Bioinformatics Identification and Experimental Verification of Disulfidptosis-Related Genes in the Progression of Osteoarthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sources
2.2. Identification of Differentially Expressed DRGs (DE-DRGs)
2.3. Function Analysis
2.4. Machine Learning
2.5. Identification of Hub Genes and Nomogram Construction
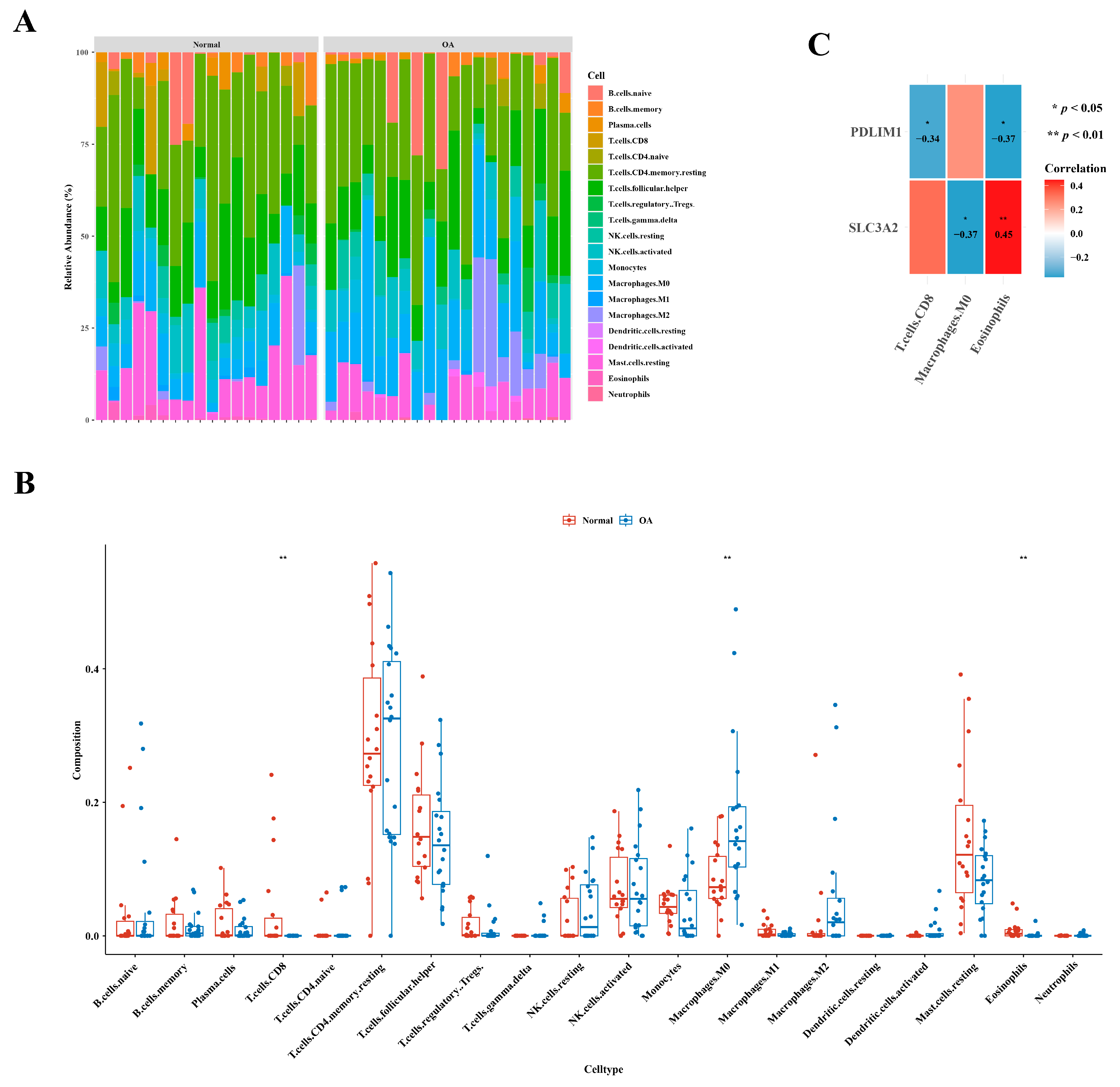
2.6. Analysis of Immune Cell Infiltration
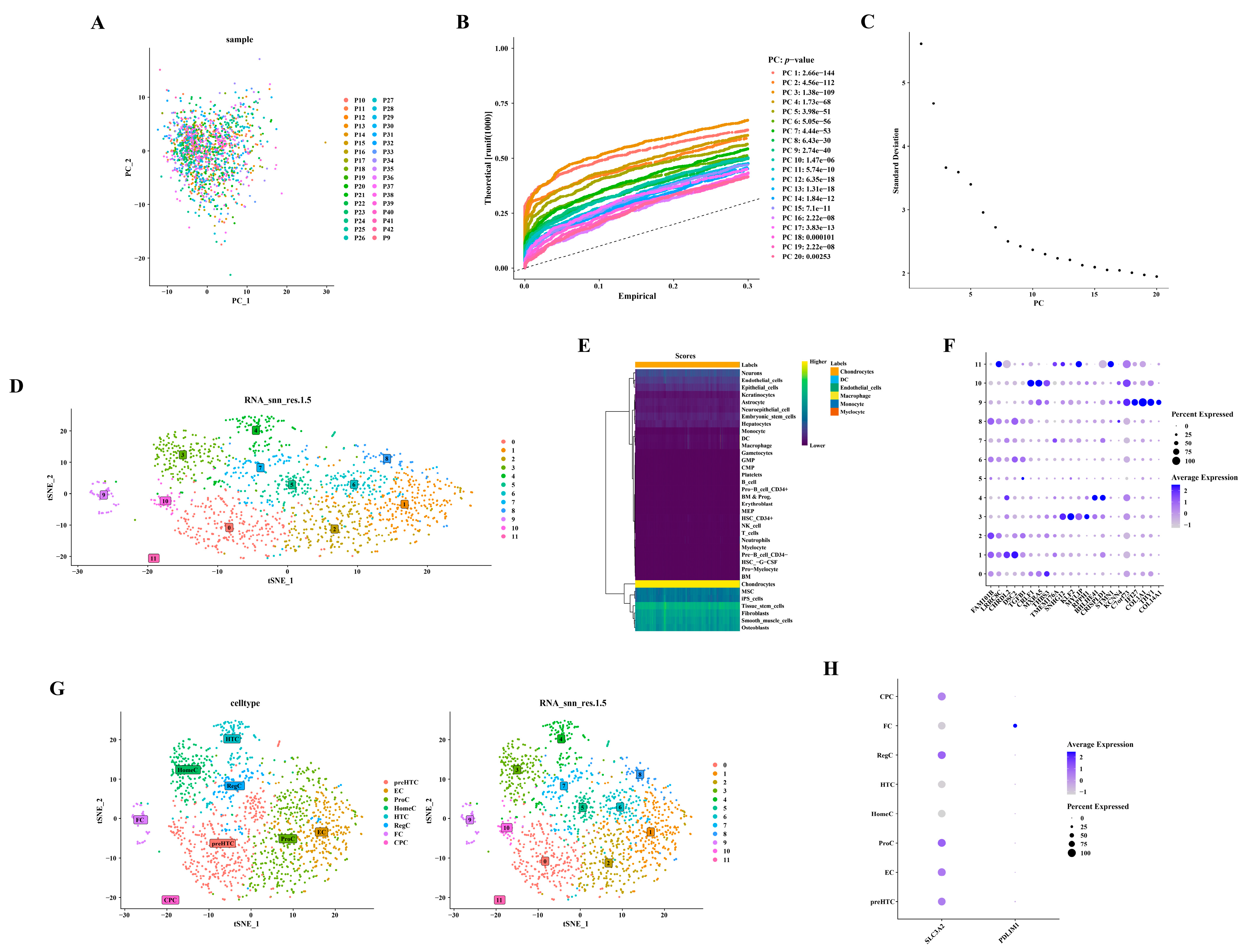
2.7. Single-Cell Analysis
2.8. Chondrocyte Culture, Treatment, and Small Interfering RNA (siRNA) Transfection
2.9. Quantitative Polymerase Chain Reaction (qPCR)
2.10. Protein Extraction and Western Blot
2.11. Statistical Analysis
3. Results
3.1. Identification of Six DE-DRGs
3.2. Definition of Hub Genes: SLC3A2 and PDLIM1
3.3. Evaluation of the Nomogram Model’s Forecasting Accuracy
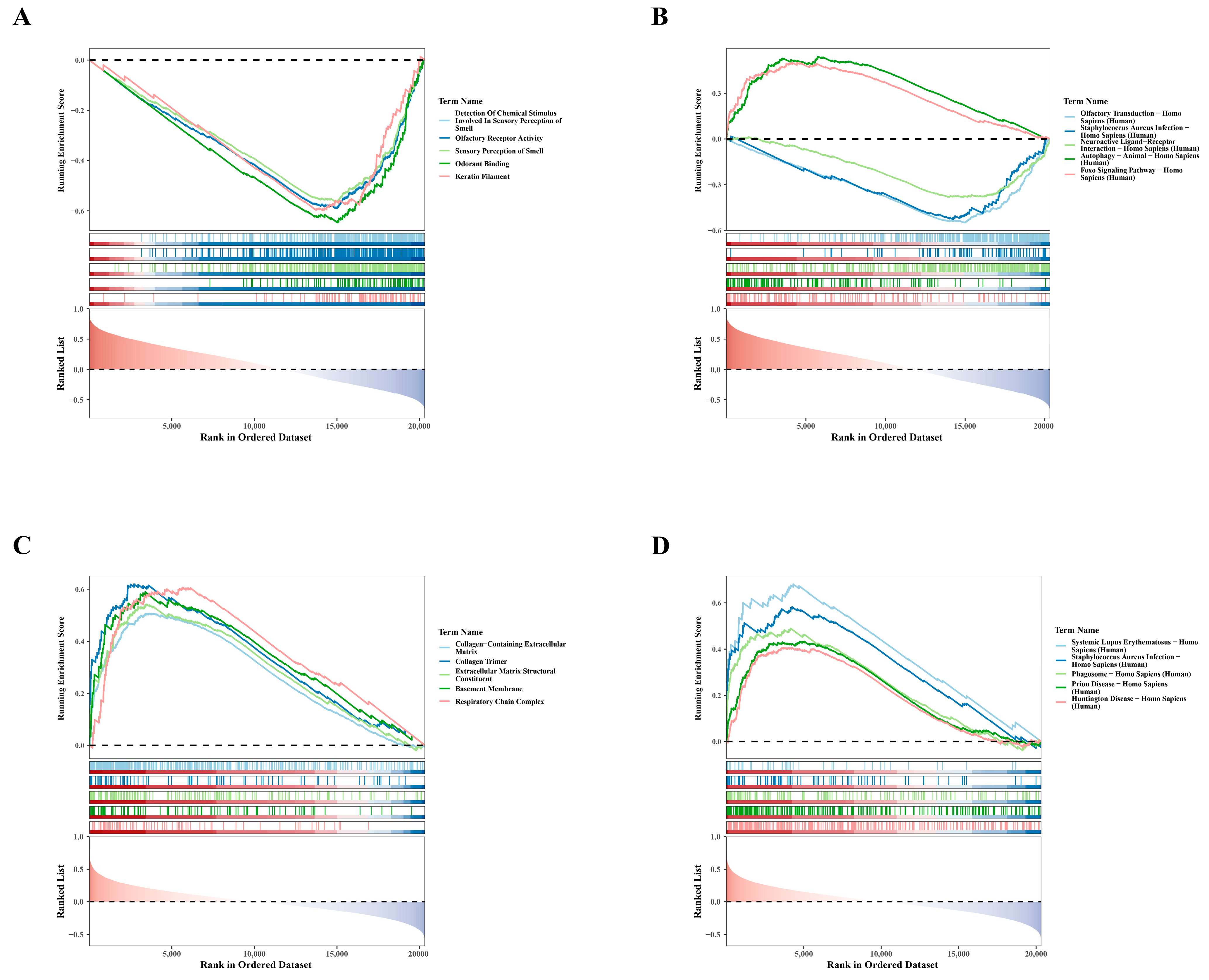
3.4. Single-Gene GSEA Analysis: SLC3A2 and PDLIM1
3.5. Identification of Three Types of Differential Immune Cells
3.6. Screening and Characterization of Ten Highly Variable Genes
3.7. Annotation of Six Cell Types from Twelve Cell Clusters
3.8. Differential Expression of the Two Hub Genes in the Differentiation Stage
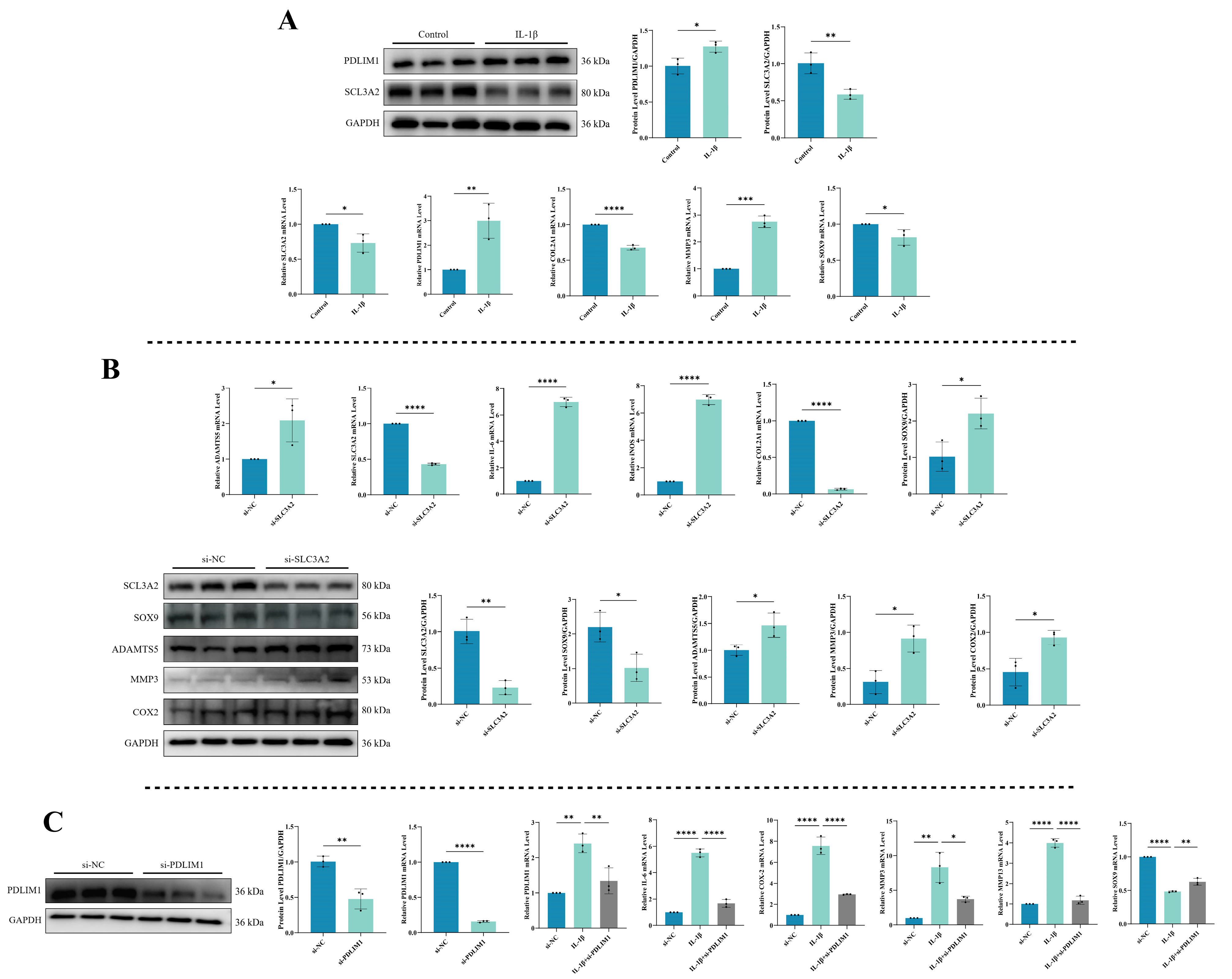
3.9. Expression Levels of SLC3A2 and PDLIM1 in Chondrocytes under Inflammatory Conditions
3.10. Impact of SLC3A2 Knockdown on Chondrocytes
3.11. Consequences of PDLIM1 Knockdown in Chondrocytes under Inflammatory Conditions
4. Discussion
4.1. Implication of Hub Genes in the Biological Processes of OA
4.2. Involvement of SLC3A2 and PDLIM1 in the Pathogenesis of OA as Hub Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hunter, D.; Bierma-Zeinstra, S. Osteoarthritis. Lancet 2019, 393, 1745–1759. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.; March, L.; Chew, M. Osteoarthritis in 2020 and beyond: A Lancet Commission. Lancet 2020, 396, 1711–1712. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, T.; McCabe, P.; McBeth, J. Update on the epidemiology, risk factors and disease outcomes of osteoarthritis. Best Pract. Res. Clin. Rheumatol. 2018, 32, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Wei, Y.; Xu, H.; Weng, J.; Qi, T.; Yu, F.; Liu, S.; Xiong, A.; Liu, P.; Zeng, H. Crosstalk between ferroptosis and chondrocytes in osteoarthritis: A systematic review of in vivo and in vitro studies. Front. Immunol. 2023, 14, 1202436. [Google Scholar] [CrossRef]
- Krakowski, P.; Karpiński, R.; Maciejewski, R.; Jonak, J.; Jurkiewicz, A. Short-Term Effects of Arthroscopic Microfracturation of Knee Chondral Defects in Osteoarthritis. Appl. Sci. 2020, 10, 8312. [Google Scholar] [CrossRef]
- Neogi, T. The epidemiology and impact of pain in osteoarthritis. Osteoarthr. Cartil. 2013, 21, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Johnson, V.L.; Hunter, D.J. The epidemiology of osteoarthritis. Best Pract. Res. Clin. Rheumatol. 2014, 28, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Reyes, C.; Garcia-Gil, M.; Elorza, J.M.; Mendez-Boo, L.; Hermosilla, E.; Javaid, M.K.; Cooper, C.; Diez-Perez, A.; Arden, N.K.; Bolibar, B.; et al. Socio-economic status and the risk of developing hand, hip or knee osteoarthritis: A region-wide ecological study. Osteoarthr. Cartil. 2015, 23, 1323–1329. [Google Scholar] [CrossRef]
- Yuan, C.; Pan, Z.; Zhao, K.; Li, J.; Sheng, Z.; Yao, X.; Liu, H.; Zhang, X.; Yang, Y.; Yu, D.; et al. Classification of four distinct osteoarthritis subtypes with a knee joint tissue transcriptome atlas. Bone Res. 2020, 8, 38. [Google Scholar] [CrossRef]
- Fischenich, K.M.; Wahlquist, J.A.; Wilmoth, R.L.; Cai, L.; Neu, C.P.; Ferguson, V.L. Human articular cartilage is orthotropic where microstructure, micromechanics, and chemistry vary with depth and split-line orientation. Osteoarthr. Cartil. 2020, 28, 1362–1372. [Google Scholar] [CrossRef]
