan, G.; Dong, X.; Wang, B.C.; Lin, J.; Yan, Y.Q.; Liu, G.H.; Akdis, M.; Akdis, C.A.; et al. Clinical, radiological, and laboratory characteristics and risk factors for severity and mortality of 289 hospitalized COVID-19 patients. Allergy 2021, 76, 533–550. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Park, M.; Hur, M.; Kim, H.; Lee, C.H.; Lee, J.H.; Kim, H.W.; Nam, M. Prognostic Utility of Procalcitonin, Presepsin, and the VACO Index for Predicting 30-day Mortality in Hospitalized COVID-19 Patients. Ann. Lab. Med. 2022, 42, 406–414. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kattner, S.; Sutharsan, S.; Berger, M.M.; Limmer, A.; Jehn, L.B.; Herbstreit, F.; Brenner, T.; Taube, C.; Bonella, F. Serum KL-6 as a Candidate Predictor of Outcome in Patients with SARS-CoV-2 Pneumonia. J. Clin. Med. 2023, 12, 6772. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- d’Alessandro, M.; Bergantini, L.; Cavallaro, D.; Gangi, S.; Cameli, P.; Conticini, E.; Siena COVID Unit Frediani, B.; Dotta, F.; Bargagli, E. Krebs von den Lungen-6 as Disease Severity Marker for COVID-19 Patients: Analytical Verification and Quality Assessment of the Tosoh AIA-360 Compared to Lumipulse G600II. Int. J. Environ. Res. Public Health 2022, 19, 2176. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Letellier, A.; Rolland-Debord, C.; Luque-Paz, D.; Milon, A.; Choinier, P.; Blin, E.; Halitim, P.; Bravais, J.; Lefèvre, G.; Parrot, A.; et al. Prognostic value of serum Krebs von den Lungen-6 (KL-6) levels in COVID-19 pneumonia. Respir. Med. Res. 2023, 84, 101054. [Google Scholar] [CrossRef] [PubMed]
