Abstract
The “Exposome” is a concept that indicates the set of exposures to which a human is subjected during their lifetime. These factors influence the health state of individuals and can drive the development of Noncommunicable Diseases (NCDs). Artificial Intelligence (AI) allows one to analyze large amounts of data in a short time. As such, several authors have used AI to study the relationship between exposome and chronic diseases. Under such premises, this study reviews the use of AI in analyzing the exposome to understand its role in the development of chronic diseases, focusing on how AI can identify patterns in exposure-related data and support prevention strategies. To achieve this, we carried out a search on multiple databases, including PubMed, ScienceDirect, and SCOPUS, from 1 January 2019 to 31 May 2023, using the MeSH terms (exposome) and (‘Artificial Intelligence’ OR ‘Machine Learning’ OR ‘Deep Learning’) to identify relevant studies on this topic. After completing the identification, screening, and eligibility assessment, a total of 18 studies were included in this literature review. According to the search, most authors used supervised or unsupervised machine learning models to study multiple exposure factors’ role in the risk of developing cardiovascular, metabolic, and chronic respiratory diseases. In some more recent studies, authors also used deep learning. Furthermore, the exposome analysis is useful to study the risk of developing neuropsychiatric disorders or evaluating pregnancy outcomes and child growth. Understanding the role of the exposome is pivotal to overcome the classic concept of a single exposure/disease. The application of AI allows one to analyze multiple environmental risks and their combined effects on health conditions. In the future, AI could be helpful in the prevention of chronic diseases, providing new diagnostic, therapeutic, and follow-up strategies.
1. Introduction
The main agents contributing to the development of the largest part of diseases known to date include genetic factors, i.e., the “genome” [], environmental factors, i.e., the “exposome” [], and infections []. The interplay of genomes and the exposome gives rise to some of the most popular Noncommunicable Diseases (NCDs). NCDs are different from the so-called Communicable Diseases (CDs), since the latter are referred to as infectious conditions, whereas NCDs include chronic diseases resulting from a combination of genetic, environmental (air pollution, climate change, etc.), sociodemographic (age, gender, etc.), and self-management (smoking, diet, physical activity, etc.) factors, and medical conditions (obesity, stress, blood pressure, etc.) []. Globally, NCDs lead to the death of 41 million people yearly (74% of all global deaths) [,], constituting a significant health burden, especially in wealthier countries [].
The main types of NCDs are represented by cardiovascular diseases, cancer, chronic respiratory diseases, and metabolic conditions, including diabetes. However, many other pathologies are present regarding gastroenterological, renal, hepatic, dermatologic, hematological, endocrine, and neurological systems [,,]. In recent decades, the genetic components of NCDs have been studied through gene sequencing and mapping to identify the genetic factors underlying such diseases. These studies enabled the understanding of gene expression and protein function in the identification of the biochemical pathways involved in the natural history of chronic diseases. Although many genes predisposing individuals to the main chronic diseases have been identified, our knowledge remains limited because, beyond genetics, it is essential to understand the interaction with environmental factors in developing the aforementioned conditions [,].
Under this light, the application of strategies aimed at reducing environmental exposure to risk factors is pivotal for disease control [,]. In this framework, the concept of the “exposome”, introduced in 2005 by Christopher Paul Wild, can be defined as the measure of all the exposures of an individual within a lifetime and how those exposures relate to health []; it is composed by three overlapping domains: general external, specific external, and internal []. The general external exposome includes measurable levels of exposure, including air pollution and meteorological factors. The specific external exposome includes information on individual exposures, including lifestyle factors (diet, physical activity, smoking, drugs, for example). Finally, the internal exposome encompasses multiple biological responses to external factors detected through molecular and omics analyses. In recent years, a new domain has emerged: the socio-exposome, i.e., the product of the interaction between health and socio-economic factors [] (see Table 1).

Table 1.
List of relevant exposures.
Notably, the exposome plays an essential role since genetic factors and enhanced clinical capabilities alone cannot explain such a rapid change in the incidence of many chronic diseases; therefore, knowledge of this aspect may explain why some people develop a disease while others, with a common genetic background and apparently similar clinical characteristics, do not. In addition, we should consider that the etiology of a disease is rarely explained by a single exposure; therefore, examining the human exposome becomes relevant to simultaneously consider multiple risk factors and more accurately estimate concurrent causes of different health outcomes. The considerable and continuously increasing data production in exposome studies has led researchers to introduce new approaches to evaluate the effect of the exposome on health as a whole, or to consider the contributions of single exposures. Among these principles, Artificial Intelligence (AI), through machine learning and deep learning, and beyond them, has begun to emerge as the elective path to follow in this regard.
AI can be understood as the part of computer science and other disciplines that analyzes complex data, and it is widely applied in nearly every sector, including the (bio-)medical world. In medicine, such data can be used in the diagnosis, treatment, and prediction of outcomes and can help to reduce diagnostic and therapeutic errors and human biases that normally occur in the clinical practice, as with experienced and skilled professionals [].
More specifically, ML is a subset of AI that enables computers to utilize large amounts of data to make predictions without being explicitly programmed to do so []. Many different ML categories are available for several applications, including Supervised Learning, Unsupervised Learning, Semi-supervised Learning, and Reinforcement Learning.
Supervised Learning involves using labeled datasets to train algorithms for accurate classification or regression tasks, most frequently in the domain of outcome prediction. Common Supervised Learning algorithms include Linear Regression, used to predict numerical values based on a linear relationship between different values; Logistic Regression (LR), which makes predictions for categorical response variables, such as “yes/no”; Random Forest (RF), which predicts a value or category by combining the results from many decision trees; decision trees, which are used for both predicting numerical values, regression, and classifying data; Naïve Bayes, which uses Bayes’ theorem to classify objects; and Neural Networks, which use Artificial Neural Networks (ANNs) to imitate the human brain’s learning process.
Unsupervised Learning applies unlabeled and unclassified datasets to make predictions without human intervention. It does normally perform Clustering (algorithms that identify patterns in data so that it can be grouped), and Association (discovering interesting relationships between features in a given dataset), as well as dimensionality reduction.
Semi-supervised Learning is a highly efficient ML approach combining labeled and unlabeled data during training.
Finally, Reinforcement Learning (RL) is an ML technique where an agent learns to take optimal actions through environmental feedback. In positive-RL, the agent is rewarded for taking actions that lead to positive outcomes, whereas in negative-RL, the agent is punished for taking actions leading to negative outcomes [].
