Identification of Transgenic Agricultural Products and Foods Using NIR Spectroscopy and Hyperspectral Imaging: A Review
Abstract
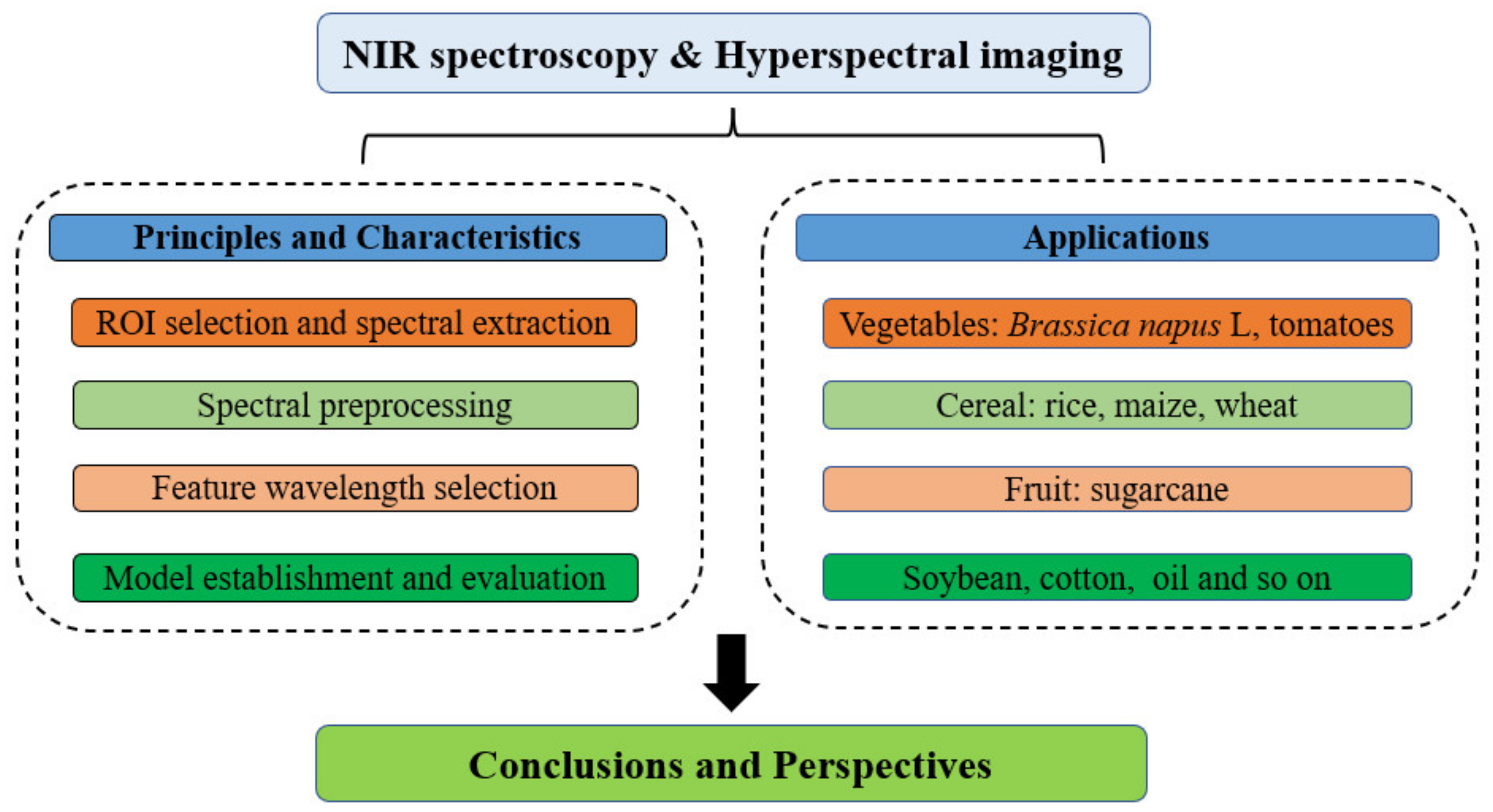
:1. Introduction
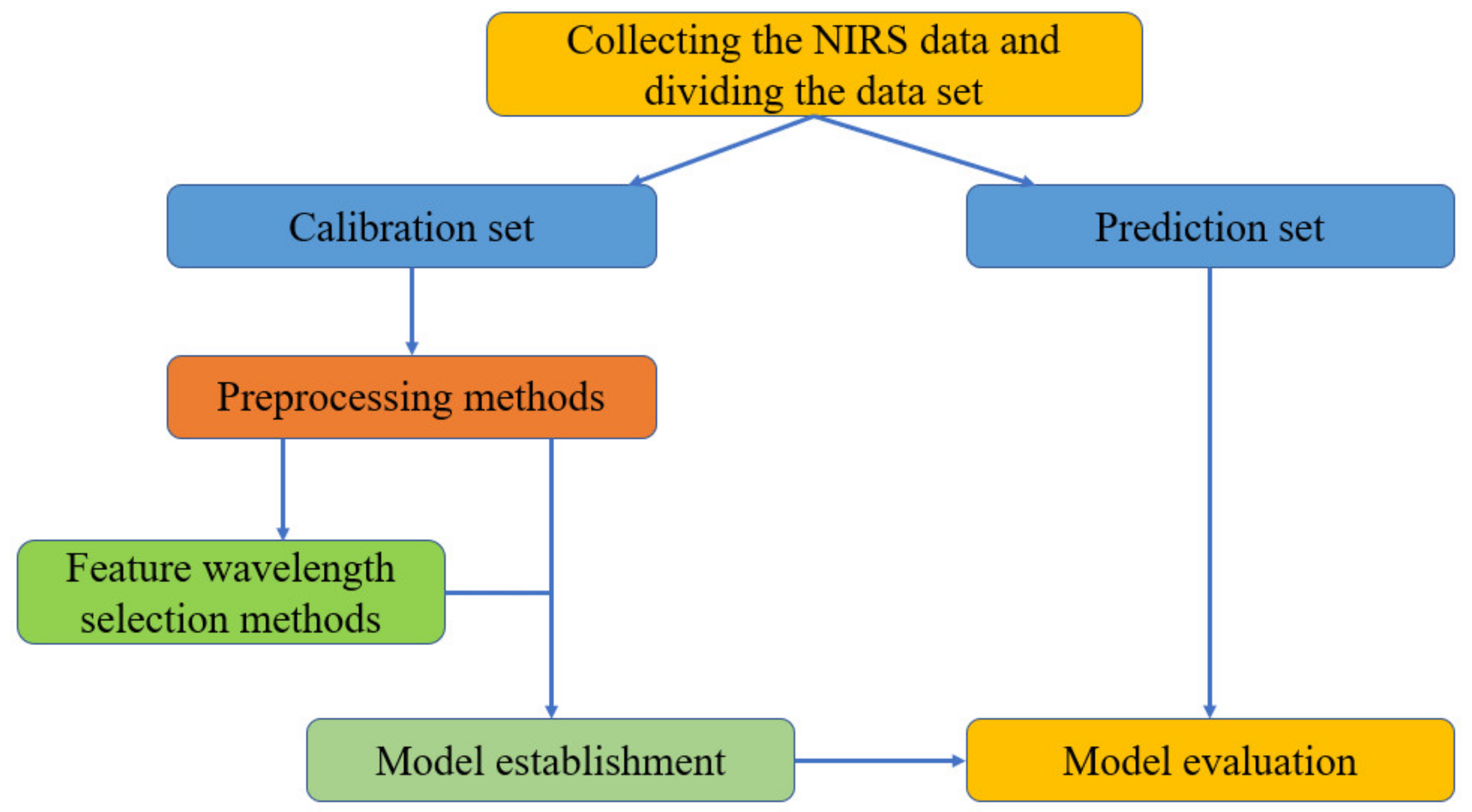
2. The Principles and Characteristics of NIRS
2.1. The Spectral Preprocessing Methods
2.2. The Feature Wavelength Selection Methods
2.3. Model Establishment and Evaluation
3. The Applications of NIRS for the Detection of Transgenic Agricultural Products and Foods
4. The Principles and Characteristics of Hyperspectral Imaging Technique
5. The Applications of HSI for the Detection of Transgenic Agricultural Products and Foods
6. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sohn, S.; Pandian, S.; Oh, Y.; Zaukuu, J.Z.; Kang, H.; Ryu, T.; Cho, W.; Cho, Y.; Shin, E.; Cho, B. An overview of near infrared spectroscopy and its applications in the detection of genetically modified organisms. Int. J. Mol. Sci. 2021, 22, 9940. [Google Scholar] [CrossRef]
- Alishahi, A.; Farahmand, H.; Prieto, N.; Cozzolino, D. Identification of transgenic foods using nir spectroscopy: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2010, 75, 1–7. [Google Scholar] [CrossRef]
- Feng, X.; Zhao, Y.; Zhang, C.; Cheng, P.; He, Y. Discrimination of transgenic maize kernel using nir hyperspectral imaging and multivariate data analysis. Sensors 2017, 17, 1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Dai, L.; Cheng, F. Classification of frozen corn seeds using hyperspectral vis/nir reflectance imaging. Molecules 2019, 24, 149. [Google Scholar] [CrossRef] [Green Version]
- Candolfi, A.; De Maesschalck, R.; Jouan-Rimbaud, D.; Hailey, P.A.; Massart, D.L. The influence of data pre-processing in the pattern recognition of excipients near-infrared spectra. J. Pharm. Biomed. Anal. 1999, 21, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Sun, D.; Zeng, X. Recent advances in wavelength selection techniques for hyperspectral image processing in the food industry. Food Bioprocess Technol. 2014, 7, 307–323. [Google Scholar] [CrossRef]
- Ramirez, C.A.M.; Greenop, M.; Ashton, L.; Rehman, I.U. Applications of machine learning in spectroscopy. Appl. Spectrosc. Rev. 2021, 8–10, 733–763. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.; Jemain, A.A. A contemporary review on data preprocessing (dp) practice strategy in atr-ftir spectrum. Chemom. Intell. Lab. Syst. 2017, 163, 64–75. [Google Scholar] [CrossRef]
- Jiao, Y.; Li, Z.; Chen, X.; Fei, S. Preprocessing methods for near-infrared spectrum calibration. J. Chemom. 2020, 34, e3306. [Google Scholar] [CrossRef]
- Rinnan, A.; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Sohn, S.; Pandian, S.; Oh, Y.; Zaukuu, J.Z.; Na, C.; Lee, Y.; Shin, E.; Kang, H.; Ryu, T.; Cho, W.; et al. Vis-NIR Spectroscopy and Machine Learning Methods for the Discrimination of Transgenic Brassica napus L. and Their Hybrids with B. juncea. Processes 2022, 10, 240. [Google Scholar] [CrossRef]
- Cheng, P.; Xuping, F.; Yong, H.; Chu, Z.; Yiying, Z.; Junfeng, X. Discrimination of transgenic maize containing the cry1ab/cry2aj and g10evo genes using near infrared spectroscopy (nir). Spectrosc. Spectr. Anal. 2018, 38, 1095–1100. [Google Scholar]
- Workman, J.J. Review of Process and Non-invasive Near-Infrared and Infrared Spectroscopy: 1993–1999. Appl. Spectrosc. Rev. 1999, 34, 1–89. [Google Scholar] [CrossRef]
- Xie, L.; Ying, Y.; Ying, T. Combination and comparison of chemometrics methods for identification of transgenic tomatoes using visible and near-infrared diffuse transmittance technique. J. Food Eng. 2007, 82, 395–401. [Google Scholar] [CrossRef]