- Cabaro, S.; D’Esposito, V.; Di Matola, T.; Sale, S.; Cennamo, M.; Terracciano, D.; Parisi, V.; Oriente, F.; Portella, G.; Beguinot, F.; et al. Cytokine signature and COVID-19 prediction models in the two waves of pandemics. Sci. Rep. 2021, 11, 20793. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mandel, M.; Harari, G.; Gurevich, M.; Achiron, A. Cytokine prediction of mortality in COVID19 patients. Cytokine 2020, 134, 155190. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Basheer, M.; Saad, E.; Kananeh, M.; Asad, L.; Khayat, O.; Badarne, A.; Abdo, Z.; Arraf, N.; Milhem, F.; Bassal, T.; et al. Cytokine Patterns in COVID-19 Patients: Which Cytokines Predict Mortality and Which Protect Against? Curr. Issues Mol. Biol. 2022, 44, 4735–4747. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Herr, C.; Mang, S.; Mozafari, B.; Guenther, K.; Speer, T.; Seibert, M.; Srikakulam, S.K.; Beisswenger, C.; Ritzmann, F.; Keller, A.; et al. Distinct Patterns of Blood Cytokines Beyond a Cytokine Storm Predict Mortality in COVID-19. J. Inflamm. Res. 2021, 14, 4651–4667. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ochoa-Ramirez, L.A.; Ramos-Payan, R.; Jimenez-Gastelum, G.R.; Rodriguez-Millan, J.; Aguilar-Medina, M.; Rios-Tostado, J.J.; Ayala-Ham, A.; Bermudez, M.; Osuna-Ramos, J.F.; Olimon-Andalon, V.; et al. The Chemokine MIG is Associated with an Increased Risk of COVID-19 Mortality in Mexican Patients. Iran. J. Immunol. 2022, 19, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Smail, S.W.; Babaei, E.; Amin, K.; Abdulahad, W.H. Serum IL-23, IL-10, and TNF-α predict in-hospital mortality in COVID-19 patients. Front. Immunol. 2023, 14, 1145840. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
| Variables | Severe COVID-19 (n = 15) | Non-Severe COVID-19 (n = 31) | p-Value |