As said, AI in general, and ML in particular, include specific tools which enable a systematic approach to adequately support the investigation towards the complex etiopathogenesis of chronic diseases and the management of these conditions that typically involve multiple characteristics (genetics, lifestyle choices, and environmental factors), differing from patient to patient and capable of contributing to their onset []; therefore, their application to studies about the exposome is quite straightforward, as the main advantage of the exposome over traditional ‘one-exposure-one-disease’ study approaches is that it provides an unprecedented conceptual framework for the study of multiple environmental hazards (urban, chemical, lifestyle, social) and their combined effects. Given the increasing availability of complex environmental health data, there is a need for more advanced statistical approaches focusing on complex mixtures of exposures; therefore, the management of chronic diseases is a task perfectly suited to innovative technologies such as AI that can speed up the development of personalized treatments []. In addition, AI could support the development of practical prevention tools, planning the management of NCDs, with key impacts for healthcare and resource optimization [,,].
Literature reviews dealing with the topic are scarce and not recent. For example, one of the main contributions to scientific knowledge in the field was published by Subramanian and co-authors in 2020 []; however, it is easy to understand that, especially within a topic that is revolutionizing scientific research in the medical field, updated evidence should be provided to promptly inform the scientific community about the latest advancements in the field.
Therefore, the present work seeks to identify the current literature contributions dealing with AI in the exposome framework, thus providing an updated perspective on AI applications’ benefits and future opportunities in studying the exposome, evaluating its implications in clinical practice to promote prevention interventions and new diagnostic, therapeutic, and follow-up strategies for chronic diseases.
2. Materials and Methods
A research study on AI applied to the exposome was conducted through a search on PubMed, ScienceDirect, and SCOPUS from 1 January 2019 to 31 May 2023 by using the Mesh terms: (“exposome”) and (“Artificial Intelligence” OR “Machine Learning” OR “Deep Learning”) according to the PRISMA guidelines []. Original articles were included in the analysis, whereas reviews, meta-analysis, proceedings, abstracts, and book chapters were excluded. Also, articles published in a language other than English were not taken into account.
In a subsequent analysis, out of the 55 unique studies observed, 18 were considered to be eligible as studies matching the criteria mentioned below were excluded.
As such, the papers excluded were those for which a subsequent analysis of the text, title, and abstract did not allow inclusion for thematic reasons, review articles, and papers written in a language other than the English language (Figure 1).
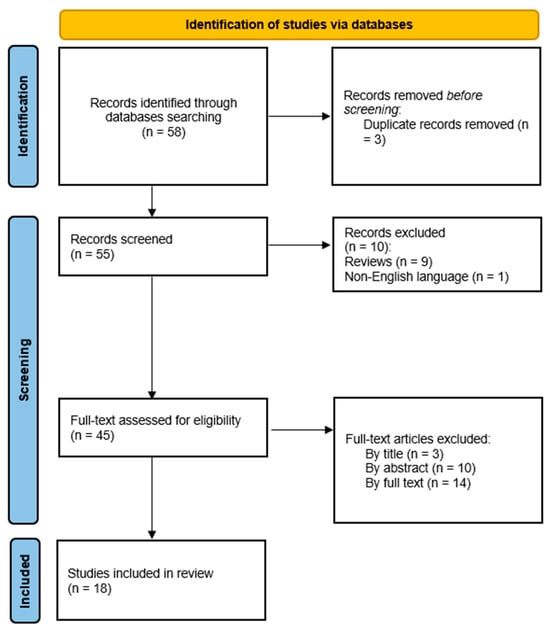
Figure 1.
Study selection overview.
3. Results and Discussion
After about four decades since the Human Genome Project takeoff, it is now clear that only a tiny percentage of chronic diseases are linked to purely genetic causes; indeed, genetic factors can play an important role in the development of CDs, but pathogenic microorganisms constitute another determining factor, and the majority of such conditions can be caused or worsened by so-called “exposures” [].
In 2005, Wild highlighted the importance of developing reliable exposure assessment tools to promote genome characterization [].
On 1 January 2020, the European Human Exposome Network (EHEN) [] was established. The EHEN is the world’s largest network of projects studying human health in relationship to the environment and includes a group of nine projects funded since 2020 for five years by the EU Horizon 2020 program for research and innovation. The EHEN research projects include the following: Human Exposomic Determinants of Immune-Mediated Diseases (HEDIMED) []; Dynamic Longitudinal Exposome Trajectories in Cardiovascular and Metabolic Non-communicable Diseases (LONGITOOLS) []; Impact of Exposome on the Course of Lung Diseases (REMEDIA) []; Advancing Tools for Human Early Life-course Exposome Research and Translation (ATHLETE) []; Exposome Project for Health and Occupational Research (EPHOR) []; Mapping Exposure-induced Immune Effects: Connecting the Exposome and the Immunome (EXIMIOUS) []; Early Environmental Quality and Life-course Mental Health Effects (EQUAL-LIFE) []; Exposome Powered Tools for Healthy Living in Urban Settings (EXPANSE) []; Human Exposome Assessment Platform (HEAP) [].
When it comes to the present work, the literature analysis here conducted found multiple associations between diseases and risk factors, which were investigated through AI algorithms.Among the projects included in the EHEN, LONGITOOLS was developed with the aim of assessing which exposures are associated with the development of cardiovascular and metabolic disorders, with these in turn being those with the highest mortality worldwide [].
Another initiative, namely FLExiGUT [], represents the first large-scale exposome study focusing on low-grade chronic inflammation. It aims to characterize environmental exposure throughout human life to assess and validate its impact on intestinal inflammation and biological processes and related diseases, including metabolic disorders, food allergies, accelerated biological aging, and gastrointestinal cancers [].
Multiple exposures and their interaction result in much stronger health effects than any single exposure []; therefore, it is necessary to seek further links between risk factors to study their impact on human health and to develop increasingly effective preventive actions []. To this extent, the knowledge of the main determinants of health has increased in recent years; however, the etiology of most NCDs, or chronic diseases, is still poorly understood.
For this reason, it is pivotal to study the effects of the different human exposures to evaluate their effects over time, especially when it comes to the appearance of undesirable effects, and also to find the starting point for prevention and health promotion projects.