- Xie, L.; Ying, Y.; Ying, T.; Yu, H.; Fu, X. Discrimination of transgenic tomatoes based on visible/near-infrared spectra. Anal. Chim. Acta 2007, 584, 379–384. [Google Scholar] [CrossRef]
- Wold, S.; Antti, H.; Lindgren, F.; öhman, J. Orthogonal signal correction of near-infrared spectra. Chemom. Intell. Lab. Syst. 1998, 44, 175–185. [Google Scholar] [CrossRef]
- Wenchao, Z.; Fang, C. Analysis of transgenic and non-transgenic rice leaves using visible/near-infrared spectroscopy. Spectrosc. Spectr. Anal. 2012, 32, 370–373. [Google Scholar]
- Hashimoto, A.; Kameoka, T. Applications of infrared spectroscopy to biochemical, food, and agricultural processes. Appl. Spectrosc. Rev. 2008, 43, 416–451. [Google Scholar] [CrossRef]
- Zinia Zaukuu, J.; Aouadi, B.; Lukács, M.; Bodor, Z.; Vitális, F.; Gillay, B.; Gillay, Z.; Friedrich, L.; Kovacs, Z. Detecting low concentrations of nitrogen-based adulterants in whey protein powder using benchtop and handheld nir spectrometers and the feasibility of scanning through plastic bag. Molecules 2020, 25, 2522. [Google Scholar] [CrossRef]
- Dai, Q.; Cheng, J.H.; Sun, D.W.; Zeng, X.A. Advances in feature selection methods for hyperspectral image processing in food industry applications: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1368–1382. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Model population analysis for variable selection. J. Chemom. 2010, 24, 418–423. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Niazi, A.; Leardi, R. Genetic algorithm in chemometrics. J. Chemom. 2012, 26, 345–351. [Google Scholar] [CrossRef]
- Depczynski, U.; Frost, V.J.; Molt, K. Genetic algorithms applied to the selection of factors in principal component regression. Anal. Chim. Acta 2000, 420, 217–227. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvão, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Centner, V.; Massart, D.L.; de Noord, O.E.; de Jong, S.; Vandeginste, B.M.; Sterna, C. Elimination of uninformative variables for multivariate calibration. Anal. Chem. 1996, 68, 3851–3858. [Google Scholar] [CrossRef]
- Feng, X.; Yin, H.; Zhang, C.; Peng, C.; He, Y. Screening of transgenic maize using near infrared spectroscopy and chemometric techniques. Span. J. Agric. Res. 2018, 16, e203. [Google Scholar] [CrossRef]
- Sohn, S.; Pandian, S.; Zaukuu, J.Z.; Oh, Y.; Park, S.; Na, C.; Shin, E.; Kang, H.; Ryu, T.; Cho, W.; et al. Discrimination of transgenic canola (brassica napus L.) And their hybrids with b. Rapa using vis-nir spectroscopy and machine learning methods. Int. J. Mol. Sci. 2022, 23, 220. [Google Scholar] [CrossRef]
- Mata, M.M.D.; Rocha, P.D.; Farias, I.K.T.D.; Silva, J.L.B.D.; Medeiros, E.P.; Silva, C.S.; Simões, S.D.S. Distinguishing cotton seed genotypes by means of vibrational spectroscopic methods (nir and raman) and chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 266, 120399. [Google Scholar] [CrossRef]
- Lee, J.H.; Choung, M. Nondestructive determination of herbicide-resistant genetically modified soybean seeds using near-infrared reflectance spectroscopy. Food Chem. 2011, 126, 368–373. [Google Scholar] [CrossRef]
- Jianguo, Z.; Yajing, W.; Zhiqin, Y.; Leiying, X.; Na, W.; Duo, C. Research on fast identification of transgenic oil based on near infrared spectroscopy. Opt. Instrum. 2020, 42, 61–66. [Google Scholar]
