Morphological and Immunocytochemical Characterization of Paclitaxel-Induced Microcells in Sk-Mel-28 Melanoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Cell Treatment
2.3. Immunocytochemistry
2.4. RNA Synthesis Detection
2.5. DNA Synthesis Detection
2.6. Flow Cytometry and Data Analysis
2.7. Microscopy
3. Results
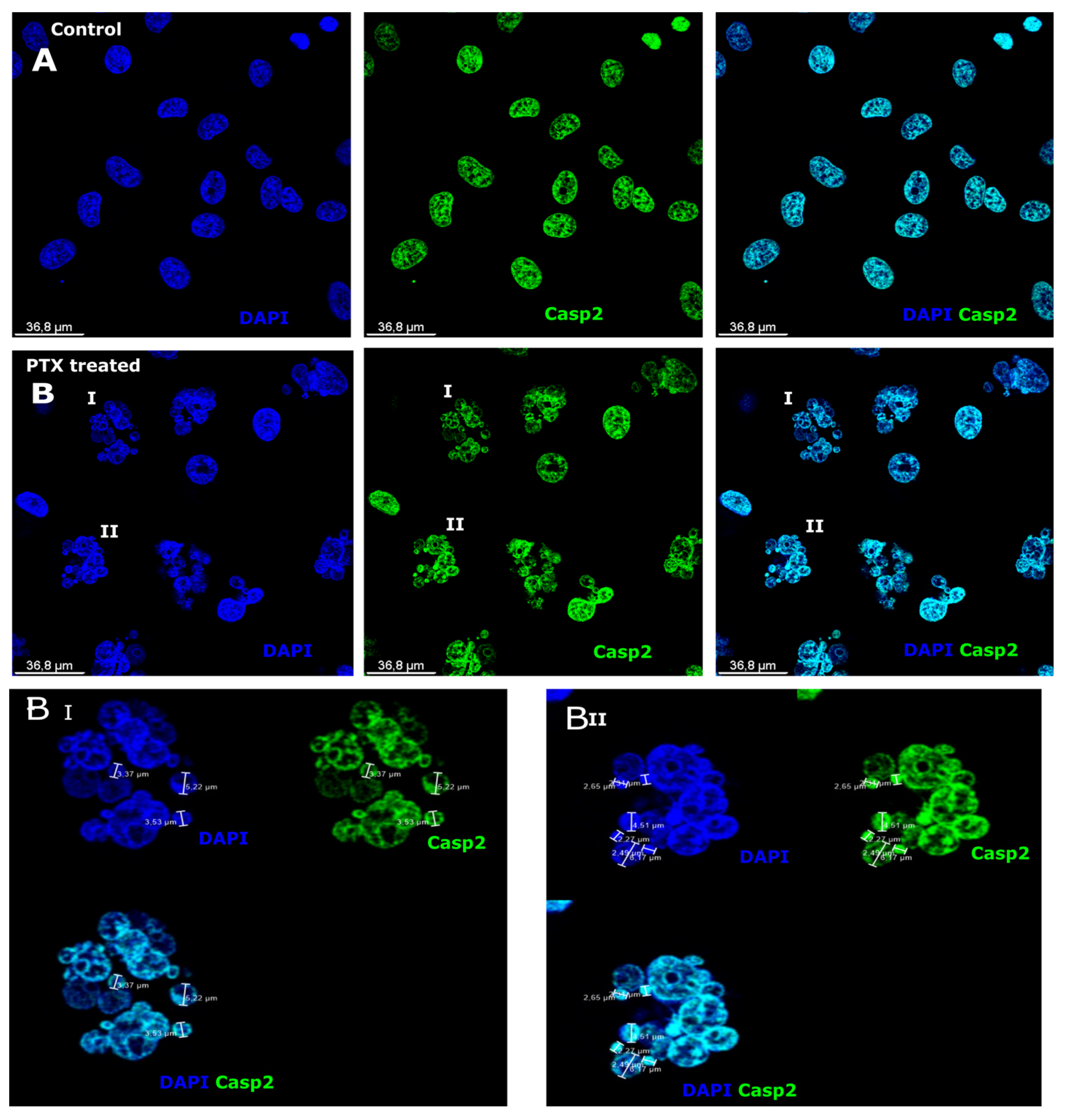
3.1. Caspase-2 Expression after Paclitaxel Treatment in Microcells
3.2. Caspase-6 Expression after Paclitaxel Treatment in Microcells
3.3. Expression of Aldehyde Dehydrogenase and Caspase-3 after Paclitaxel Treatment in Microcells
3.4. SOX2 and Nanog Expression after Paclitaxel Treatment in Microcells
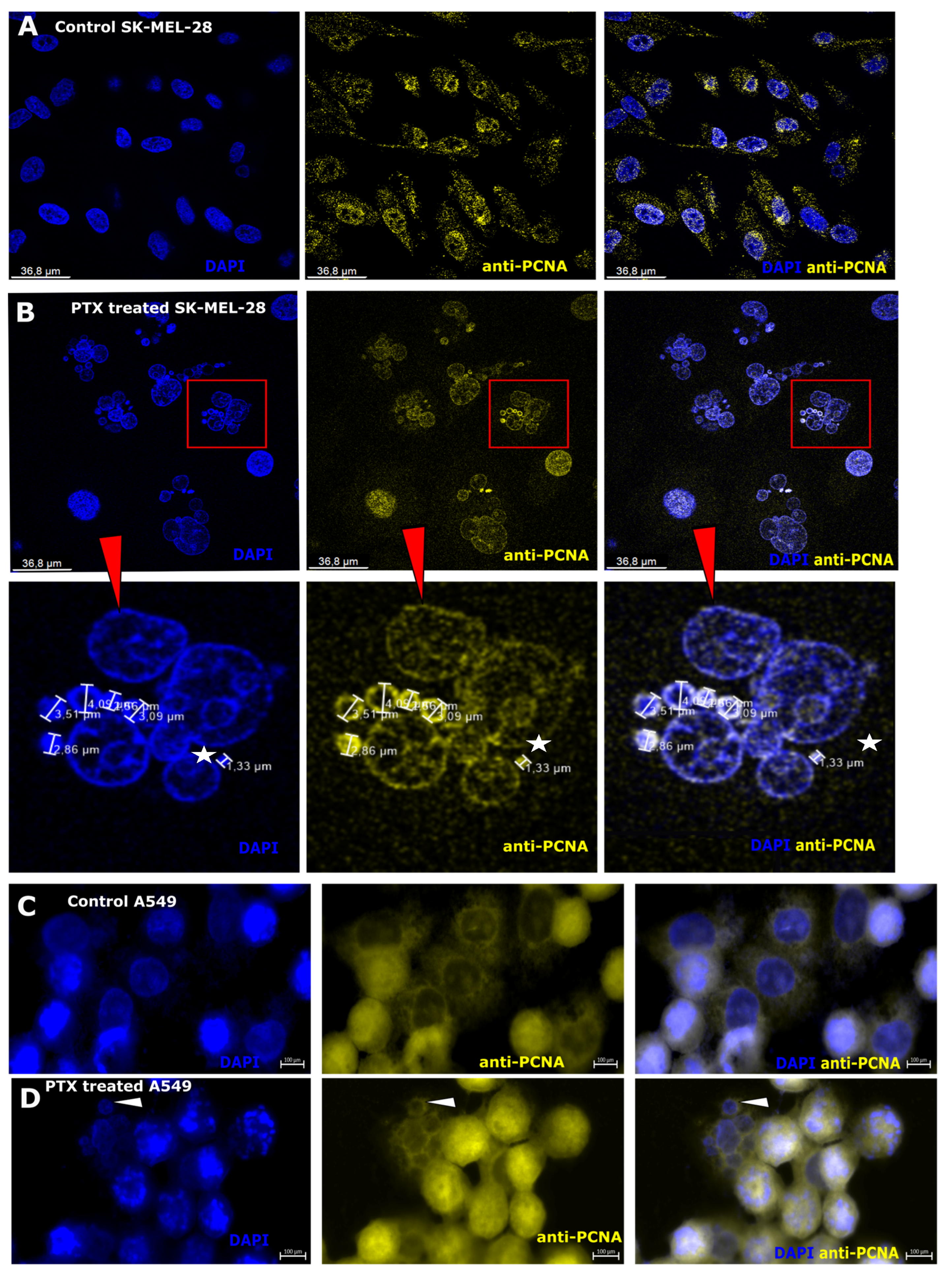
3.5. Nuclear Antigen Expression after Paclitaxel Treatment in Microcells
3.6. Ability of Microcells to Regeneration of DNA
3.7. RNA Synthesis in Microcells after Paclitaxel Treatment
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malihi, P.D.; Morikado, M.; Welter, L.; Carlsson, A.; Velasco, C.R.; Kolatkar, A.; Rodriguez-Lee, M.; Hicks, J.; Kuhn, P.; Liu, S.T.; et al. Abstract 1576: Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg. Sci. Phys. Oncol. 2018, 4, 1576. [Google Scholar] [CrossRef]
- De Francesco, E.M.; Sotgia, F.; Lisanti, M.P. Cancer stem cells (CSCs): Metabolic strategies for their identification and eradication. Biochem. J. 2018, 475, 1611–1634. [Google Scholar] [CrossRef] [PubMed]