For this purpose, it was stated that “The Human Exposome Assessment Platform (HEAP) is a research resource for the integrated and efficient management and analysis of human exposome data. The primary goal of HEAP is to enable global collaborative research on exposure to cost-effective health interventions” [].
The articles and results included in our investigation are shown in Table 2, with a summary of the main characteristics of the AI models employed depicted in Table 3.

Table 2.
Included articles and their characteristics (ANN: Artificial Neural Network; BMI: body mass index; CNN: Convolutional Neural Networks; GNB: Gaussian Naive Bayes; KNN: K-Nearest Neighbor; LASSO: Least Absolute Shrinkage and Selection Operator Regression; LR: logistic regression; ML: Machine Learning; MLR: Multiple Linear Regression; PLS: Partial Least Squares; PLS-DA: Partial Least Squares Discriminant Analysis; RF: Random Forest; SVM: Support Vector Machine, T2D: type 2 diabetes).

Table 3.
Main models employed in studies included in present analysis.
Regarding cardiovascular diseases, the main studies here included analyze inputs like air pollution, poor food quality, neighborhood, sleep disturbances, and family history, in turn being associated with conditions such as obesity or the presence of high BMI [], diabetes [], stroke, and coronary heart disease [].
In particular, Ohanyan et al. [] performed a cross-sectional analysis using data from 14,829 participants from the Occupational and Environmental Health Cohort study and 86 different environmental factors. The exposures were derived linking the individual home addresses based on geolocalization. Authors applied different ML approaches (sparse-group Partial Least Squares, Bayesian Model Averaging, penalized regression using the Minimax Concave Penalty, Generalized Additive Model-based boosted RF, Extreme Gradient Boosting, and Multiple Linear Regression) to characterize the obesogenic urban exposome, demonstrating the role of neighborhood socioeconomic position, urbanicity, and air pollution []; these data have been assessed by self-reported questionnaires.
On the same data, the authors applied Least Absolute Shrinkage and Selection Operator (LASSO), RF, and ANN models to study the association between type 2 diabetes (T2D) and the urban exposome, derived as specified above []; in this way, they identified neighborhood socio-economic and socio-demographic characteristics and surface temperature to be strongly associated with T2D risk, and also in this case used self-reported questionnaires [].
Lee et al. [] used an exposure-level association study (ExWAS) to examine all exposure–outcome associations to better understand environmental effects on cardiovascular health. Broad and comprehensive questionnaire-based exposome data that cross-sectionally encompass internal and external exposures at work and home have been collected within a North Carolina-based cohort. The authors applied the deletion–substitution–addition (DSA) variable selection algorithm to build up a final multi-exposure model []. Analyses showed new associations between blood type A (Rh-) with heart attack, paint exposures with stroke, exposure to biohazardous materials with arrhythmia, and higher paternal education level with a reduced risk of multiple CVD outcomes. In multiple exposure models, sleep disorders and smoking are confirmed as important risk factors [].
Asthma and chronic obstructive pulmonary disease (COPD) [,] are highly debilitating chronic respiratory conditions, on which some AI-based models have been used to develop causation patterns. Among the main players, air pollution is notoriously a common factor in various chronic diseases.
De Vito et al. [] developed an IoT AQMS architecture called MONICA (MONItoraggio Coooperativo della qualità dell’Aria or “Cooperative Air Quality Monitoring”) based on a hybrid network of low-cost portable devices using electrochemical sensor arrays for air quality assessment and exposome monitoring. The platform has demonstrated good accuracy, maintaining good performance over the long term. However, an annual recalibration routine seems to be the minimum requirement to ensure performance [].
Other projects to assess the most frequent risk factors for development or exacerbations of chronic respiratory diseases have been started, including the REMEDIA project [] and the PROMESA cohort study []. According to the evidence, the risk factors most frequently involved in the pathogenesis and exacerbation of respiratory diseases include air pollution (both indoor and outdoor) and cigarette smoking. Sharing common etiopathological pathways, allergic diseases have also been investigated with the support of AI. Notably, ML has been used in this specific field [] with the ambitious challenge of achieving precise characterizations of allergic endotypes allergy medicine, understanding allergic multimorbidity relationships, contextualizing the impact of exposure and ancestry/genetic risks, achieving viable multi-omic integration, and using this information to develop patient cohorts and refined clinical trials.
In their study of 19 asthma patients, Bae et al. [] evaluated the correlation between asthma exacerbation probability and patient exposure to indoor environmental factors. The air quality data points were obtained through a laser light-scattering sensor and the peak expository flow rate (PEFR) measurements were used to evaluate lung function. The authors overcome the limitations of existing predictive models by using two regression models, obtaining an increase in the accuracy of the classification model of 11.5–18.4% [], but it is worth stating that the extremely small sample size could have led to spurious results in terms of the generalizability of the model, overall.
The supervised ML approach was also used to assess infective respiratory disease, primarily COVID-19 outcomes. Ren et al. [] used six geostatistical models and two ML methods (RF, Extreme Gradient Boosting) to assess socio-exposomic associations with COVID-19 outcomes. The authors considered 84 heterogeneous environmental, demographic, and socio-economic factors, retrieved based on the living area of people included in the study according to existing exposure-related databases. This study revealed a strong correlation between COVID-19 mortality and historical exposures to NO2, population density, and the percentage of minors and those with less than a high school education [].
The EUGEI study [] and the Equal Life project [] were conducted to assess how certain types of exposures were associated with mental health and cognitive development disorders. Notably, Pries et al. analyzed factors such as cannabis use, noise pollution, birth in the winter months, and physical and emotional abuse, derived from existing, reliable datasets, as possible risk factors for schizophrenia, using algorithms such as LASSO, Ridge, LR, and Gaussian Naive Bayes (GNB), among which LASSO, Ridge, and LR had the best predictive performances.
Overall, it has been found that social inequalities, abuse, and stress can lead to the onset of psychogenic and also biological disorders in children. These factors can also determine early exposure to cigarette smoking, alcohol, unhealthy diets, attention disorders, cognitive and language delays, and the development of schizophrenia [].
The EU-funded Equal Life project studied combined exposures and their effects on children’s mental and cognitive health based on data from eight birth cohorts and three school studies (N = 240,000) linked to exposure data. The project has enabled the development and use of the exposome concept by integrating internal, physical, and social exposomes and aims to propose the best supportive environments for all children [].
The relationship between phenotypes and chemicals and genes has always aroused great interest from a biomedical point of view; for this reason, tools such as Phexpo have been developed to predict chemical–phenotypic relationships and to better understand these relationships [,].