|---|---|---|---|
| Age (mean ± SD) | 71.9 ± 8.8 | 65.2 ± 13.6 | 0.744 |
| Age category | 0.051 | ||
| <40 years | 0 (0%) | 2 (6.5%) | |
| 40–59 years | 1 (6.7%) | 11 (35.5%) | |
| ≥60 years | 14 (93.3%) | 18 (58.1%) | |
| Gender | 0.293 | ||
| Men | 7 (46.7%) | 21 (68.8%) | |
| Women | 8 (53.3%) | 10 (31.3%) | |
| Place of origin | 0.311 | ||
| Rural | 2 (13.3%) | 10 (31.3%) | |
| Urban | 13 (86.7%) | 21 (68.8%) | |
| COVID-19 vaccination | Yes: 4 (26.7%) | Yes: 11 (34.4%) | 0.793 |
| Smoking | 0.329 | ||
| Yes | 0 (0%) | 4 (12.9%) | |
| No | 9 (60.0%) | 15 (48.4%) | |
| Past smoker | 5 (33.3%) | 9 (29.0%) | |
| Days since symptom onset (mean ± SD) | 7.9 ± 5.6 | 4.9 ± 3.9 | 0.076 |
| Days of hospitalization (mean ± SD) | 11.1 ± 5.3 | 9.5 ± 6.6 | 0.782 |
| Oxygen saturation (mean ± SD) | 80.9 ± 7.7 | 96.2 ± 2.8 | <0.001 |
| Variables | Normal Range | Severe COVID-19 (n = 15) | Nonsevere COVID-19 (n = 31) | p-Value |
|---|---|---|---|---|
| WBC (×103/L) | 4.0–10.0 | 1.45 (3.0) | 9.10 (4.5) | <0.001 |
| Hemoglobin (g/dL) | 12.0–16.0 | 11.26 ± 1.74 | 12.45 ± 1.88 | 0.026 |
| Neutrophils (×103/L) | 2.0–7.0 | 8.12 ± 3.91 | 6.74 ± 4.34 | 0.797 |
| Lymphocytes (×103/L) | 1.0–3.0 | 2.30 ± 0.76 | 1.83 ± 0.87 | 0.020 |
| Platelets (×103/μL) | 150–400 | 209.80 ± 110.12 | 262.94 ± 138.68 | 0.341 |
| ESR (mm/h) | <20 | 58 (46) | 51 (39) | 0.286 |
| Fibrinogen (mg/dL) | 200–400 | 662.00 ± 140.76 | 475.07 ± 228.91 | 0.002 |
| CRP (mg/L) | <5 | 51 (40) | 18 (31) | <0.001 |
| LDH | 100–250 | 446 (213) | 277 (209) | <0.001 |
| AST (U/L) | 0–40 | 57.00 ± 29.57 | 24.90 ± 6.51 | <0.001 |
| ALT (U/L) | 0–40 | 48.07 ± 27.11 | 25.19 ± 16.24 | 0.004 |
