1. Introduction
The concept of Industry 4.0 has initiated a revolution in a number of industrial sectors. While digitalization emerges as a crucial step towards modernization in its own right [
1], it also serves as the foundation to promote intellectualization. As of late, digitalization has gained significant attention and has had a profound impact on various process industries [
2]. The rise of digital technologies has provided plenty of valuable tools to effectively manage voluminous data and make informed decisions in the production process. Given the progression of sensors [
3], virtualization technology [
4], and modelling [
5], as well as the proliferation of computing power, simulation has emerged as a ubiquitous research methodology. By leveraging in silico modelling, comprehensive and precise digital replicas of entire manufacturing systems or product lifecycles can be constructed. With these technical prerequisites, the notion of digital twin (DT) attracted increasing interest from both academia and industry [
6].
In 2003, Grieves introduced the notion of digital representation for tangible products in the context of product lifecycle management, which was an early exploration of what we now know as the digital twin [
7]. In response to the growing demands of the aerospace industry, NASA gave a comprehensive definition of digital twin in 2012. This definition presented an integrated multi-scale and multi-physics model, supported by physical models, feedback from sensors, and historical data, all of which work in concert to simulate the complete lifecycle of a physical system [
8]. The Gartner Group’s 2018 report recognized the digital twin as one of the top ten most promising technological trends of the time [
9]. Subsequently, digital twins have been implemented in a wide range of applications, including but not limited to, manufacturing [
10], medicine [
11] and the process industry [
12].
The concept of the digital twin introduced a novel paradigm of Cyber-Physical System (CPS), which operates on a foundation of multi-scale digital twins in conjunction with a physical manufacturing system. The fundamental constituents of a digital-twin-driven manufacturing system include a physical manufacturing system, corresponding digital twin, a Human–Machine Interface (HMI) application, and bidirectional data channels for seamless data exchange. A multitude of online sensors are embedded within the physical manufacturing system, allowing for the continuous monitoring and tracking of its status. The collected data are synchronized with the digital twin, which operate autonomously to simulate the current or future status of the manufacturing system. The HMI application is tasked with displaying sensor data and the digital twin’s outputs in a coherent manner. Furthermore, the system operators possess the capability to alter manufacturing system parameters, subsequently triggering updates to both the digital twin and the predicted data that are in need of recalculation.
Despite the potential advantages of the digital twin, its implementation in the realm of biomanufacturing is facing a host of challenges. To achieve optimal levels of productivity, it is incumbent upon both the academic and industrial sectors to undertake the comprehensive digitization of biomanufacturing processes. Although the efficacy of digital twins has been demonstrated in myriad domains, their applications in biomanufacturing remains incipient owing to the complexities of biological systems [
13] and the stringent safety requirements governing this field [
14].
Udugama et al. [
15] elaborated a three-tiered approach to constructing bioprocess digital twins, involving the digital model, the digital shadow, and ultimately, the digital twin. Whereas the digital model solely offers static prognoses of the bioprocesses, and the digital shadow merely reflects their actual status, the digital twin furnishes dynamic predictions based on mathematical models, executing self-iteration to enhance performance through the continued analysis of process data.
The domain of biomanufacturing has seen a proliferation of modelling methodologies, including computational fluid dynamics (CFD) [
16], kinetics [
17], and metabolic models [
18], which have been extensively studied. Additionally, the feasibility of digital twins has been explored and validated through a series of studies focused on HIV-Gag VLP production in HEK293 cells [
19,
20,
21], demonstrating their potential in optimizing processes and achieving autonomous process control. Furthermore, Park et al. [
22] have discussed the essential technologies required to realize the bioprocess digital twin for Chinese hamster ovary (CHO) cell culture. However, despite these promising developments, there remains several critical areas that require further attention. For instance, there is a dearth of research concerning the updating of model parameters according to process data. Moreover, existing research is primarily focused on specific scenarios within narrow scopes, with a lack of emphasis on interoperability and reusability, hindering their adoption in biomanufacturing.
As an exploratory foray, we present BioDT, a digital-twin-based cell culture framework, which serves as an example of a digital-twin-based biomanufacturing system and offers a reference for future research. It is noteworthy that all components of this framework are open-source, enabling researchers to adapt it to diverse research scenarios. Furthermore, to elucidate the operational mechanisms of this framework, we designed a CHO cell culture microbioreactor based on it. Our design encompasses a 3D-printed automated microbioreactor, a bioprocess digital twin, and an HMI application. The microbioreactor is designed to support in-place cell density monitoring, and it is specifically adapted to accommodate T25 flasks, a widely used cell culture vessel. The bioprocess digital twin was designed to take advantage of hybrid programming where C++ was used to encode the kinetic formulae of mass transfer, cell growth, and metabolism, while calculations and simulations were carried out in a Python program. The HMI application was also implemented in Python, and it is used to display process trends and provide users with an interface to interact with the system. The digital twin was validated by comparing the experimental data of cell density, glucose, and lactic acid concentrations with those predicted by the digital twin in CHO cell culture. To further illustrate the versatility of the system, we presented a case study of manual media change and automated glucose feeding. Results indicate that the digital twin provided reliable reference values of metabolite concentrations, and showed its potential in Advanced Process Control (APC).