Recently, Zhao et al. [] used RF to develop a prediction model based on the annotation of chemicals in human blood []. The study was conducted on a sample of 7858 substances selected from the US EPA ToxCast chemical list, which includes more than 9000 compounds, including industrial chemicals, pesticides, consumer product ingredients, and pharmaceuticals. The objective was to develop an ML model to predict blood concentrations of chemicals and prioritize chemicals of health concern. In this specific scenario, RF outperformed the ANN and Support Vector Regression (SVR) models; the chemical compounds most commonly represented in this framework include food additives and pesticides rather than widely monitored environmental pollutants.
Moreover, some studies have been conducted which apply supervised ML models to analyze the presence of chemicals in adipose tissue and the association with the risk of developing endometriosis [] or an alteration of the metabolites in breast milk and the subsequent risk of alterations in the neurological development of the child [].
Matta et al. [] analyzed a dataset from a case–control study conducted in France to identify associations between mixtures of organochlorine persistent organic pollutants (POPs) and endometriosis. The five models tested revealed the POPs most associated with endometriosis: octachlorodibenzofuran, cis-heptachlor epoxide, polychlorinated biphenyl 77, and trans-nonachlor. These compounds were analyzed using gas chromatography coupled with high-resolution mass spectrometry. All models showed excellent classification performances. Amongst them, regularized LR provided a good trade-off between the interpretability of traditional statistical approaches and the classification ability of ML approaches [].
Li et al. [] analyzed the predictive potential for the human milk metabolome and exposome in infants at risk of neurodevelopmental delay in a retrospective cohort study of 82 mother–infant pairs. Milk samples were collected before 9 months of age and neurocognitive development was assessed using the Ages and Stages-2 (ASQ-2) questionnaire. The metabolomic analysis of endogenous metabolites was performed using a UFLC XR HPLC system (LC-20AD, Shimadzu) coupled with an Qtrap 5500 triple quadrupole mass spectrometer. In total, 453 metabolites and 61 environmental chemicals present in breast milk were identified. ML detected changes in deoxysphingolipids, phospholipids, glycosphingolipids, plasmalogens, and acylcarnitines present in the milk of mothers with infants at risk of delayed neurocognitive development [].
Instead, Louis et al. [] used an unsupervised model on 50 women with uncomplicated pregnancies [] with the objective of evaluating how some chemical substances (EDC) change in the serum and the urine for each trimester of pregnancy since, as we know, the exposures that pregnant women encounter during this critical and sensitive window of fetal development can impact maternal and infant health. Four chemical clusters included 80 compounds, of which six increased, 63 steadily decreased, and 11 reflected inconsistent patterns during pregnancy. Overall, concentrations tended to decrease during pregnancy for persistent EDCs, whereas an inverse trend was observed for many non-persistent chemicals. Gas chromatography interfaced with high-resolution mass spectrometry (HRGC-HRMS) was used to quantify most persistent EDCs in serum (polychlorinated biphenyls, polybrominated diphenyl ethers, and organochlorine pesticides; Table 1), whereas perfluoroalkyl and polyfluoroalkyl substances (PFASs) were measured using high-performance liquid chromatography–tandem mass spectrometry (HPLC-MS/MS). Various classes of non-persistent EDCs including phthalate metabolites, environmental phenols, organophosphate pesticide metabolites, hydroxylated polycyclic aromatic hydrocarbons, phytoestrogens, perchlorate, and other related anions were quantified in urine by HPLC-MS/MS. The explained variance was highest for five persistent chemicals: polybrominated diphenyl ethers #191 (51%) and #126 (47%), hexachlorobenzene (46%), p,p’-dichloro-diphenyl—dichloroethylene (46%), and o, p’-dichloro-diphenyl-dichloroethane (36%). The concentrations of many EDCs are not stable during pregnancy and reflect varying patterns depending on their persistence, highlighting the importance of timed biological sample collection []. Dietary, iatrogenic, or environmental exposures could also alter erythrocyte energy and redox metabolism, influencing the quality of red blood cell preservation and posttransfusion efficacy []. For this reason, Nemkov et al. [] studied blood donor exposomes, derived from existing datasets, and data regarding the use of common medications in 250 healthy volunteers in the Recipient Epidemiology and Donor Evaluation Study III Red Blood Cell-Omics Study (REDS-III RBC-Omics). By carrying out pharmacological screening on 1366 drugs, it emerged that approximately 65% of these had an impact on the metabolism of erythrocytes. Specifically, the authors focus on the anti-acid ranitidine, which had a substantial effect on markers of the quality of preservation of red blood cells in vitro [].
The RF algorithm was successfully used to evaluate the impact of pharmaceuticals and surfactants on the resistome and microbiota on both hospital wastewater (HWW) and municipal wastewater (UWW) [] in 126 WW samples (UWW, HWW, and WW mixed) collected in a French city for approximately four years (34 months with different treatments for H and UWW and 11 months with H and UWW mixed 1:2 HWW:UWW in a single system) []. After filtration for microorganisms, water samples were subjected to DNA extraction, and DNA concentration was estimated through fluorimetric quantification, then subjected to quantitative PCR and 16S rRNA analysis. On the other hand, chemical data were treated using solid-phase extraction (SPE) and liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS), used in particular to measure antibiotics like ciprofloxacin, sulfamethoxazole, and vancomycin, and the pharmaceutical carbamazepine. In addition, heavy metals (Zn, Cu, Ni, Pb, Cr, Gd, Hg, As, and Cd) were measured with inductively coupled plasma combined with atomic emission spectroscopy (ICP-AES).
The differences between the H and UWW signatures are consistent with the differences in antibiotic exposure in the two frameworks. Furthermore, hospital WW contains significantly greater loads of antimicrobial resistance genes, mobile genetic elements, and integrons [].
Supervised models, mainly the RF model, were also selected to analyze which longitudinal exposures had the most significant predictive value for self-perceived health in a cohort of 3419 Doetinchem participants over 30 years []. This study considers many exposome models from different domains: demographic, lifestyle, environmental, and biological. More specifically, exposome variables include the measurements related to the physical environment outside and inside the participants’ home, whereas biological internal measurements were performed by trained staff. Other variables were collected through self-reported approaches.
The RF model’s ability to discriminate between poor and good self-perceived health was deemed acceptable. The other 87 exposures contributed little to the performance.