- Rueff, J.; Rodrigues, A.S. Cancer drug resistance: A brief overview from a genetic viewpoint. Methods Mol. Biol. 2016, 1395, 1–18. [Google Scholar] [CrossRef]
- Gay, L.; Baker, A.M.; Graham, T.A. Tumour Cell Heterogeneity. F1000Research 2016, 5, 238. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Jin, X.; Kim, H. Cancer stem cells and differentiation therapy. Tumor Biol. 2017, 39, 1010428317729933. [Google Scholar] [CrossRef] [PubMed]
- Galofré, C.; Geyik, Ö.G.; Asensio, E.; Wangsa, D.; Hirsch, D.; Parra, C.; Saez, J.; Mollà, M.; Yüce, Z.; Castells, A.; et al. Tetraploidy-associated genetic heterogeneity confers chemo-radiotherapy resistance to colorectal cancer cells. Cancers 2020, 12, 1118. [Google Scholar] [CrossRef]
- Yu, Z.; Pestellc, T.G.; Lisantic, M.P.; Pestell, R.G. Cancer Stem Cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef] [PubMed]
- Mamun, M.A.; Mannoor, K.; Cao, J.; Qadri, F.; Song, X. SOX2 in cancer stemness: Tumor malignancy and therapeutic potentials. J. Mol. Cell Biol. 2020, 12, 85–98. [Google Scholar] [CrossRef]
- Islam, F.; Gopalan, V.; Lam, A.K.Y. Cancer stem cells: Role in tumor progression and treatment resistance. In Oncogenomics: From Basic Research to Precision Medicine; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 77–87. ISBN 9780128117859. [Google Scholar]