2. Materials and Methods
2.1. Cell Culture and Sampling
Our laboratory preserved the cell line of FreeStyle™ CHO (Thermo Fisher Scientific, Waltham, MA, USA) for suspension culture. The culture was conducted in a T25 flask (Thermo Fisher Scientific, Waltham, USA) at 37 °C and 8% CO
2 within an incubator using FreeStyle™ CHO Expression Media (Thermo Fisher Scientific, Waltham, USA), supplemented with 8 mM glutamine. The culture volume was 10 mL. Given the weaker shear stress present in rocking T25 flasks compared to other culture vessels [
23], 0.2% of the Anti-Clumping Agent (Thermo Fisher Scientific, Waltham, USA) was incorporated into the media to prevent cell clumping. The T25 flask was secured onto a 3D-printed platform driven by a stepper motor at a predetermined speed and angle. The in-place measurement of cell density was executed every 15 min using a pair of near-infrared emitter and receiver, while glucose and lactic acid concentration levels were sampled every 12 h with a biological analyzer (Sieman Technology, Shenzhen, China).
2.2. 3D-Printed Automated Microbioreactor with Real-Time Cell Density Measurement
Many automated miniature-scale devices have been developed for cell or bacterial cultures. One notable example is eVOLVER, presented by Wong et al. [
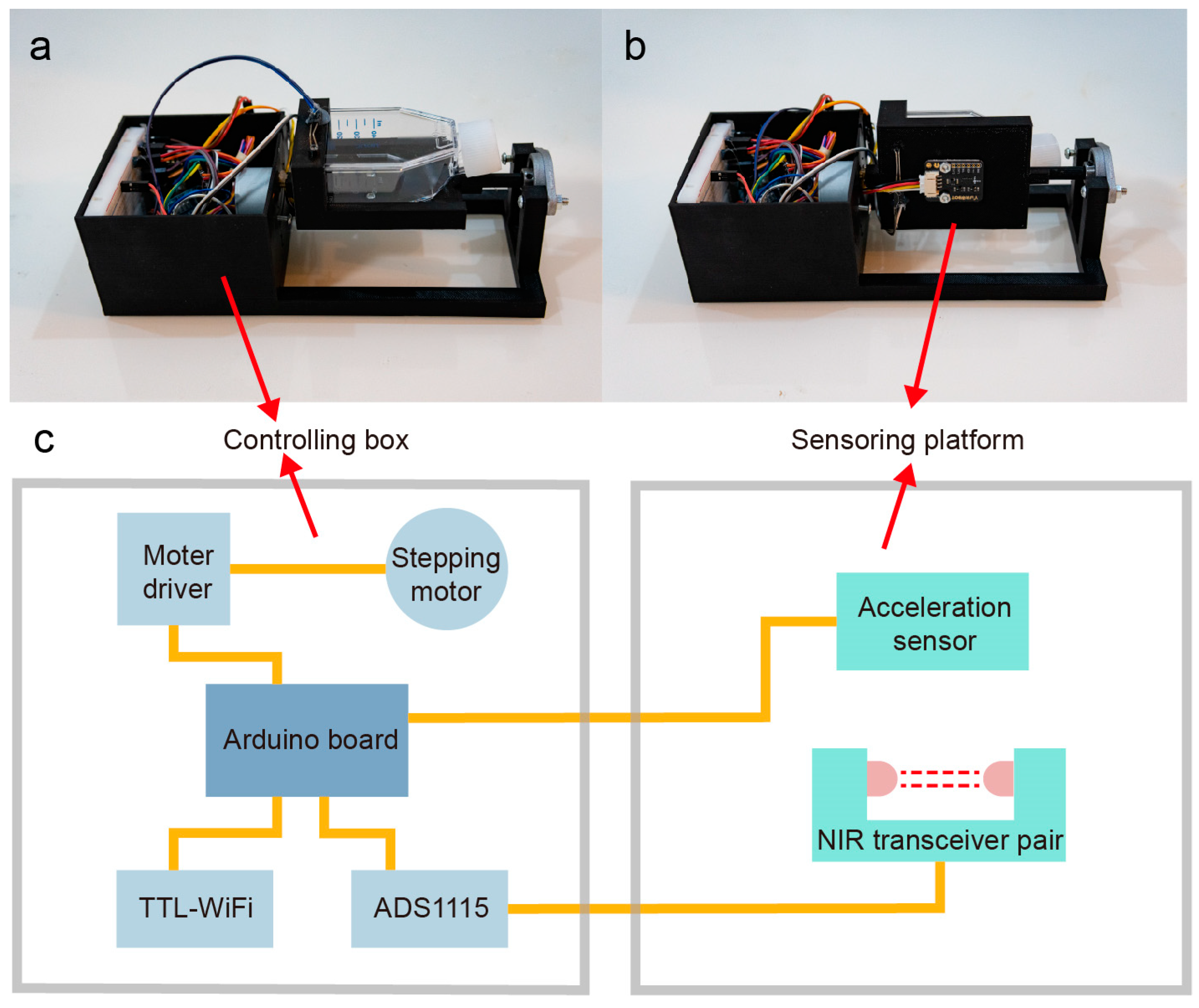
24], which enables fine-tuning of a few process parameters and supports high-throughput operation. However, despite these advancements, most cell culture devices merely record process parameters and fail to meet the crucial requirement of real-time communication with models, which is a fundamental characteristic of the digital twin. In this study, we designed and 3D-printed an automated microbioreactor for the cultivation of CHO cells in T25 flasks (
Figure 1a–c). Our design incorporates the capability of adjusting the rocking speed and angle, as well as monitoring the cell density in place. Importantly, the device is also equipped with a Wi-Fi chip that enables data exchange with a digital twin, thus facilitating the continuous updating of process parameters and monitoring of system status, supporting the implementation of the digital twin in terms of hardware.