| Urea (mg/dL) | 15–45 | 84 (61) | 42 (33) | <0.001 |
| Creatinine (mg/dL) | 0.6–1.2 | 1.28 ± 0.86 | 1.09 ± 0.97 | 0.083 |
| Blood glucose (mg/dL) | 70–140 | 173.69 ± 102.21 | 126.03 ± 94.93 | 0.203 |
| D-dimers (μg/mL) | 0.0–0.5 | 1.82 ± 1.74 | 0.91 ± 1.58 | 0.035 |
| Variables | Severe COVID-19 (n = 15) | Nonsevere COVID-19 (n = 31) | p-Value |
|---|---|---|---|
| NLR | 8.22 ± 4.89 | 5.49 ± 6.75 | 0.057 |
| CXCL | 408.13 ± 83.52 | 298.07 ± 106.28 | <0.001 |
| IL-1β | 14.78 ± 29.63 | 6.91 ± 4.81 | 0.152 |
| IL-6 | 30.35 ± 17.01 | 11.87 ± 7.54 | 0.001 |
| IL-10 | 13.45 ± 13.85 | 6.37 ± 1.11 | 0.006 |
| IL-17A | 16.42 ± 38.01 | 4.15 ± 8.93 | 0.092 |
| MCP | 439.11 ± 248.02 | 240.67 ± 112.36 | 0.004 |
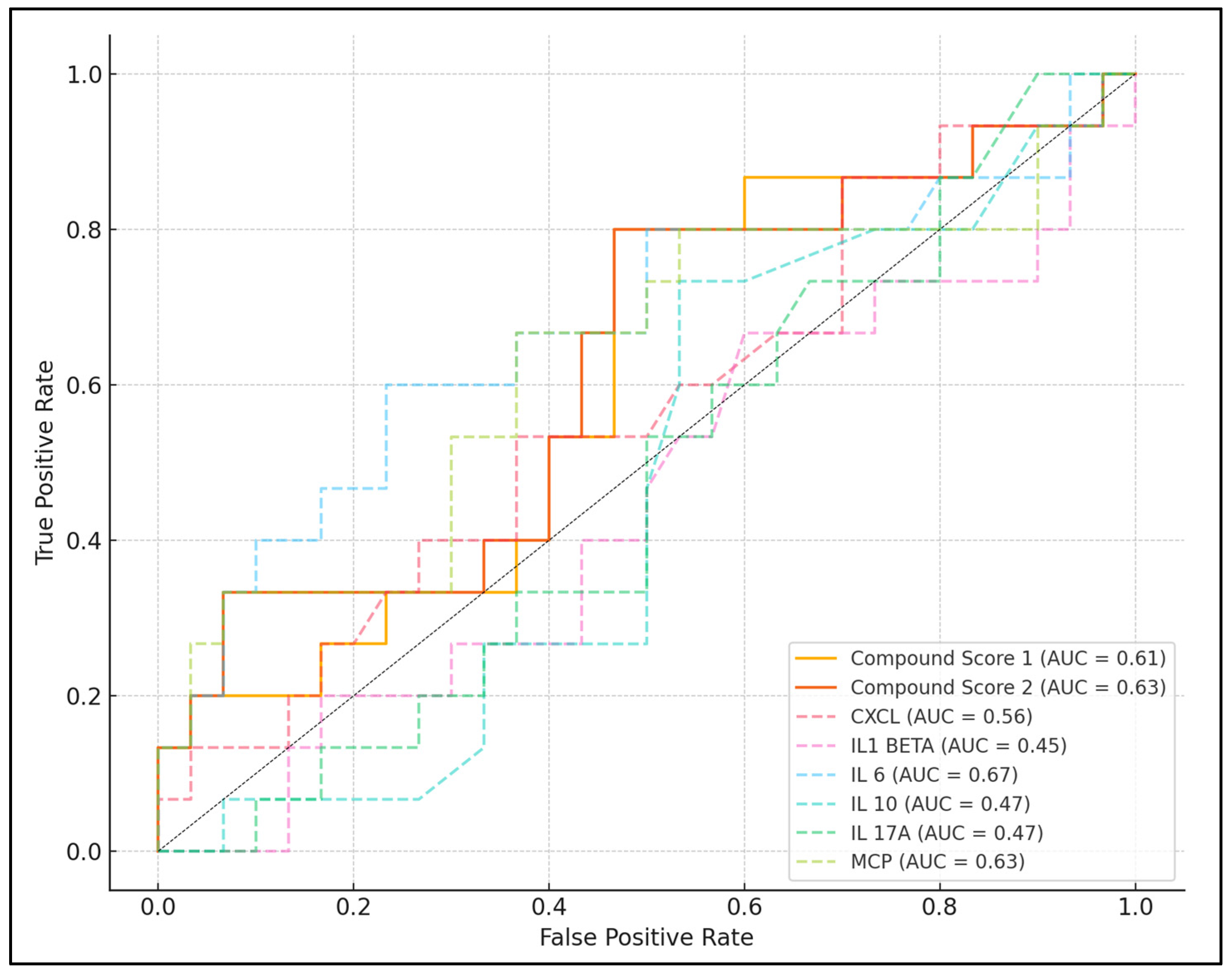
| Laboratory Parameter | Best Cutoff Value | Sensitivity | Specificity | AUC | p-Value |
|---|---|---|---|---|---|
| NLR | 6.91 | 73.33% | 61.29% | 0.503 | 0.237 |
| CXCL | 342.04 | 66.67% | 64.52% | 0.564 | 0.191 |
| IL-1β | 8.80 | 50.00% | 76.67% | 0.448 | 0.583 |
| IL-6 | 18.74 | 73.33% | 86.67% | 0.672 | <0.001 |
| IL-10 | 7.98 | 80.00% | 33.33% | 0.467 | 0.714 |
| IL-17A | 14.66 | 66.67% | 63.33% | 0.472 | 0.536 |
| MCP | 335.68 | 86.67% | 76.67% | 0.629 | 0.001 |
| Compound score 1 | 549.71 | 80.00% | 53.33% | 0.609 | 0.006 |
| Compound score 2 | 768.30 | 86.67% | 63.33% | 0.627 | <0.001 |
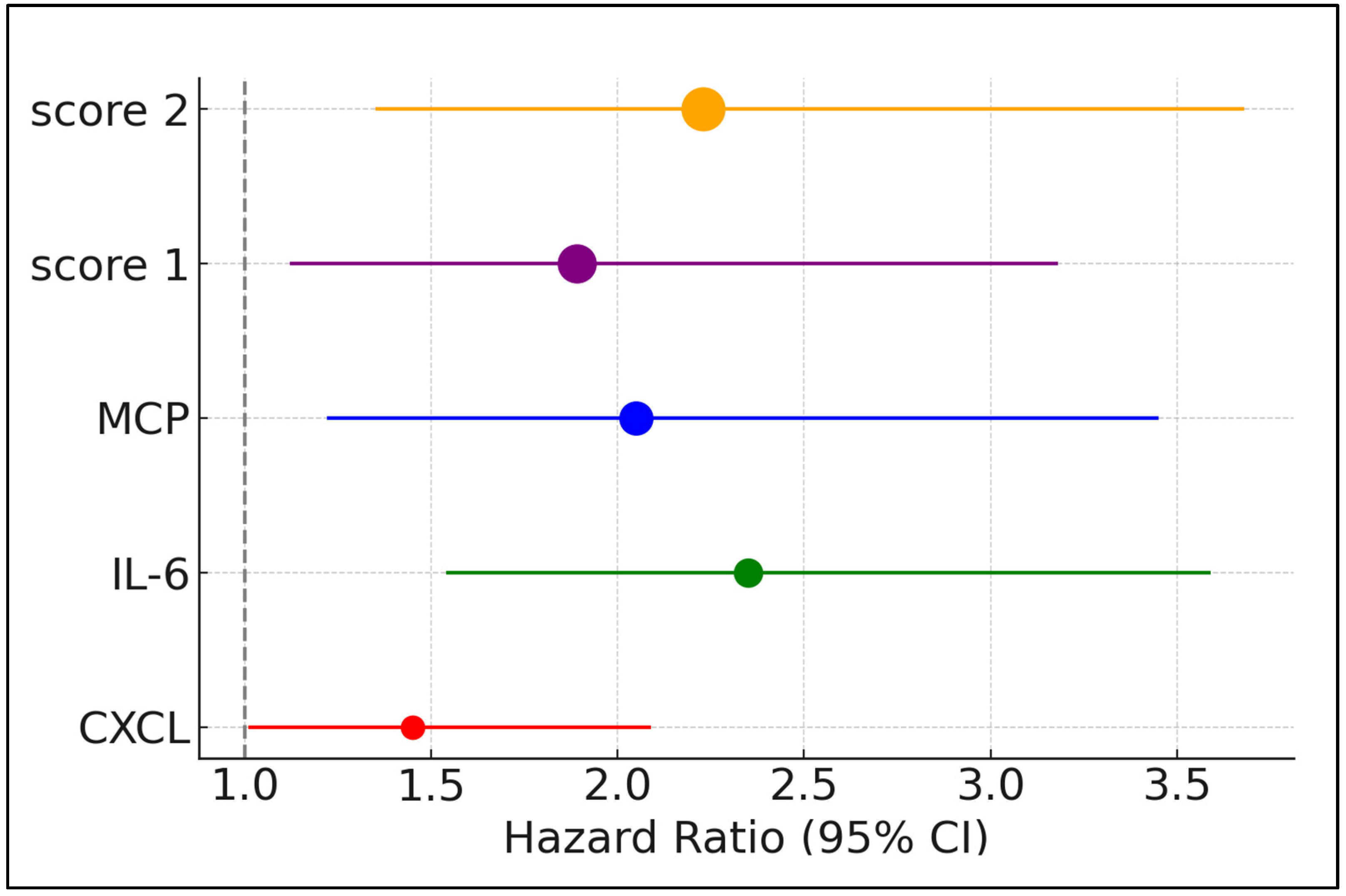
| Factors above the Best Cutoff | Hazard Ratio | 95% CI Lower | 95% CI Upper | p-Value |
|---|---|---|---|---|
| NLR | 1.23 | 0.85 | 1.78 | 0.299 |
| CXCL | 1.45 | 1.01 | 2.09 | 0.041 |
| IL-1β | 0.88 | 0.45 | 1.72 | 0.710 |
| IL-6 | 2.35 | 1.54 | 3.59 | 0.014 |
| IL-10 | 1.12 | 0.69 | 1.82 | 0.652 |