- Zhang, J.; Dai, L.; Cheng, F. Identification of corn seeds with different freezing damage degree based on hyperspectral reflectance imaging and deep learning method. Food Anal. Meth. 2021, 14, 389–400. [Google Scholar] [CrossRef]
- Jiang, W.; Furong, H.; Caihuan, H.; Jun, Z.; Xingdan, C. Study on near infrared spectroscopy of transgenic soybean identification based on principal component analysis and neural network. Spectrosc. Spectr. Anal. 2013, 33, 1537–1541. [Google Scholar]
- Hattori, T.; Murakami, S.; Mukai, M.; Yamada, T.; Hirochika, H.; Ike, M.; Tokuyasu, K.; Suzuki, S.; Sakamoto, M.; Umezawa, T. Rapid analysis of transgenic rice straw using near-infrared spectroscopy. Plant Biotechnol. 2012, 29, 359–366. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Ying, Y.; Ying, T. Rapid determination of ethylene content in tomatoes using visible and short-wave near-infrared spectroscopy and wavelength selection. Chemom. Intell. Lab. Syst. 2009, 97, 141–145. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yang, J.; Xu, J.; Zhang, X.; Wu, C.; Lin, T.; Ying, Y. Deep learning for vibrational spectral analysis: Recent progress and a practical guide. Anal. Chim. Acta 2019, 1081, 6–17. [Google Scholar] [CrossRef]
- Zhang, X.; Lin, T.; Xu, J.; Luo, X.; Ying, Y. Deepspectra: An end-to-end deep learning approach for quantitative spectral analysis. Anal. Chim. Acta 2019, 1058, 48–57. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, J.; Lin, T.; Ying, Y. Food and agro-product quality evaluation based on spectroscopy and deep learning: A review. Trends Food Sci. Technol. 2021, 112, 431–441. [Google Scholar] [CrossRef]
- Prieto, N.; Roehe, R.; Lavín, P.; Batten, G.; Andrés, S. Application of near infrared reflectance spectroscopy to predict meat and meat products quality: A review. Meat Sci. 2009, 83, 175–186. [Google Scholar] [CrossRef]
- Qu, J.H.; Liu, D.; Cheng, J.H.; Sun, D.W.; Ma, J.; Pu, H.; Zeng, X.A. Applications of near-infrared spectroscopy in food safety evaluation and control: A review of recent research advances. Crit. Rev. Food Sci. Nutr. 2015, 55, 1939–1954. [Google Scholar] [CrossRef]
- Fu, X.; Ying, Y. Food safety evaluation based on near infrared spectroscopy and imaging: A review. Crit. Rev. Food Sci. Nutr. 2016, 56, 1913–1924. [Google Scholar] [CrossRef]
- Huang, H.; Yu, H.; Xu, H.; Ying, Y. Near infrared spectroscopy for on/in-line monitoring of quality in foods and beverages: A review. J. Food Eng. 2008, 87, 303–313. [Google Scholar] [CrossRef]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive measurement of fruit and vegetable quality by means of nir spectroscopy: A review. Postharvest Biol. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Chandrasekaran, I.; Panigrahi, S.S.; Ravikanth, L.; Singh, C.B. Potential of near-infrared (nir) spectroscopy and hyperspectral imaging for quality and safety assessment of fruits: An overview. Food Anal. Methods 2019, 12, 2438–2458. [Google Scholar] [CrossRef]
- Ge, W.; Zhang, L.; Li, X.; Zhang, C.; Sun, M.; An, D.; Wu, J. Applying multimodal data fusion based on manifold learning with nuclear magnetic resonance (nmr) and near infrared spectroscopy (nirs) to maize haploid identification. Biosyst. Eng. 2021, 210, 299–309. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, L.; An, D.; Zhang, X.; Zhu, D. Identification of maize seed varieties based on near infrared reflectance spectroscopy and chemometrics. Int. J. Agric. Biol. Eng. 2018, 11, 177–183. [Google Scholar] [CrossRef]