- Salmina, K.; Huna, A.; Kalejs, M.; Pjanova, D.; Scherthan, H.; Cragg, M.S.; Erenpreisa, J. The Cancer Aneuploidy Paradox: In the Light of Evolution. Genes 2019, 10, 83. [Google Scholar] [CrossRef]
- Yan, S.; Wu, G. Could ALDH2*2 be the reason for low incidence and mortality of ovarian cancer for East Asia women? Oncotarget 2018, 9, 12503–12512. [Google Scholar] [CrossRef]
- Vassalli, G. Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef]
- Wang, S.; Liu, X.; Chen, Y.; Zhan, X.; Wu, T.; Chen, B.; Sun, G.; Yan, S.; Xu, L. The role of SOX2 overexpression in prognosis of patients with solid tumors: A meta-analysis and system review. Medicine 2020, 99, e19604. [Google Scholar] [CrossRef]
- Yaghobi, Z.; Movassaghpour, A.; Talebi, M.; Abdoli Shadbad, M.; Hajiasgharzadeh, K.; Pourvahdani, S.; Baradaran, B. The role of CD44 in cancer chemoresistance: A concise review. Eur. J. Pharmacol. 2021, 903, 174147. [Google Scholar] [CrossRef]
- Johnson-Arbor, K.; Dubey, R. Doxorubicin. NCBI Bookshelf. A Service of the National Library of Medicine, National Institutes of Health. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2018. Available online: https://www.ncbi.nlm.nih.gov/books/NBK459232/?report=printable (accessed on 11 October 2020).