- Yang, J.; Hu, S.; Bian, Y.; Yao, J.; Wang, D.; Liu, X.; Guo, Z.; Zhang, S.; Peng, L. Targeting Cell Death: Pyroptosis, Ferroptosis, Apoptosis and Necroptosis in Osteoarthritis. Front. Cell Dev. Biol. 2021, 9, 789948. [Google Scholar] [CrossRef]
- Liu, S.; Pan, Y.; Li, T.; Zou, M.; Liu, W.; Li, Q.; Wan, H.; Peng, J.; Hao, L. The Role of Regulated Programmed Cell Death in Osteoarthritis: From Pathogenesis to Therapy. Int. J. Mol. Sci. 2023, 24, 5364. [Google Scholar] [CrossRef]
- Cao, S.; Wei, Y.; Yue, Y.; Xiong, A.; Zeng, H. Zooming in and Out of Programmed Cell Death in Osteoarthritis: A Scientometric and Visualized Analysis. J. Inflamm. Res. 2024, 17, 2479–2498. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Nie, L.; Zhang, Y.; Yan, Y.; Wang, C.; Colic, M.; Olszewski, K.; Horbath, A.; Chen, X.; Lei, G.; et al. Actin cytoskeleton vulnerability to disulfide stress mediates disulfidptosis. Nat. Cell Biol. 2023, 25, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, W.; Li, Y.; Ma, L.; Ji, Z. Leveraging a disulfidptosis-based signature to improve the survival and drug sensitivity of bladder cancer patients. Front. Immunol. 2023, 14, 1198878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, T.; Ji, K.; Cao, S.; Ai, J.; Pan, J.; Cao, Y.; Yang, Y.; Jing, L.; Sun, J.H. Development and experimental validation of a machine learning-based disulfidptosis-related ferroptosis score for hepatocellular carcinoma. Apoptosis Int. J. Program. Cell Death 2023, 29, 103–120. [Google Scholar] [CrossRef]
- Liu, F.; Yuan, D.; Liu, X.; Zhuo, S.; Liu, X.; Sheng, H.; Sha, M.; Ye, J.; Yu, H. A demonstration based on multi-omics transcriptome sequencing data revealed disulfidptosis heterogeneity within the tumor microenvironment of esophageal squamous cell carcinoma. Discover. Oncol. 2023, 14, 96. [Google Scholar] [CrossRef]