Johnson et al. [] used Convolutional Neural Networks (CNN) to classify self-reported wellbeing from multimodal datasets, including urban environmental factors (e.g., noise, nearby people counting), body reactions (physiological reactions including EDA, HR, HRV, body temperature, BVP, and movement) and individuals’ perceived responses (e.g., self-referential valence) in urban settings. The study was conducted on a sample of 40 participants. The sensing kit built and used for the project includes a sensing edge, namely Enviro-Edge, featured ten embedded sensors for air quality, was linked to a customized smartphone app (EnvBodySens2) collecting accelerometer data, and used Bluetooth Low Energy (BLE) for self-report labels, noise, date and time, as well as GPS traces. Finally, body data were collected by means of E4 Empatica. The results showed that the electrodermal activity frequency (EDA) and heart rate variability (HRV) are markedly influenced by an environment’s level of particulate matter [].
The exposome also plays a role in the pathogenesis and course of skin diseases; Patella et al. [] evaluated the impact of air pollutants, measured through environmental control units discharged in specific sites in an area, and weather changes on patients with atopic dermatitis, demonstrating that changes in weather and air pollution have a significant effect on skin reactivity and symptoms in AD patients, increasing the severity of dermatitis.
Furthermore, the skin microbiome regularly comes into contact with cosmetic agents, pollutants, and topical compounds, such as skin-care products and medical ointments. These substances, part of the skin exposome, can be processed into toxic compounds responsible for rashes and neoplasms. To predict all possible metabolic reactions that may occur to these chemicals from our skin microbiome, along with information about the respective reaction centers, metabolic enzymes, microbial species that carry these enzymes, and also the skin sites that host these species, a tool called SkinBug [] was proposed. Jaiswal et al. [] developed SkinBug using a database of metabolic enzymes, reactions, and substrates from 900 bacterial species from 19 different skin sites. This tool uses ML and neural networks to predict the xenobiotic metabolism of the skin microbiome with a binary accuracy of up to 90.0% [].
In an international team effort, coordinated by Atehortúa [], a model, based on ML, was developed to estimate the risk of developing cardiovascular disease (CVD) and type 2 diabetes (T2D) based on exposome factors. The study made use of data coming from 13,764 individuals, equally distributed between cases and controls, with a prevalence of CVD patients (5348 vs. 1534 for T2D). Using the popular Extreme Gradient Boost ensemble model (XGBoost), the authors reached an area under the curve of the ROC of 0.78 ± 0.01 for CVD and a slightly lower result (0.77 ± 0.01) for T2D.
Finally, Dong and collaborators [] tried to shed light on the pre-conception risk factors for childhood atopic problems, using XGBoost, genetic algorithms, and logistic regression to analyze data from a dataset populated by Singapore-based mother–child pairs (1151 overall). With this approach, the authors discovered that pre-conception alcohol consumption and maternal depressive symptoms were clear risk factors for the subsequent development of eczema and rhinitis. In addition, higher maternal blood neopterin and child blood dimethylglycine were found to be protective against early childhood wheezing. In addition, after birth, early infections were seen to be key drivers of atopic problems, including atopic eczema and rhinitis.
The analysis of the studies included in this review highlights the possibility of using AI to identify complex associations between environmental exposures and NCDs by integrating multidimensional datasets, which are difficult to interpret using traditional statistical methods.
However, the studies also reported significant gaps such as the heterogeneity of the exposome data, this representing an intrinsic bias.
For future research, in order to improve the analysis of the exposome in developing chronic diseases through AI algorithms, significant challenges must be confronted regarding data integration, standardization, and the appropriate choice of the analytical model.
Additionally, the limited use of longitudinal study designs decreased the ability to infer causal relationships between environmental factors and disease outcomes. The need for more standardization across studies in terms of data collection methods and AI model selection also hinders the reproducibility and generalizability of their findings. Addressing these weaknesses will be crucial for advancing the reliability and impact of AI in medical research.
4. Conclusions
All the exposures of an individual within a lifetime and how those exposures relate to health define the exposome. Understanding how multiple environmental exposures act together over time is the goal to reach, overcoming the classic concept of one single exposure/disease, particularly in the development of NCDs, for which has been proven that, beyond genetics, it is essential to identify the interaction with environmental factors, to plan prevention and management strategies. In such a framework, our review highlighted how AI can be effectively used in analyzing multiple environmental hazards and their combined effects on health conditions. Notably, the European Human Exposome Network (EHEN) projects and others used AI to integrate multiple environmental exposure data with clinical data, providing prevention interventions and resource-optimization tools. Therefore, applying AI to the exposome helps to expand our understanding of NCDs and improves health outcomes. In fact, AI can play an essential role in identifying specific biomarkers related to NCDs, integrating genomic data and environmental factors, allowing for early disease diagnosis, and developing targeted diagnostic tools. Thanks to AI, personalized intervention strategies can be formulated, thus improving therapeutic effectiveness in treating chronic diseases. Nevertheless, by studying the complex interaction between environmental exposures and genetic susceptibilities, AI can facilitate the development of more effective and individualized treatments. They can be used in AI algorithms in public education programs that allow people to undertake informed health promotion actions. If implemented, these interventions aim to effectively prevent or mitigate health risks and improve the outcomes of non-communicable diseases, radically transforming the management approach of NCDs, offering many opportunities to promote prevention interventions and new diagnostic, therapeutic, and follow-up strategies for chronic diseases.