- Chemocare. Doxorubicin (Adriamycin, Rubex) Chemotherapy Drug Information. In NCBI. 2020. Available online: http://chemocare.com/about/default.aspx (accessed on 11 October 2020).
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.M.; Jang, M.O.; Jin, Y.W.; Lee, E.K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149–158. [Google Scholar] [CrossRef]
- Farrar, M.C.; Jacobs, T.F. Paclitaxel. In NCBI Bookshelf. 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK536917/ (accessed on 11 October 2020).
- Zhang, D.; Yang, R.; Wang, S.; Zheng, D. Paclitaxel: New uses for an old drug. Drug Des. Devel. Ther. 2014, 8, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Hadzic, T.; Aykin-Burns, N.; Zhu, Y.; Coleman, M.C.; Leick, K.; Jacobson, G.M.; Spitz, D.R. Paclitaxel combined with inhibitors of glucose and hydroperoxide metabolism enhances breast cancer cell killing via H2O2-mediated oxidative stress. Free Radic. Biol. Med. 2010, 48, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Minotti, G.; Menna, P.; Saponiero, A.; Licata, S.; Calafiore, A.M.; Teodori, G.; Gianni, L. Paclitaxel and docetaxel enhance the metabolism of doxorubicin to toxic species in human myocardium. Clin. Cancer Res. 2001, 7, 1511–1515. [Google Scholar] [PubMed]
- Içduygu, F.M.; Samli, H.; Özgöz, A.; Vatansever, B.; Oztürk, K.H.; Akgün, E. Possibility of paclitaxel to induce the stemness-related characteristics of prostate cancer cells. Adv. Clin. Exp. Med. 2021, 30, 1283–1291. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Vallette, F.M.; Olivier, C.; Lézot, F.; Oliver, L.; Cochonneau, D.; Lalier, L.; Cartron, P.F.; Heymann, D. Dormant, quiescent, tolerant and persister cells: Four synonyms for the same target in cancer. Biochem. Pharmacol. 2019, 162, 169–176. [Google Scholar] [CrossRef]
- Fiorillo, M.; Sotgia, F.; Lisanti, M.P. “Energetic” Cancer Stem Cells (e-CSCs): A New Hyper-Metabolic and Proliferative Tumor Cell Phenotype, Driven by Mitochondrial Energy. Front. Oncol. 2019, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- Inde, Z.; Dixon, S.J. The impact of non-genetic heterogeneity on cancer cell death. Crit. Rev. Biochem. Mol. Biol. 2017, 53, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Ju, F.; Atyah, M.M.; Horstmann, N.; Gul, S.; Vago, R.; Bruns, C.J.; Zhao, Y.; Dong, Q.Z.; Ren, N. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res. Ther. 2022, 13, 233. [Google Scholar] [CrossRef]
- Lozach, P.Y. Cell Biology of Viral Infections. Cell 2020, 9, 2431. [Google Scholar] [CrossRef] [PubMed]
- Gaillochet, C.; Lohmann, J.U. The never-ending story: From pluripotency to plant developmental plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef]
- Piña-Sánchez, P.; Chávez-González, A.; Ruiz-Tachiquín, M.; Vadillo, E.; Monroy-García, A.; Montesinos, J.J.; Grajales, R.; Gutiérrez de la Barrera, M.; Mayani, H. Cancer Biology, Epidemiology, and Treatment in the 21st Century: Current Status and Future Challenges From a Biomedical Perspective. Cancer Control 2021, 28, 10732748211038735. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Maheswaran, S.; Mcdermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; Wong, K.; et al. A chromatin-mediated reversible drug tolerant state in cancer cell subpopulations. Cell 2011, 141, 69–80. [Google Scholar] [CrossRef]