- Huang, J.; Zhang, J.; Zhang, F.; Lu, S.; Guo, S.; Shi, R.; Zhai, Y.; Gao, Y.; Tao, X.; Jin, Z.; et al. Identification of a disulfidptosis-related genes signature for prognostic implication in lung adenocarcinoma. Comput. Biol. Med. 2023, 165, 107402. [Google Scholar] [CrossRef]
- Ma, S.; Wang, D.; Xie, D. Identification of disulfidptosis-related genes and subgroups in Alzheimer’s disease. Front. Aging Neurosci. 2023, 15, 1236490. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhang, C.; Ni, S.; Ma, M.; Zhang, X.; Sang, W.; Lv, T.; Qian, Z.; Yi, C.; Yu, B. NFATc1-mediated expression of SLC7A11 drives sensitivity to TXNRD1 inhibitors in osteoclast precursors. Redox Biol. 2023, 63, 102711. [Google Scholar] [CrossRef]
- Sasaki, E.; Yamamoto, H.; Asari, T.; Matsuta, R.; Ota, S.; Kimura, Y.; Sasaki, S.; Ishibashi, K.; Yamamoto, Y.; Kami, K.; et al. Metabolomics with severity of radiographic knee osteoarthritis and early phase synovitis in middle-aged women from the Iwaki Health Promotion Project: A cross-sectional study. Arthritis Res. Ther. 2022, 24, 145. [Google Scholar] [CrossRef]
- Zhao, S.; Wang, L.; Ding, W.; Ye, B.; Cheng, C.; Shao, J.; Liu, J.; Zhou, H. Crosstalk of disulfidptosis-related subtypes, establishment of a prognostic signature and immune infiltration characteristics in bladder cancer based on a machine learning survival framework. Front. Endocrinol. 2023, 14, 1180404. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gustavsson, E.K.; Zhang, D.; Reynolds, R.H.; Garcia-Ruiz, S.; Ryten, M. ggtranscript: An R package for the visualization and interpretation of transcript isoforms using ggplot2. Bioinformatics 2022, 38, 3844–3846. [Google Scholar] [CrossRef]