- Qiu, G.; Lü, E.; Lu, H.; Xu, S.; Zeng, F.; Shui, Q. Single-kernel ft-nir spectroscopy for detecting supersweet corn (Zea mays L. Saccharata sturt) seed viability with multivariate data analysis. Sensors 2018, 18, 1010. [Google Scholar] [CrossRef] [Green Version]
- Egesel, C.Ö.; Kahrıman, F.; Ekinci, N.; Kavdır, O.; Büyükcan, M.B. Analysis of fatty acids in kernel, flour, and oil samples of maize by nir spectroscopy using conventional regression methods. Cereal Chem. J. 2016, 93, 487–492. [Google Scholar] [CrossRef]
- Rosales, A.; Galicia, L.; Oviedo, E.; Islas, C.; Palacios-Rojas, N. Near-infrared reflectance spectroscopy (nirs) for protein, tryptophan, and lysine evaluation in quality protein maize (qpm) breeding programs. J. Agric. Food Chem. 2011, 59, 10781–10786. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Qin, X. Rapid quantitative analysis of corn starch adulteration in konjac glucomannan by chemometrics-assisted ft-nir spectroscopy. Food Anal. Methods 2016, 9, 61–67. [Google Scholar] [CrossRef]
- Levasseur-Garcia, C.; Bailly, S.; Kleiber, D.; Bailly, J.; Pan, L. Assessing risk of fumonisin contamination in maize using near-infrared spectroscopy. J. Chem. 2015, 2015, 485864. [Google Scholar] [CrossRef] [Green Version]
- Darnell, R.E.; Harvey, J.J.; Fox, G.P.; Fletcher, M.T.; Wainaina, J.; Wanjuki, I.; Turner, W.J. Nirs calibration of aflatoxin in maize. Aust. J. Chem. 2018, 71, 868. [Google Scholar] [CrossRef]
- Jia, S.; Yang, L.; An, D.; Liu, Z.; Yan, Y.; Li, S.; Zhang, X.; Zhu, D.; Gu, J. Feasibility of analyzing frost-damaged and non-viable maize kernels based on near infrared spectroscopy and chemometrics. J. Cereal Sci. 2016, 69, 145–150. [Google Scholar] [CrossRef]
- Rady, A.; Adedeji, A. Assessing different processed meats for adulterants using visible-near-infrared spectroscopy. Meat Sci. 2018, 136, 59–67. [Google Scholar] [CrossRef]
- Pullanagari, R.R.; Yule, I.J.; Agnew, M. On-line prediction of lamb fatty acid composition by visible near infrared spectroscopy. Meat Sci. 2015, 100, 156–163. [Google Scholar] [CrossRef]
- Xie, L.; Ying, Y.; Ying, T. Quantification of chlorophyll content and classification of nontransgenic and transgenic tomato leaves using visible/near-infrared diffuse reflectance spectroscopy. J. Agric. Food Chem. 2007, 55, 4645–4650. [Google Scholar] [CrossRef]
- Xie, L.; Ying, Y.; Ying, T.; Tian, H.; Niu, X.; Fu, X. Application of vis/nir diffuse reflectance spectroscopy to the detection and identification of transgenic tomato leaf. Spectrosc. Spectr. Anal. 2008, 28, 1062–1066. [Google Scholar]
- Long, Z.; Shan-Shan, W.; Yan-Fei, D.; Jia-Rong, P.; Cheng, Z. Discrimination of transgenic rice based on near infrared reflectance spectroscopy and partial least squares regression discriminant analysis. Rice Sci. 2015, 22, 245–249. [Google Scholar] [CrossRef] [Green Version]
- Hao, Y.; Geng, P.; Wu, W.; Wen, Q.; Rao, M. Identification of rice varieties and transgenic characteristics based on near-infrared diffuse reflectance spectroscopy and chemometrics. Molecules 2019, 24, 4568. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Chen, J.; Pan, T.; Wang, J.; Cao, G. Vis-nir wavelength selection for non-destructive discriminant analysis of breed screening of transgenic sugarcane. Anal. Methods 2014, 6, 8810–8816. [Google Scholar] [CrossRef]
- Guisong, L.; Haosong, G.; Tao, P.; Wang, J.; Cao, G. Vis-nir spectroscopic pattern recognition combined with sg smoothing applied to breed screening of transgenic sugarcane. Spectrosc. Spectr. Anal. 2014, 34, 2701–2706. [Google Scholar]