- Risom, T.; Langer, E.M.; Chapman, M.P.; Rantala, J.; Fields, A.J.; Boniface, C.; Alvarez, M.J.; Kendsersky, N.D.; Pelz, C.R.; Johnson-Camacho, K.; et al. Differentiation-state plasticity is a targetable resistance mechanism in basal-like breast cancer. Nat. Commun. 2018, 9, 3815. [Google Scholar] [CrossRef]
- Klener, P.; Klanova, M. Drug resistance in non-hodgkin lymphomas. Int. J. Mol. Sci. 2020, 21, 2081. [Google Scholar] [CrossRef]
- Bell, C.C.; Gilan, O. Principles and mechanisms of non-genetic resistance in cancer. Br. J. Cancer 2020, 122, 465–472. [Google Scholar] [CrossRef]
- Rehman, S.K.; Haynes, J.; Collignon, E.; Brown, K.R.; Wang, Y.; Nixon, A.M.L.; Bruce, J.P.; Wintersinger, J.A.; Singh Mer, A.; Lo, E.B.L.; et al. Colorectal Cancer Cells Enter a Diapause-like DTP State to Survive Chemotherapy. Cell 2021, 184, 226–242.e21. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Knasmueller, S.; Bolognesi, C.; Holland, N.; Bonassi, S.; Kirsch-Volders, M. Micronuclei as biomarkers of DNA damage, aneuploidy, inducers of chromosomal hypermutation and as sources of pro-inflammatory DNA in humans. Mutat. Res. Mutat. Res. 2020, 786, 108342. [Google Scholar] [CrossRef] [PubMed]
- Buiķis, I.; Harju, L.; Freivalds, T. Origin of microcells in the human sarcoma cell line HT-1080. Anal. Cell. Pathol. 1999, 18, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Buiķis, I.; Freivalds, T.; Harju, L.; Nikulshin, S. Sporosis: A Cytological Mechanisms of Differentiation of the Human Sarcoma Line HT-1080 Cells. Acta Med. Litu. 2002, 9, 18–21. [Google Scholar]
- Bema, D.; Freivalds, T.; Buikis, I.; Harju, L. Microcells development and endocytosis ability morphological and quantitative characterization in HeLa cancer cells. In Proceedings of the 14th Nordic-Baltic Conference on Biomedical Engineering and Medical Physics: NBC, Riga, Latvia, 16–20 June 2008; Springer: Berlin/Heidelberg, Germnay, 2008; pp. 598–601. [Google Scholar] [CrossRef]
- Simsone, Z.; Freivalds, T.; Bēma, D.; Miķelsone, I.; Patetko, L.; Bērziņš, J.; Harju, L.; Buiķis, I. Cancer microcell initiation and determination. BMC Cancer 2021, 21, 1087. [Google Scholar] [CrossRef]
- Mu, L.; Huang, K.; Hu, Y.; Yan, C.; Li, X.; Tao, D.; Gong, J.; Qin, J. Small-sized colorectal cancer cells harbor metastatic tumorinitiating cells. Oncotarget 2017, 8, 107907–107919. [Google Scholar] [CrossRef]
- Brier, J.; Jayanti, L.D. Cluster of differentiation (cd) antigens. In Immunology Guidebook; Cruse, J.M., Lewis, R.E., Huan, W., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 47–124. ISBN 9789896540821. [Google Scholar]
- Walcher, L.; Kistenmacher, A.K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer Stem Cells—Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Freivalds, T.; Simsone, Z.; Kudaba, I.; Berzins, J. Prognostic and Predictive Significance of Breast Cancer Stem Cells. Acta Chir. Latv. 2012, 11, 122–125. [Google Scholar] [CrossRef]
- Zhou, N.; Wu, X.; Yang, B.; Yang, X.; Zhang, D.; Qing, G. Stem cell characteristics of dormant cells and cisplatin-induced effects on the stemness of epithelial ovarian cancer cells. Mol. Med. Rep. 2014, 10, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Recasens, A.; Munoz, L. Targeting Cancer Cell Dormancy. Trends Pharmacol. Sci. 2019, 40, 128–141. [Google Scholar] [CrossRef]
- Abcam. ab228561 RNA Synthesis Assay Kit. 2020. Available online: https://www.abcam.com/ps/products/228/ab228561/documents/RNA-Synthesis-Assay-protocol-book-v2-ab228561(website).pdf (accessed on 14 March 2021).