- Gu, Z.; Hübschmann, D. Make Interactive Complex Heatmaps in R. Bioinformatics 2022, 38, 1460–1462. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Ghahghaei, S.; Linnell, K.J. The effect of load on spatial attention depends on preview: Evidence from a reading study. Vis. Res. 2018, 149, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Tatsumi, K.; Igarashi, N.; Mengxue, X. Prediction of plant-level tomato biomass and yield using machine learning with unmanned aerial vehicle imagery. Plant Methods 2021, 17, 77. [Google Scholar] [CrossRef] [PubMed]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.C.; Müller, M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Sachs, M.C. plotROC: A Tool for Plotting ROC Curves. J. Stat. Softw. 2017, 79, 1–19. [Google Scholar] [CrossRef]
- Sturm, G.; Finotello, F.; List, M. Immunedeconv: An R Package for Unified Access to Computational Methods for Estimating Immune Cell Fractions from Bulk RNA-Sequencing Data. Methods Mol. Biol. 2020, 2120, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Aibar, S.; González-Blas, C.B.; Moerman, T.; Huynh-Thu, V.A.; Imrichova, H.; Hulselmans, G.; Rambow, F.; Marine, J.-C.; Geurts, P.; Aerts, J.; et al. SCENIC: Single-cell regulatory network inference and clustering. Nat. Methods 2017, 14, 1083–1086. [Google Scholar] [CrossRef] [PubMed]
- Karsdal, M.A.; Michaelis, M.; Ladel, C.; Siebuhr, A.S.; Bihlet, A.R.; Andersen, J.R.; Guehring, H.; Christiansen, C.; Bay-Jensen, A.C.; Kraus, V.B. Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: Lessons learned from failures and opportunities for the future. Osteoarthr. Cartil. 2016, 24, 2013–2021. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Chen, S.; Xie, Z.; Shen, S.; Xu, W.; Chen, W.; Li, X.; Wu, Y.; Li, L.; Liu, B.; et al. TGFβ attenuates cartilage extracellular matrix degradation via enhancing FBXO6-mediated MMP14 ubiquitination. Ann. Rheum. Dis. 2020, 79, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, Y. Targeting Autophagy in Aging and Aging-Related Cardiovascular Diseases. Trends Pharmacol. Sci. 2018, 39, 1064–1076. [Google Scholar] [CrossRef] [PubMed]