- Yafeng, Z.; Qian, S.; Wenjin, W.; Zhentian, H.; Zongying, Z.; Jiashuang, A.; Jin, D.; Xin, D.; Chenggui, H.; Jialin, Y.; et al. Fast discrimination of varieties of transgene wheat based on bionimetic pattern recognition and near infrared spectra. Spectrosc. Spectr. Anal. 2010, 30, 924–928. [Google Scholar]
- Luna, A.S.; Da Silva, A.P.; Pinho, J.S.A.; Ferré, J.; Boqué, R. Rapid characterization of transgenic and non-transgenic soybean oils by chemometric methods using nir spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 100, 115–119. [Google Scholar] [CrossRef]
- Gowen, A.; Odonnell, C.; Cullen, P.; Downey, G.; Frias, J. Hyperspectral imaging–an emerging process analytical tool for food quality and safety control. Trends Food Sci. Technol. 2007, 18, 590–598. [Google Scholar] [CrossRef]
- Mahesh, S.; Jayas, D.S.; Paliwal, J.; White, N.D.G. Hyperspectral imaging to classify and monitor quality of agricultural materials. J. Stored Prod. Res. 2015, 61, 17–26. [Google Scholar] [CrossRef]
- Qin, J.; Chao, K.; Kim, M.S.; Lu, R.; Burks, T.F. Hyperspectral and multispectral imaging for evaluating food safety and quality. J. Food Eng. 2013, 118, 157–171. [Google Scholar] [CrossRef]
- Dale, L.M.; Thewis, A.; Boudry, C.; Rotar, I.; Dardenne, P.; Baeten, V.; Pierna, J.A.F. Hyperspectral imaging applications in agriculture and agro-food product quality and safety control: A review. Appl. Spectrosc. Rev. 2013, 48, 142–159. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Elmasry, G.; Sun, D.; Allen, P. Prediction of some quality attributes of lamb meat using near-infrared hyperspectral imaging and multivariate analysis. Anal. Chim. Acta 2012, 714, 57–67. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, L.; Cheng, F. Corn seed variety classification based on hyperspectral reflectance imaging and deep convolutional neural network. J. Food Meas. Charact. 2021, 15, 484–494. [Google Scholar] [CrossRef]
- Wang, L.; Sun, D.; Pu, H.; Zhu, Z. Application of hyperspectral imaging to discriminate the variety of maize seeds. Food Anal. Methods 2016, 9, 225–234. [Google Scholar] [CrossRef]
- Wakholi, C.; Kandpal, L.M.; Lee, H.; Bae, H.; Park, E.; Kim, M.S.; Mo, C.; Lee, W.; Cho, B. Rapid assessment of corn seed viability using short wave infrared line-scan hyperspectral imaging and chemometrics. Sens. Actuators B Chem. 2018, 255, 498–507. [Google Scholar] [CrossRef]
- Ambrose, A.; Kandpal, L.M.; Kim, M.S.; Lee, W.; Cho, B. High speed measurement of corn seed viability using hyperspectral imaging. Infrared Phys. Technol. 2016, 75, 173–179. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, S.; Wu, J.; Zhang, C.; Xu, F.; Yang, X.; Li, J. Application of long-wave near infrared hyperspectral imaging for determination of moisture content of single maize seed. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 254, 119666. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Zhao, W.; Wang, Q.; Zhang, M.; Zhu, Q. Prediction of moisture content uniformity using hyperspectral imaging technology during the drying of maize kernel. Int. Agrophys. 2015, 29, 39–46. [Google Scholar] [CrossRef] [Green Version]
- Weinstock, B.A.; Janni, J.; Hagen, L.; Wright, S. Prediction of oil and oleic acid concentrations in individual corn (Zea mays L.) Kernels using near-infrared reflectance hyperspectral imaging and multivariate analysis. Appl. Spectrosc. 2006, 60, 9–16. [Google Scholar] [CrossRef]
- Qiao, M.; Xu, Y.; Xia, G.; Su, Y.; Lu, B.; Gao, X.; Fan, H. Determination of hardness for maize kernels based on hyperspectral imaging. Food Chem. 2022, 366, 130559. [Google Scholar] [CrossRef]