- Ando, K.; Parsons, M.J.; Shah, R.B.; Charendoff, C.I.; Paris, S.L.; Liu, P.H.; Fassio, S.R.; Rohrman, B.A.; Thompson, R.; Oberst, A.; et al. NPM1 directs PIDDosome-dependent caspase-2 activation in the nucleolus. J. Cell Biol. 2017, 216, 1795–1810. [Google Scholar] [CrossRef] [PubMed]
- Brown-Suedel, A.N.; Bouchier-Hayes, L. Caspase-2 Substrates: To Apoptosis, Cell Cycle Control, and Beyond. Front. Cell Dev. Biol. 2020, 8, 610022. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, C.; Shu, G.L.; Zheng, T.S.; Flavell, R.A.; Clark, E.A. Caspase 6 Regulates B Cell Activation and Differentiation into Plasma Cells. J. Immunol. 2008, 181, 6810–6819. [Google Scholar] [CrossRef] [PubMed]
- Toumia, I.B.; Sobeh, M.; Ponassi, M.; Banelli, B.; Dameriha, A.; Wink, M.; Chekir Ghedira, L.; Rosano, C. A Methanol Extract of Scabiosa atropurpurea Enhances Doxorubicin Cytotoxicity against Resistant Colorectal Cancer Cells In Vitro. Molecules 2020, 25, 5265. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Nie, Q.; Tang, C.; Zhang, L. Nanog induced intermediate state in regulating stem cell differentiation and reprogramming. BMC Syst. Biol. 2018, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Peng, A.; Xu, X.; Wang, C.; Yang, J.; Wang, S.; Dai, J.; Ye, L. EZH2 promotes DNA replication by stabilizing interaction of POLδ and PCNA via methylation-mediated PCNA trimerization. Epigenet. Chromatin 2018, 11, 44. [Google Scholar] [CrossRef]
- Freivalds, T.; Buikis, I.; Harju, L. Sporosis: An Ambiguous Mode of the Cell Death. Biochem. Soc. Trans. 1996, 24, 595S. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, T.; Wang, Z.; Yi, F.; Li, C.; Guo, W.; Xu, H.; Cui, H.; Dong, X.; Liu, J.; et al. Post-translational modifications of pcna in control of dna synthesis and dna damage tolerance-the implications in carcinogenesis. Int. J. Biol. Sci. 2021, 17, 4047–4059. [Google Scholar] [CrossRef] [PubMed]
- Golubnitschaja, O.; Topolcan, O.; Kucera, R.; Costigliola, V. 10th Anniversary of the European Association for Predictive, Preventive and Personalised (3P) Medicine-EPMA World Congress Supplement 2020. EPMA J. 2020, 11 (Suppl. S1), 1–133. [Google Scholar] [CrossRef]
- Simsone, Z.; Freivalds, T.; Bēma, D.; Harju, L.; Bērziņš, J.; Buiķis, I. Microcells as a possible predictive factor for tumour treatment efficiency. EPMA J. Popul. 2020, 11, 78–90. [Google Scholar] [CrossRef]
- Ye, X.; Ling, B.; Xu, H.; Li, G.; Zhao, X.; Xu, J.; Liu, J.; Liu, L. Clinical significance of high expression of proliferating cell nuclear antigen in non-small cell lung cancer. Medicine 2020, 99, e19755. [Google Scholar] [CrossRef] [PubMed]
- Technologies Molecular Probes by Life. Click-iT TM EdU Flow Cytometry Assay Kits. In Mol. Probes by Life Technol. 2009. Available online: https://www.thermofisher.com/order/catalog/product/C10419#/C10419 (accessed on 14 March 2021).