Author Contributions
Conceptualization, G.M. and S.G.; methodology, S.I., S.B., A.T., E.Z. and S.G.; software, S.B. and A.T.; validation, S.I., E.Z. and G.M.; formal analysis, S.B. and A.T.; investigation, S.I., S.B. and A.T.; resources, G.M. and S.G.; data curation, S.I. and S.B.; writing—original draft preparation, S.I., S.B. and E.Z.; writing—review and editing, S.B. and A.T.; visualization, G.M. and S.G.; supervision, G.M. and S.G.; project administration, G.M. and S.G.; funding acquisition, S.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gonzaga-Jauregui, C.; Lupski, J.R.; Gibbs, R.A. Human Genome Sequencing in Health and Disease. Annu. Rev. Med. 2012, 63, 35–61. [Google Scholar] [CrossRef] [PubMed]
- Wild, C.P. Complementing the Genome with an “Exposome”: The Outstanding Challenge of Environmental Exposure Measurement in Molecular Epidemiology. Cancer Epidemiol. Biomark. Prev. 2005, 14, 1847–1850. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F. Infectious Disease in an Era of Global Change. Nat. Rev. Microbiol. 2022, 20, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Peters, R.; Ee, N.; Peters, J.; Beckett, N.; Booth, A.; Rockwood, K.; Anstey, K.J. Common Risk Factors for Major Noncommunicable Disease, a Systematic Overview of Reviews and Commentary: The Implied Potential for Targeted Risk Reduction. Ther. Adv. Chronic Dis. 2019, 10, 2040622319880392. [Google Scholar] [CrossRef] [PubMed]
- Budreviciute, A.; Damiati, S.; Sabir, D.K.; Onder, K.; Schuller-Goetzburg, P.; Plakys, G.; Katileviciute, A.; Khoja, S.; Kodzius, R. Management and Prevention Strategies for Non-Communicable Diseases (NCDs) and Their Risk Factors. Front. Public Health 2020, 788, 574111. [Google Scholar] [CrossRef]
- Chaker, L.; Falla, A.; van der Lee, S.J.; Muka, T.; Imo, D.; Jaspers, L.; Colpani, V.; Mendis, S.; Chowdhury, R.; Bramer, W.M. The Global Impact of Non-Communicable Diseases on Macro-Economic Productivity: A Systematic Review. Eur. J. Epidemiol. 2015, 30, 357–395. [Google Scholar] [CrossRef]
- Benziger, C.P.; Roth, G.A.; Moran, A.E. The Global Burden of Disease Study and the Preventable Burden of NCD. Glob. Heart 2016, 11, 393–397. [Google Scholar] [CrossRef]
- Vineis, P.; Robinson, O.; Chadeau-Hyam, M.; Dehghan, A.; Mudway, I.; Dagnino, S. What Is New in the Exposome? Environ. Int. 2020, 143, 105887. [Google Scholar] [CrossRef]
- Senier, L.; Brown, P.; Shostak, S.; Hanna, B. The Socio-Exposome: Advancing Exposure Science and Environmental Justice in a Postgenomic Era. Environ. Sociol. 2017, 3, 107–121. [Google Scholar] [CrossRef]
- Vermeulen, R.; Schymanski, E.L.; Barabási, A.-L.; Miller, G.W. The Exposome and Health: Where Chemistry Meets Biology. Science 2020, 367, 392–396. [Google Scholar] [CrossRef]
- Sillé, F.; Karakitsios, S.; Kleensang, A.; Koehler, K.; Maertens, A.; Miller, G.W.; Prasse, C.; Quiros-Alcala, L.; Ramachandran, G.; Hartung, T. The Exposome: A New Approach for Risk Assessment. Altern. Anim. Exp. ALTEX 2020, 37, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Liu, X.; Zheng, Y.; He, X.; Hart, J.; James, P.; Laden, F.; Chen, Y.; Bian, J. Methodological Challenges in Spatial and Contextual Exposome-Health Studies. Crit. Rev. Environ. Sci. Technol. 2022, 53, 827–846. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.; Maitre, L.; Warembourg, C.; Agier, L.; Richiardi, L.; Basagaña, X.; Vrijheid, M. Applying the Exposome Concept in Birth Cohort Research: A Review of Statistical Approaches. Eur. J. Epidemiol. 2020, 35, 193–204. [Google Scholar] [CrossRef]
- Rowe, M. An Introduction to Machine Learning for Clinicians. Acad. Med. 2019, 94, 1433–1436. [Google Scholar] [CrossRef]
- Nuzzi, R.; Boscia, G.; Marolo, P.; Ricardi, F. The Impact of Artificial Intelligence and Deep Learning in Eye Diseases: A Review. Front. Med. 2021, 8, 710329. [Google Scholar] [CrossRef]
- Jiang, F.; Jiang, Y.; Zhi, H.; Dong, Y.; Li, H.; Ma, S.; Wang, Y.; Dong, Q.; Shen, H.; Wang, Y. Artificial Intelligence in Healthcare: Past, Present and Future. Stroke Vasc. Neurol. 2017, 2, 230–243. [Google Scholar] [CrossRef]
- Babel, A.; Taneja, R.; Mondello Malvestiti, F.; Monaco, A.; Donde, S. Artificial Intelligence Solutions to Increase Medication Adherence in Patients With Non-Communicable Diseases. Front. Digit. Health 2021, 3, 669869. [Google Scholar] [CrossRef] [PubMed]
- Lavanya, J.M.S.; Subbulakshmi, P. Machine Learning Techniques for the Prediction of Non-Communicable Diseases. In Proceedings of the 2023 International Conference on Artificial Intelligence and Knowledge Discovery in Concurrent Engineering (ICECONF), Chennai, India, 5–7 January 2023; IEEE: Piscataway, NJ, USA, 2023; pp. 1–8. [Google Scholar]
- Allegra, A.; Tonacci, A.; Sciaccotta, R.; Genovese, S.; Musolino, C.; Pioggia, G.; Gangemi, S. Machine Learning and Deep Learning Applications in Multiple Myeloma Diagnosis, Prognosis, and Treatment Selection. Cancers 2022, 14, 606. [Google Scholar] [CrossRef] [PubMed]
- Murdaca, G.; Banchero, S.; Tonacci, A.; Nencioni, A.; Monacelli, F.; Gangemi, S. Vitamin D and Folate as Predictors of MMSE in Alzheimer’s Disease: A Machine Learning Analysis. Diagnostics 2021, 11, 940. [Google Scholar] [CrossRef]
- Subramanian, M.; Wojtusciszyn, A.; Favre, L.; Boughorbel, S.; Shan, J.; Letaief, K.B.; Pitteloud, N.; Chouchane, L. Precision medicine in the era of artificial intelligence: Implications in chronic disease management. J. Transl. Med. 2020, 18, 472. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Prisma Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Hartung, T. A Call for a Human Exposome Project. ALTEX Altern. Anim. Exp. 2023, 40, 4–33. [Google Scholar] [CrossRef]
- Home—The European Human Exposome Network (EHEN). Available online: https://www.humanexposome.eu/ (accessed on 5 December 2023).
- Home—Hedimed. Available online: https://www.hedimed.eu/ (accessed on 5 December 2023).