- Williams, P.J.; Kucheryavskiy, S. Classification of maize kernels using nir hyperspectral imaging. Food Chem. 2016, 209, 131–138. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, B.; Wang, Z.; Cheng, F. Application of hyperspectral imaging in the detection of aflatoxin b1 on corn seed. J. Food Meas. Charact. 2022, 16, 448–460. [Google Scholar] [CrossRef]
- Tao, F.; Yao, H.; Hruska, Z.; Kincaid, R.; Rajasekaran, K.; Bhatnagar, D. A novel hyperspectral-based approach for identification of maize kernels infected with diverse aspergillus flavus fungi. Biosyst. Eng. 2020, 200, 415–430. [Google Scholar] [CrossRef]
- Rocha, P.D.; Medeiros, E.P.; Silva, C.S.; Da Silva Simões, S. Chemometric strategies for near infrared hyperspectral imaging analysis: Classification of cotton seed genotypes. Anal. Methods 2021, 13, 5065–5074. [Google Scholar] [CrossRef]
- Feng, X.; Yu, C.; Chen, Y.; Peng, J.; Ye, L.; Shen, T.; Wen, H.; He, Y. Non-destructive determination of shikimic acid concentration in transgenic maize exhibiting glyphosate tolerance using chlorophyll fluorescence and hyperspectral imaging. Front. Plant Sci. 2018, 9, 468. [Google Scholar] [CrossRef]
- Wang, H.L.; Yang, X.D.; Zhang, C.; Guo, D.Q.; Bao, Y.D.; He, Y.; Liu, F. Fast identification of transgenic soybean varieties based near infrared hyperspectral imaging technology. Spectrosc. Spectr. Anal. 2016, 36, 1843–1847. [Google Scholar]
- Xuping, F.; Cheng, P.; Chu, Z.; Xiaodan, L.; Tingting, S.; Yong, H.; Junfeng, X. A simple and efficient method for crispr/cas9-induced rice mutant screening. Spectrosc. Spectr. Anal. 2018, 38, 570–574. [Google Scholar]
- Lian, F.; Xu, D.; Fu, M.; Ge, H.; Jiang, Y.; Zhang, Y. Identification of transgenic ingredients in maize using terahertz spectra. IEEE Trans. Terahertz Sci. Technol. 2017, 7, 378–384. [Google Scholar] [CrossRef]
- Liu, J.; Li, Z.; Hu, F.; Chen, T.; Zhu, A.; Du, Y.; Xin, H. Method for identifying transgenic cottons based on terahertz spectra and wlda. Opt. Int. J. Light Electron Opt. 2015, 126, 1872–1877. [Google Scholar] [CrossRef]
- Shen, X.; Li, B.; Li, X.; Long, Y. Identification of transgenic and non-transgenic cotton seed based on terahertz range spectroscopy. Trans. Chin. Soc. Agric. Eng. 2017, 33, 288–292. [Google Scholar]
- Fang, H.; Zhang, Z.; Wang, H.; Yang, X.; He, Y.; Bao, Y. Identification of transgenic soybean varieties using mid-infrared spectroscopy. Spectrosc. Spectr. Anal. 2017, 37, 760–765. [Google Scholar]
- Liu, X.; Yu, Y.; Bai, X.; Li, X.; Zhang, J.; Wang, D. Rapid identification of insecticide- and herbicide-tolerant genetically modified maize using mid-infrared spectroscopy. Processes 2023, 11, 90. [Google Scholar] [CrossRef]

| Methods | Advantages | Disadvantages | |
|---|---|---|---|
| Protein-based methods | Western blot | Reliable results | Difficult to use, destructive, time-consuming (2 days) |
| ELISA | Reliable results | Moderate to use, destructive, time-consuming (30–90 min) | |
| Lateral flow strip | Simple to use, quick testing (10 min) | destructive | |
| DNA-based methods | Southern blot | Reliable results | Difficult to use, destructive, time-consuming (6 h) |
| Qualitative PCR | Reliable results | Difficult to use, destructive, time-consuming (1.5 h) | |
| Real time PCR | Reliable results | Difficult to use, destructive, time-consuming (1 day) | |
| Microscopy | Classical microscopy | Results visualization | Difficult to use, destructive, time-consuming (1 day) |