- Henry, N.L.; Hayes, D.F. Cancer biomarkers. Mol. Oncol. 2012, 6, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Aponte, P.M.; Caicedo, A. Stemness in Cancer: Stem Cells, Cancer Stem Cells, and Their Microenvironment. Stem Cells Int. 2017, 2017, 5619472. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Festjens, N.; Declercq, W.; Berghe, T.V.; Vandenabeele, P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007, 14, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Herr, I.; Debatin, K.M. Cellular stress response and apoptosis in cancer therapy. Blood 2001, 98, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Krysko, D.V.; Vanden Berghe, T.; D’Herde, K.; Vandenabeele, P. Apoptosis and necrosis: Detection, discrimination and phagocytosis. Methods 2008, 44, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Karki, R.; Vogel, P.; Kanneganti, T.D. Caspase-6 Is a Key Regulator of Innate Immunity, Inflammasome Activation, and Host Defense. Cell 2020, 181, 674–687.e13. [Google Scholar] [CrossRef]
- Feng, X.; Tian, L.; Zhang, Z.; Yu, Y.; Cheng, J.; Gong, Y.; Li, C.Y.; Huang, Q. Caspase 3 in dying tumor cells mediates post-irradiation angiogenesis. Oncotarget 2015, 6, 32353–32367. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Okabayashi, T.; Yokota, S.I.; Kan, Y.; Ogino, J.; Yamashita, T.; Fujii, N.; Jimbow, K. Increased caspase-2 activity is associated with induction of apoptosis in IFN-β sensitive melanoma cell lines. J. Interf. Cytokine Res. 2010, 30, 349–357. [Google Scholar] [CrossRef]
- O’Byrne, K.J.; Richard, D.J. Nucleolar caspase-2: Protecting us from DNA damage. J. Cell Biol. 2017, 216, 1521–1523. [Google Scholar] [CrossRef]
- Paroni, G.; Henderson, C.; Schneider, C.; Brancolini, C. Caspase-2 can trigger cytochrome c release and apoptosis from the nucleus. J. Biol. Chem. 2002, 277, 15147–15161. [Google Scholar] [CrossRef] [PubMed]
- Capo-Chichi, C.D.; Cai, K.Q.; Xu, X.X. Overexpression and cytoplasmic localization of caspase-6 is associated with lamin A degradation in set of ovarian cancers. Biomark. Res. 2018, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Balderstone, L.A.; Dawson, J.C.; Welman, A.; Serrels, A.; Wedge, S.R.; Brunton, V.G. Development of a fluorescence-based cellular apoptosis reporter. Methods Appl. Fluoresc. 2019, 7, 015001. [Google Scholar] [CrossRef] [PubMed]
- Novak, D.; Hüser, L.; Elton, J.J.; Umansky, V.; Altevogt, P.; Utikal, J. SOX2 in development and cancer biology. Semin. Cancer Biol. 2020, 67, 74–82. [Google Scholar] [CrossRef]
- Zhang, S.; Xiong, X.; Sun, Y. Functional characterization of SOX2 as an anticancer target. Signal Transduct. Target. Ther. 2020, 5, 135. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, T.; Lengerke, C. SOX2 protein biochemistry in stemness, reprogramming, and cancer: The PI3K/AKT/SOX2 axis and beyond. Oncogene 2020, 39, 278–292. [Google Scholar] [CrossRef]
- Zamzam, Y.; Abdelmonem Zamzam, Y.; Aboalsoud, M.; Harras, H. The Utility of SOX2 and AGR2 Biomarkers as Early Predictors of Tamoxifen Resistance in ER-Positive Breast Cancer Patients. Int. J. Surg. Oncol. 2021, 2021, 9947540. [Google Scholar] [CrossRef]
- Gu, T.-T.; Liu, S.-Y.; Zheng, P.-S. Cytoplasmic NANOG-Positive Stromal Cells Promote Human Cervical Cancer Progression. Am. J. Pathol. 2012, 181, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Yosefzon, Y.; Soteriou, D.; Feldman, A.; Kostic, L.; Koren, E.; Brown, S.; Ankawa, R.; Sedov, E.; Glaser, F.; Fuchs, Y. Caspase-3 Regulates YAP-Dependent Cell Proliferation and Organ Size. Mol. Cell 2018, 70, 573–587.e4. [Google Scholar] [CrossRef]