| IL-17A | 1.57 | 0.93 | 2.66 | 0.196 |
| MCP | 2.05 | 1.22 | 3.45 | 0.007 |
| Compound score 1 | 1.89 | 1.12 | 3.18 | 0.017 |
| Compound score 2 | 2.23 | 1.35 | 3.68 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cîrjaliu, R.-E.; Tofolean, I.-T.; Tofolean, D.-E.; Chisoi, A.; Oancea, C.; Vastag, E.; Marc, M.; Bratosin, F.; Rosca, O.; Fildan, A.-P. Predictive Value and Diagnostic Potential of IL-10, IL-17A, IL1-β, IL-6, CXCL, and MCP for Severe COVID-19 and COVID-19 Mortality. Biomedicines 2024, 12, 1532. https://doi.org/10.3390/biomedicines12071532
Cîrjaliu R-E, Tofolean I-T, Tofolean D-E, Chisoi A, Oancea C, Vastag E, Marc M, Bratosin F, Rosca O, Fildan A-P. Predictive Value and Diagnostic Potential of IL-10, IL-17A, IL1-β, IL-6, CXCL, and MCP for Severe COVID-19 and COVID-19 Mortality. Biomedicines. 2024; 12(7):1532. https://doi.org/10.3390/biomedicines12071532
Chicago/Turabian StyleCîrjaliu, Roxana-Elena, Ioan-Tiberiu Tofolean, Doina-Ecaterina Tofolean, Anca Chisoi, Cristian Oancea, Emanuela Vastag, Monica Marc, Felix Bratosin, Ovidiu Rosca, and Ariadna-Petronela Fildan. 2024. "Predictive Value and Diagnostic Potential of IL-10, IL-17A, IL1-β, IL-6, CXCL, and MCP for Severe COVID-19 and COVID-19 Mortality" Biomedicines 12, no. 7: 1532. https://doi.org/10.3390/biomedicines12071532
APA StyleCîrjaliu, R.-E., Tofolean, I.-T., Tofolean, D.-E., Chisoi, A., Oancea, C., Vastag, E., Marc, M., Bratosin, F., Rosca, O., & Fildan, A.-P. (2024). Predictive Value and Diagnostic Potential of IL-10, IL-17A, IL1-β, IL-6, CXCL, and MCP for Severe COVID-19 and COVID-19 Mortality. Biomedicines, 12(7), 1532. https://doi.org/10.3390/biomedicines12071532