| Chromatography | HPLS, GC-MS | Reliable results | Difficult to use, destructive, time-consuming (1–2 days) |
| Spectroscopy-based methods | NIRS | Non-destructive, Quick testing (Less than 1 min), Easy to use | Model-reliable |
| Hyperspectral Imaging | Non-destructive, Quick testing (Less than 5 min), Moderate to use | Model-reliable | |
| MIRS | Non-destructive Quick testing (Less than 5 min), Easy to use | Model-reliable, Not widely used | |
| Terahertz Spectroscopy | Non-destructive, Quick testing (Less than 15 min), Moderate to use | Model-reliable, Not widely used | |
| Author | Object | Preprocessing Methods | Models | Results | Reference |
|---|---|---|---|---|---|
| Soo-In Sohn et al. | Transgenic Brassica napus L. | SG, smoothing filter, SNV, Normalization | LDA, CNN, GBT, SVM, RF | The highest accuracy of the combination of SG and SVM was 100%. | [28] |
| Soo-In Sohn et al. | Transgenic Brassica napus L. | Normalization, SNV, SG | LDA, Deep Learning, SVM, GLM, DT, NB, FLM, RF | 99.4% classification accuracy for SNV and SVM, 99.1% classification accuracy for SG and deep learning | [11] |
| Lijuan Xie et al. | Transgenic Tomatoes | MSC, 1st and 2nd derivatives | DA, PLS-DA | PLS-DA with the classification accuracy of 100% | [15] |
| Lijuan Xie et al. | Transgenic Tomatoes | MSC, SG 1st, 2nd | SIMCA, DPLS | DPLS with the classification accuracy of 100% | [14] |
| Lijuan Xie et al. | Chlorophyll Content of Transgenic Tomato Leaves | MSC, 1st and 2nd derivatives | PLS-DA | PLS-DA with the classification accuracy of 100% | [57] |
| Lijuan Xie et al. | Transgenic tomato leaf | MSC, 1st and 2nd derivatives | DA, PLS | With the classification accuracy of 89.7% | [58] |
| Lijuan Xie et al. | ethylene content in tomatoes | SNV, MSC, 1st and 2nd derivatives | PLSR, SMLR | PLSR and SMLR can determine the ethylene content in tomato. | [35] |
| Wenchao Zhu et al. | Leaves of transgenic rice, SPAD in leaf | MSC, OSC | LS-SVM | SPA-LS-SVM method can quickly identify transgenic rice leaves and accurately predict the SPAD value. | [17] |
| Takefumi Hattori et al. | Transgenic rice straw | 1st and 2nd derivatives, SNV | PLSR | SNV-PLSR obtained a strong correlation between laboratory wet chemistry values and NIR predicted values. | [34] |
| Long Zhang et al. | Transgenic Rice | SNV, PCA | PLS-DA | The correct classification rate of the validation test was 100.0%. | [59] |
| Yong Hao et al. | Transgenic Rice | NWS, SNV, MSC, SG 1st-Derivative | PLS-DA, SVM | Model achieved good analytical results with 100% accuracy rate. | [60] |
| Mayara Macedo da Mata et al. | Transgenic cotton | SNV, 1st derivative | PLS-DA | NIR and Raman prediction sets had classification errors of 2.23% and 0.0%, respectively | [29] |
| Jin Hwan Lee et al. | Transgenic soybean | 1st, 2nd derivatives | PLS-DA | 2nd derivatives and PLSDA had results with 97% accuracy | [30] |
| Jiang Wu et al. | Transgenic soybean | SNV | BPNN | BPNN had 100% identification rate | [33] |
| Xuping Feng et al. | Transgenic maize | SG smoothing | KNN, SIMCA, NBC, ELM, RBFNN | The classification rates of full-spectrum and the feature wavelength were 100% and 90.83% in ELM model. | [27] |
| Cheng Peng et al. | Transgenic maize | SG smoothing | PLS, SVM | The accuracy of the SVM model based on full-band spectra of transgenic maize powder was 90.625%. | [12] |
| Haosong Guo et al. | Transgenic sugarcane | SG, MW | LDA | The corresponding validation recognition rates of transgenic and non-transgenic samples achieved 99.1% and 98.0%, respectively. | [61] |