- Lu, E.M.-C.; Ratnayake, J.; Rich, A.M. Assessment of proliferating cell nuclear antigen (PCNA) expression at the invading front of oral squamous cell carcinoma. BMC Oral Health 2019, 19, 233. [Google Scholar] [CrossRef]
- Malkas, L.H.; Herbert, B.S.; Abdel-Aziz, W.; Dobrolecki, L.E.; Liu, Y.; Agarwal, B.; Hoelz, D.; Badve, S.; Schnaper, L.; Arnold, R.J.; et al. A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. Proc. Natl. Acad. Sci. USA 2006, 103, 19472–19477. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kong, W.; Liu, B.; Zhang, X. Proliferating cell nuclear antigen promotes cell proliferation and tumorigenesis by up-regulating STAT3 in non-small cell lung cancer. Biomed. Pharmacother. 2018, 104, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Hickey, R.J.; Malkas, L.H. Therapeutic Targeting of DNA Replication Stress in Cancer. Genes 2023, 14, 1346. [Google Scholar] [CrossRef] [PubMed]






| Antibody | Description | Antibody Emission and Excitation | Dilution/Used Concentration | Product No and Manufacturer |
|---|---|---|---|---|
| ALDH2 | Primary antibody: rabbit anti-human Rabbit polyclonal | 1:75 | 158-201549-T08-200, Sino Biological by Nordic BioSite, Täby, Sweden | |
| SOX2 | Primary antibody: mouse anti-human antibody Mouse monoclonal | 1:100 | TA302025, Origene, Rockville, MD, USA | |
| Anti-PCNA | Primary antibody: rabbit anti-human, rabbit recombinant monoclonal | 1:100 | ab92552, Abcam, Amsterdam, The Netherlands | |
| Nanog | Primary antibody: rabbit anti-human, rabbit polyclonal | 1:100 | 14295-1-AP, Proteintech Europe, Manchester, UK | |
| Caspase-2 | Primary antibody: rabbit anti-human, rabbit polyclonal | FITC Conjugated; Ex.494 nm/Em.518 nm | 1:100 | BS-5802R-FITC, Bioss antibodies/Nordic BioSite, Täby, Sweden |
| Caspase-6 | Primary antibody: rabbit anti-human, rabbit polyclonal | ALEXA FLUOR® 555 conjugated Ex.553 nm/Em.568 nm | 1:100 | BS-0151R-A555, Bioss antibodies/Nordic BioSite, Täby, Sweden |
| Caspase-3 | Primary antibody: mouse anti-human, mouse monoclonal | 1:100 | GTX13586, Biolegend/GeneTex Inc., Tampere, Finland | |
| Alexa fluor 488 | Secondary antibody: goat anti-rabbit Igg H&L | Ex.495 nm/Em.519 nm | 1:1000 | ab150077 Abcam, Carlsbad, CA, USA |
| Secondary antibody: goat anti-mouse Igg H&L | Ex.495 nm/Em.519 nm | 1:1000 | ab150113 Abcam, Carlsbad, CA, USA | |
| Alexa fluor 594 | Secondary antibody: goat anti-mouse Igg H&L | Ex.590 nm/Em.617 nm | 1:1000 | ab150116 Abcam, Carlsbad, CA, USA |
| Secondary antibody: goat anti-rabbit Igg H&L | Ex.590 nm/Em.617 nm | 1:1000 | ab150088 Abcam, Carlsbad, CA, USA |
| Microcells | ||
|---|---|---|
| Microcell Count | Mean (EdU/PI) | |
| Sk-Mel-28_EdU/PI (control) | 84 | 20.6 |
| Sk-Mel-28 PTX 2_EdU/PI. | 442 | 18.7 |
| All cells | ||
| Sk-Mel-28_EdU/PI. | 10,000 | 0.329 |
| SkMel-28 PTX 2_Ed_ PI. | 10,000 | 0.937 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simsone, Z.; Feivalds, T.; Harju, L.; Miķelsone, I.; Blāķe, I.; Bērziņš, J.; Buiķis, I. Morphological and Immunocytochemical Characterization of Paclitaxel-Induced Microcells in Sk-Mel-28 Melanoma Cells. Biomedicines 2024, 12, 1576. https://doi.org/10.3390/biomedicines12071576
Simsone Z, Feivalds T, Harju L, Miķelsone I, Blāķe I, Bērziņš J, Buiķis I. Morphological and Immunocytochemical Characterization of Paclitaxel-Induced Microcells in Sk-Mel-28 Melanoma Cells. Biomedicines. 2024; 12(7):1576. https://doi.org/10.3390/biomedicines12071576
Chicago/Turabian StyleSimsone, Zane, Tālivaldis Feivalds, Līga Harju, Indra Miķelsone, Ilze Blāķe, Juris Bērziņš, and Indulis Buiķis. 2024. "Morphological and Immunocytochemical Characterization of Paclitaxel-Induced Microcells in Sk-Mel-28 Melanoma Cells" Biomedicines 12, no. 7: 1576. https://doi.org/10.3390/biomedicines12071576