- Caramés, B.; Hasegawa, A.; Taniguchi, N.; Miyaki, S.; Blanco, F.J.; Lotz, M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann. Rheum. Dis. 2012, 71, 575–581. [Google Scholar] [CrossRef]
- Feng, L.; Feng, C.; Wang, C.X.; Xu, D.Y.; Chen, J.J.; Huang, J.F.; Tan, P.L.; Shen, J.M. Circulating microRNA let-7e is decreased in knee osteoarthritis, accompanied by elevated apoptosis and reduced autophagy. Int. J. Mol. Med. 2020, 45, 1464–1476. [Google Scholar] [CrossRef]
- Shapiro, I.M.; Layfield, R.; Lotz, M.; Settembre, C.; Whitehouse, C. Boning up on autophagy: The role of autophagy in skeletal biology. Autophagy 2014, 10, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Lai, Y.; Chen, S.; Zhou, C.; Tao, C.; Fu, X.; Li, J.; Tong, W.; Tian, H.; Shao, Z.; et al. Kindlin-2 preserves integrity of the articular cartilage to protect against osteoarthritis. Nat. Aging 2022, 2, 332–347. [Google Scholar] [CrossRef] [PubMed]
- Fort, J.; de la Ballina, L.R.; Burghardt, H.E.; Ferrer-Costa, C.; Turnay, J.; Ferrer-Orta, C.; Usón., I.; Zorzano, A.; Fernández-Recio, J.; Orozco, M.; et al. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J. Biol. Chem. 2007, 282, 31444–31452. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xiao, Y.; Xu, D.; Huili, Y.; Nie, S.; Li, H.; Guan, X.; Cao, F. Prognosis Prediction of Disulfidptosis-Related Genes in Bladder Cancer and a Comprehensive Analysis of Immunotherapy. Crit. Rev. Eukaryot. Gene Expr. 2023, 33, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Yang, H.; Wu, X.; Zhou, K.; Wang, S. The expression and prognostic value of disulfidptosis progress in lung adenocarcinoma. Aging 2023, 15, 7741–7759. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Deng, Z.; Yu, B.; Liu, H.; Yang, Z.; Zeng, A.; Fu, M. Identification of SLC3A2 as a Potential Therapeutic Target of Osteoarthritis Involved in Ferroptosis by Integrating Bioinformatics, Clinical Factors and Experiments. Cells 2022, 11, 3430. [Google Scholar] [CrossRef]
- Zhou, J.K.; Fan, X.; Cheng, J.; Liu, W.; Peng, Y. PDLIM1: Structure, function and implication in cancer. Cell Stress 2021, 5, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Bauer, K.; Kratzer, M.; Otte, M.; de Quintana, K.L.; Hagmann, J.; Arnold, G.J.; Eckerskorn, C.; Lottspeich, F.; Siess, W. Human CLP36, a PDZ-domain and LIM-domain protein, binds to alpha-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood 2000, 96, 4236–4245. [Google Scholar] [CrossRef] [PubMed]
- Joos, H.; Albrecht, W.; Laufer, S.; Reichel, H.; Brenner, R.E. IL-1beta regulates FHL2 and other cytoskeleton-related genes in human chondrocytes. Mol. Med. 2008, 14, 150–159. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, Q.; Zhang, R.; Chen, L.; Gao, S.; Wang, Y.; Li, T.; Xue, J. Direct Knockdown of PDZ and LIM Domain 1 Using an Adenoviral Delivery System Accelerates Osteogenesis and Fracture Healing in Mice. Hum. Gene Ther. 2023, 34, 649–661. [Google Scholar] [CrossRef]
- Yao, Q.; Wu, X.; Tao, C.; Gong, W.; Chen, M.; Qu, M.; Zhong, Y.; He, T.; Chen, S.; Xiao, G. Osteoarthritis: Pathogenic signaling pathways and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 56. [Google Scholar] [CrossRef] [PubMed]
- Ye, B.; Yu, M.; Yue, M.; Yin, M.; Zhang, C.; Wang, Q.; Ding, X.; Shen, W.; Zhao, Z. Role of PDLIM1 in hepatic stellate cell activation and liver fibrosis progression. Sci. Rep. 2023, 13, 10946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, J.; Wang, X.; Sun, Y.; Yang, T.; Shen, X.; Yang, X.; Shi, H.; Sun, X.; Xin, A. Loss of ACTL7A causes small head sperm by defective acrosome-acroplaxome-manchette complex. Reprod. Biol. Endocrinol. 