- Ronkainen, J.; Nedelec, R.; Atehortua, A.; Balkhiyarova, Z.; Cascarano, A.; Dang, V.N.; Elhakeem, A.; Van Enckevort, E.; Soares, A.G.; Haakma, S. LongITools: Dynamic Longitudinal Exposome Trajectories in Cardiovascular and Metabolic Noncommunicable Diseases. Environ. Epidemiol. 2022, 6, e184. [Google Scholar] [CrossRef]
- Benjdir, M.; Audureau, É.; Beresniak, A.; Coll, P.; Epaud, R.; Fiedler, K.; Jacquemin, B.; Niddam, L.; Pandis, S.N.; Pohlmann, G. Assessing the Impact of Exposome on the Course of Chronic Obstructive Pulmonary Disease and Cystc Fibrosis: The REMEDIA European Project Approach. Environ. Epidemiol. 2021, 5, e165. [Google Scholar] [CrossRef]
- Vrijheid, M.; Basagaña, X.; Gonzalez, J.R.; Jaddoe, V.W.V.; Jensen, G.; Keun, H.C.; McEachan, R.R.C.; Porcel, J.; Siroux, V.; Swertz, M.A.; et al. Advancing Tools for Human Early Lifecourse Exposome Research and Translation (ATHLETE): Project Overview. Environ. Epidemiol. 2021, 5, e166. [Google Scholar] [CrossRef] [PubMed]
- EPHOR—EPHOR Project. Available online: https://www.ephor-project.eu/ (accessed on 5 December 2023).
- Ronsmans, S.; Hougaard, K.S.; Nawrot, T.S.; Plusquin, M.; Huaux, F.; Cruz, M.J.; Moldovan, H.; Verpaele, S.; Jayapala, M.; Tunney, M. The EXIMIOUS Project—Mapping Exposure-Induced Immune Effects: Connecting the Exposome and the Immunome. Environ. Epidemiol. 2022, 6, e193. [Google Scholar] [CrossRef]
- Van Kamp, I.; Waye, K.P.; Kanninen, K.; Gulliver, J.; Bozzon, A.; Psyllidis, A.; Boshuizen, H.; Selander, J.; van den Hazel, P.; Brambilla, M. Early Environmental Quality and Life-Course Mental Health Effects: The Equal-Life Project. Environ. Epidemiol. 2022, 6, e183. [Google Scholar] [CrossRef]
- Vlaanderen, J.; De Hoogh, K.; Hoek, G.; Peters, A.; Probst-Hensch, N.; Scalbert, A.; Melén, E.; Tonne, C.; De Wit, G.A.; Chadeau-Hyam, M.; et al. Developing the Building Blocks to Elucidate the Impact of the Urban Exposome on Cardiometabolic-Pulmonary Disease: The EU EXPANSE Project. Environ. Epidemiol. 2021, 5, e162. [Google Scholar] [CrossRef] [PubMed]
- Martinez, R.M.; Müller, H.; Negru, S.; Ormenisan, A.; Mühr, L.S.A.; Zhang, X.; Møller, F.T.; Clements, M.S.; Kozlakidis, Z.; Pimenoff, V.N. Human Exposome Assessment Platform. Environ. Epidemiol. 2021, 5, e182. [Google Scholar] [CrossRef]
- Pero-Gascon, R.; Hemeryck, L.Y.; Poma, G.; Falony, G.; Nawrot, T.S.; Raes, J.; Vanhaecke, L.; De Boevre, M.; Covaci, A.; De Saeger, S. FLEXiGUT: Rationale for Exposomics Associations with Chronic Low-Grade Gut Inflammation. Environ. Int. 2022, 158, 106906. [Google Scholar] [CrossRef]
- Fine, L.J.; Philogene, G.S.; Gramling, R.; Coups, E.J.; Sinha, S. Prevalence of Multiple Chronic Disease Risk Factors: 2001 National Health Interview Survey. Am. J. Prev. Med. 2004, 27, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.O.; Slater, M.; Kottke, T.E. Preventing Chronic Disease Risk Factors: Rationale and Feasibility. Medicina 2008, 44, 745. [Google Scholar] [CrossRef] [PubMed]
- Ohanyan, H.; Portengen, L.; Huss, A.; Traini, E.; Beulens, J.W.J.; Hoek, G.; Lakerveld, J.; Vermeulen, R. Machine Learning Approaches to Characterize the Obesogenic Urban Exposome. Environ. Int. 2022, 158, 107015. [Google Scholar] [CrossRef] [PubMed]
- Ohanyan, H.; Portengen, L.; Kaplani, O.; Huss, A.; Hoek, G.; Beulens, J.W.J.; Lakerveld, J.; Vermeulen, R. Associations between the Urban Exposome and Type 2 Diabetes: Results from Penalised Regression by Least Absolute Shrinkage and Selection Operator and Random Forest Models. Environ. Int. 2022, 170, 107592. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Akhtari, F.; House, J.S.; Simpson Jr, R.J.; Schmitt, C.P.; Fargo, D.C.; Schurman, S.H.; Hall, J.E.; Motsinger-Reif, A.A. Questionnaire-Based Exposome-Wide Association Studies (ExWAS) Reveal Expected and Novel Risk Factors Associated with Cardiovascular Outcomes in the Personalized Environment and Genes Study. Environ. Res. 2022, 212, 113463. [Google Scholar] [CrossRef]
- Bae, W.D.; Alkobaisi, S.; Horak, M.; Park, C.-S.; Kim, S.; Davidson, J. Predicting Health Risks of Adult Asthmatics Susceptible to Indoor Air Quality Using Improved Logistic and Quantile Regression Models. Life 2022, 12, 1631. [Google Scholar] [CrossRef]
- Ren, X.; Mi, Z.; Georgopoulos, P.G. Socioexposomics of COVID-19 across New Jersey: A Comparison of Geostatistical and Machine Learning Approaches. J. Expo. Sci. Environ. Epidemiol. 2023, 34, 197–207. [Google Scholar] [CrossRef]
- Pries, L.-K.; Lage-Castellanos, A.; Delespaul, P.; Kenis, G.; Luykx, J.J.; Lin, B.D.; Richards, A.L.; Akdede, B.; Binbay, T.; Altinyazar, V. Estimating Exposome Score for Schizophrenia Using Predictive Modeling Approach in Two Independent Samples: The Results from the EUGEI Study. Schizophr. Bull. 2019, 45, 960–965. [Google Scholar] [CrossRef]