| Guisong Liu et al. | Transgenic sugarcane | SG | PCA, LDA, HCA | The optimal SG-PCA-LDA model for positive and negative samples were 94.3% and 96.0%, respectively, and that of the optimal SG-PCA-HCA model for positive and negative samples were 92.5% and 98.0%, respectively. | [62] |
| Yafeng Zhai et al. | Transgenic wheat | Normalization | BPR | A model for identification of wheat varieties was developed using PCA combined with biomimetic pattern recognition method. | [63] |
| Aderval S. Lunaet al. | Transgenic soybean oils | MC, MSC, OSC, SG 1st, 2nd derivatives | SVM-DA, PLS-DA | The classification rate of SVM-DA was 100% in the training group and 100% and 90% in the validation group for non-GMO and GMO soybean oil samples. In PLS-DA model, the classification rates were 95% and 100% for the training group and 100% and 80% for the validation group of non-GMO and GM soybean oil samples, respectively. | [64] |
| Jianguo Zhu et al. | Transgenic oils | MSC, first derivative (FD), MWS, SG1 preprocessing | SVM | MSC had the best prediction performance with the accuracy rate of 91.6%. The prediction accuracy of SVM was improved to 98.3% by using the SPA algorithm. | [31] |
| Author | Object | Preprocessing Methods | Models | Results | Reference |
|---|---|---|---|---|---|
| Priscilla Dantas Rocha et al. | Transgenic cotton seed | SNV, SG smoothing | PLS-DA | The specificity and sensitivity values of the different methods ranged from 0.78–0.92 and 0.62–0.93, respectively. | [81] |
| Xuping Feng et al. | Transgenic maize | WT, MSC, SNV | SVM, PLS-DA | SVM and PLS-DA models could obtain good performance with almost 100% accuracy. | [3] |
| Xuping Feng et al. | Shikimic acid concentration in transgenic maize plant | SNV, MSC, WT, SG smoothing | PLS-DA, PLSR, RF | A coefficient of determination value of 0.79 for the calibration set and a coefficient of determination of 0.82 for the prediction set. | [82] |
| Hailong Wang et al. | Transgenic soybeans | MA | PLS-DA | The results showed that hyperspectral imaging techniques could be used for the identification of non-GM soybeans. | [83] |
| Xuping Feng et al. | Transgenic rice | WT | RBFNN, KNN, ELM | The RBFNN models based on the 24 feature bands extracted from the 2nd derivative achieved the accuracy of 92.25% and 89.50% of the modeling set and prediction set, respectively. | [84] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, Z.; Pu, Y.; Wang, J.; Tang, B.; Dai, L.; Yu, S.; Chen, R. Identification of Transgenic Agricultural Products and Foods Using NIR Spectroscopy and Hyperspectral Imaging: A Review. Processes 2023, 11, 651. https://doi.org/10.3390/pr11030651
Zhang J, Liu Z, Pu Y, Wang J, Tang B, Dai L, Yu S, Chen R. Identification of Transgenic Agricultural Products and Foods Using NIR Spectroscopy and Hyperspectral Imaging: A Review. Processes. 2023; 11(3):651. https://doi.org/10.3390/pr11030651
Chicago/Turabian StyleZhang, Jun, Zihao Liu, Yaoyuan Pu, Jiajun Wang, Binman Tang, Limin Dai, Shuihua Yu, and Ruqing Chen. 2023. "Identification of Transgenic Agricultural Products and Foods Using NIR Spectroscopy and Hyperspectral Imaging: A Review" Processes 11, no. 3: 651. https://doi.org/10.3390/pr11030651
APA StyleZhang, J., Liu, Z., Pu, Y., Wang, J., Tang, B., Dai, L., Yu, S., & Chen, R. (2023). Identification of Transgenic Agricultural Products and Foods Using NIR Spectroscopy and Hyperspectral Imaging: A Review. Processes, 11(3), 651. https://doi.org/10.3390/pr11030651