2023, 21, 82. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Wang, H.; Jia, P.; Zhao, H.; Liu, C.; Liu, W.; Song, Z.; Xu, Z.; Yang, L.; Wang, Y.; et al. Autophagy regulates spermatid differentiation via degradation of PDLIM1. Autophagy 2016, 12, 1575–1592. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, J.K.; Wang, K.; Chen, H.; Qin, S.; Liu, J.; Luo, M.; Chen, Y.; Jiang, J.; Zhou, L.; et al. PDLIM1 Inhibits Tumor Metastasis Through Activating Hippo Signaling in Hepatocellular Carcinoma. Hepatology 2020, 71, 1643–1659. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, S.; Wei, Y.; Yue, Y.; Wang, D.; Xiong, A.; Yang, J.; Zeng, H. Bioinformatics Identification and Experimental Verification of Disulfidptosis-Related Genes in the Progression of Osteoarthritis. Biomedicines 2024, 12, 1840. https://doi.org/10.3390/biomedicines12081840
Cao S, Wei Y, Yue Y, Wang D, Xiong A, Yang J, Zeng H. Bioinformatics Identification and Experimental Verification of Disulfidptosis-Related Genes in the Progression of Osteoarthritis. Biomedicines. 2024; 12(8):1840. https://doi.org/10.3390/biomedicines12081840
Chicago/Turabian StyleCao, Siyang, Yihao Wei, Yaohang Yue, Deli Wang, Ao Xiong, Jun Yang, and Hui Zeng. 2024. "Bioinformatics Identification and Experimental Verification of Disulfidptosis-Related Genes in the Progression of Osteoarthritis" Biomedicines 12, no. 8: 1840. https://doi.org/10.3390/biomedicines12081840
APA StyleCao, S., Wei, Y., Yue, Y., Wang, D., Xiong, A., Yang, J., & Zeng, H. (2024). Bioinformatics Identification and Experimental Verification of Disulfidptosis-Related Genes in the Progression of Osteoarthritis. Biomedicines, 12(8), 1840. https://doi.org/10.3390/biomedicines12081840