- Zhao, F.; Li, L.; Lin, P.; Chen, Y.; Xing, S.; Du, H.; Wang, Z.; Yang, J.; Huan, T.; Long, C. HExpPredict: In Vivo Exposure Prediction of Human Blood Exposome Using a Random Forest Model and Its Application in Chemical Risk Prioritization. Environ. Health Perspect. 2023, 131, 037009. [Google Scholar] [CrossRef]
- Matta, K.; Vigneau, E.; Cariou, V.; Mouret, D.; Ploteau, S.; Le Bizec, B.; Antignac, J.-P.; Cano-Sancho, G. Associations between Persistent Organic Pollutants and Endometriosis: A Multipollutant Assessment Using Machine Learning Algorithms. Environ. Pollut. 2020, 260, 114066. [Google Scholar] [CrossRef]
- Li, K.; Bertrand, K.; Naviaux, J.C.; Monk, J.M.; Wells, A.; Wang, L.; Lingampelly, S.S.; Naviaux, R.K.; Chambers, C. Metabolomic and Exposomic Biomarkers of Risk of Future Neurodevelopmental Delay in Human Milk. Pediatr. Res. 2023, 93, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- Louis, G.M.B.; Yeung, E.; Kannan, K.; Maisog, J.; Zhang, C.; Grantz, K.L.; Sundaram, R. Patterns and Variability of Endocrine Disrupting Chemicals during Pregnancy: Implications for Understanding the Exposome of Normal Pregnancy. Epidemiology 2019, 30, S65. [Google Scholar] [CrossRef] [PubMed]
- Nemkov, T.; Stefanoni, D.; Bordbar, A. Recipient Epidemiology and Donor Evaluation Study III Red Blood Cell–Omics (REDS-III RBC-Omics) Study. Blood Donor Exposome and Impact of Common Drugs on Red Blood Cell Metabolism. JCI Insight 2021, 6, 146175. [Google Scholar] [CrossRef] [PubMed]
- Buelow, E.; Rico, A.; Gaschet, M.; Lourenço, J.; Kennedy, S.P.; Wiest, L.; Ploy, M.-C.; Dagot, C. Hospital Discharges in Urban Sanitation Systems: Long-Term Monitoring of Wastewater Resistome and Microbiota in Relationship to Their Eco-Exposome. Water Res. X 2020, 7, 100045. [Google Scholar] [CrossRef] [PubMed]
- Loef, B.; Wong, A.; Janssen, N.A.H.; Strak, M.; Hoekstra, J.; Picavet, H.S.J.; Boshuizen, H.C.; Verschuren, W.M.; Herber, G.-C.M. Using Random Forest to Identify Longitudinal Predictors of Health in a 30-Year Cohort Study. Sci. Rep. 2022, 12, 10372. [Google Scholar] [CrossRef]
- Johnson, T.; Kanjo, E.; Woodward, K. DigitalExposome: Quantifying Impact of Urban Environment on Wellbeing Using Sensor Fusion and Deep Learning. Comput. Urban Sci. 2023, 3, 14. [Google Scholar] [CrossRef]
- Patella, V.; Florio, G.; Palmieri, M.; Bousquet, J.; Tonacci, A.; Giuliano, A.; Gangemi, S. Atopic Dermatitis Severity during Exposure to Air Pollutants and Weather Changes with an Artificial Neural Network (ANN) Analysis. Pediatr. Allergy Immunol. 2020, 31, 938–945. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Agarwal, S.M.; Thodum, P.; Sharma, V.K. SkinBug: An Artificial Intelligence Approach to Predict Human Skin Microbiome-Mediated Metabolism of Biotics and Xenobiotics. Iscience 2021, 24, 101925. [Google Scholar] [CrossRef]
- Atehortúa, A.; Gkontra, P.; Camacho, M.; Diaz, O.; Bulgheroni, M.; Simonetti, V.; Chadeau-Hyam, M.; Felix, J.F.; Sebert, S.; Lekadir, K. Cardiometabolic risk estimation using exposome data and machine learning. Int. J. Med. Inform. 2023, 179, 105209. [Google Scholar] [CrossRef]
- Dong, Y.; Lau, H.X.; Suaini, N.H.A.; Kee, M.Z.L.; Ooi, D.S.Q.; Shek, L.P.; Lee, B.W.; Godfrey, K.M.; Tham, E.H.; Ong, M.E.H.; et al. A machine-learning exploration of the exposome from preconception in early childhood atopic eczema, rhinitis and wheeze development. Environ. Res. 2024, 250, 118523. [Google Scholar] [CrossRef]
- Marín, D.; Orozco, L.Y.; Narváez, D.M.; Ortiz-Trujillo, I.C.; Molina, F.J.; Ramos, C.D.; Rodriguez-Villamizar, L.; Bangdiwala, S.I.; Morales, O.; Cuellar, M. Characterization of the External Exposome and Its Contribution to the Clinical Respiratory and Early Biological Effects in Children: The PROMESA Cohort Study Protocol. PLoS ONE 2023, 18, e0278836. [Google Scholar] [CrossRef] [PubMed]
- De Vito, S.; Esposito, E.; Massera, E.; Formisano, F.; Fattoruso, G.; Ferlito, S.; Del Giudice, A.; D’Elia, G.; Salvato, M.; Polichetti, T. Crowdsensing IoT Architecture for Pervasive Air Quality and Exposome Monitoring: Design, Development, Calibration, and Long-Term Validation. Sensors 2021, 21, 5219. [Google Scholar] [CrossRef] [PubMed]
- Shamji, M.H.; Ollert, M.; Adcock, I.M.; Bennett, O.; Favaro, A.; Sarama, R.; Riggioni, C.; Annesi-Maesano, I.; Custovic, A.; Fontanella, S. EAACI Guidelines on Environmental Science in Allergic Diseases and Asthma–Leveraging Artificial Intelligence and Machine Learning to Develop a Causality Model in Exposomics. Allergy 2023, 78, 1742–1757. [Google Scholar] [CrossRef] [PubMed]
- Hawthorne, C.; Lopez-Campos, G.H. Integration of Annotated Phenotype, Gene and Chemical Text Data to Advance Exposome Informatics. Stud. Health Technol. Inform. 2022, 294, 870–871. [Google Scholar]
- Hawthorne, C.; Simpson, D.A.; Devereux, B.; López-Campos, G. Phexpo: A Package for Bidirectional Enrichment Analysis of Phenotypes and Chemicals. JAMIA Open 2020, 3, 173–177. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).