The 3D model of the device was designed using Autodesk Fusion 360 and realized by a 3D printer (Creative 3D) with PLA filament. All electronic components were connected to an Arduino UNO board (Smart Projects, Pescara, Italy) as the micro controller. The controller board consisted of five modules: (1) a Wi-Fi chip for communication with the HMI, enabling data transmission and receipt of commands. (2) A stepping motor for rocking the T25 flask, which was powered and controlled by an encoder board wired to the Arduino. The motor allowed for adjustable rocking speeds from 40 to 80 rpm and angles of 0–90° (typical range is 7–15°). (3) An acceleration sensor for monitoring the rotation speed and angle of the flask by detecting the vertical component of gravity. However, the rocking action was not based on the real-time output from the acceleration sensor in a feedback loop manner. Instead, the acceleration sensor was only used to adjust the initial position of the flask before rocking commenced. Afterwards, the rocking angle and velocity were open-loop-controlled by taking advantage of the stepper motor’s precise discrete motion. During cell density measurements, the motor firstly levels the flask, then rotates it by 90° (
Figure 2a), collecting the liquid at the bottom of the flask. After the optical measurement, the flask is rotated back to its initial position and the rocking resumes. (4) A coupled 940 nm near-infrared emitter and receiver (
Figure 2b) were utilized to measure the intensity of light passing through the media, thereby assessing the optical density of the culture. (5) A 16-bit analog–digital (AD) converter was used to convert the analog signal to amplified digital signal (
Figure 2d), which can be interpreted by Arduino. The optical density was converted to cell density using a predefined calibration curve (
Figure 2c). Since signals can fluctuate during sampling, which can lead to noise and inaccuracies in the data, a Kalman filter was utilized to reduce the noise. The Kalman filter is designed to analyze the current and past values of a signal and calculate the optimal estimate of the signal’s true value.
2.3. Bioprocess Simulation
For the modelling part, we introduce Pyrex, a Python library for bioreaction computing currently remaining in development, which we anticipate to be a valuable resource for researchers. To provide a preview of its capabilities, we present a simplified version of it in the context of CHO cell culture. To accurately track the CHO cell growth and metabolism, we incorporated a set of commonly used kinetics equations with parameters obtained through batch culture experiments and the literature.
The cell growth rate is calculated using a logistic equation (Equation (1)). The initial value of
of 0.05 h
−1 is acquired from the literature [
25], and it is updated along with the sensor data. The maximum cell density,
is set to 8 × 10
6 according to batch culture experiments. It should be noted that the use of the logistic equation is for demonstration purposes only. Other types of empirical or mechanistic equations can be readily incorporated.
where
is the specific growth rate (cell/cell);
is the maximum growth rate (cell/cell);
is the cell density (cell/mL);
is the maximum cell density (cell/mL).
The consumption rate of glucose and the generation of lactic acid are carried out using Equations (2) and (3). The initial values of
and
were estimated from batch experiments, which were -2.06 × 10
−13 L/g/cell/h and 0.46, respectively.
where
is the concentration of glucose (g/L);
is the concentration of lactic acid (g/L);
is the specific glucose consumption rate (L/g/cell/h);
is the yield coefficient of lactate to glucose (g/g).
As these two parameters continuously evolve with the environmental conditions and the physiological status of the cells, they were re-evaluated by the digital twin using the process data, as described later.
To consider the variations in dissolved oxygen during cell culture, the oxygen transfer rate and the oxygen uptake rate were calculated, respectively. The relationship between the rocking speed and the volumetric mass transfer coefficient,
, was previously determined by Liu et al. [
23] at an interval of 10 rpm and an amplitude of 7°. We use a polynomial function, Equation (4), to fit these data to estimate the
at any rocking speed between 10 and 80 rpm. The oxygen saturation concentration was set to 0.25 mol/L, i.e., 92% of that in equilibrium with air at atmosphere pressure, as the incubator has 8% of CO
2 in it. The value of
was set to 3.2 × 10
−13 mol/cell/h according to Deshpande et al. [
26]. The value of dissolved oxygen was then calculated with Equation (5).
where
is the volumetric mass transfer coefficient (h
−1);
is the rocking speed;
is the dissolved oxygen concentration (mmol/L);
is the equilibrium oxygen concentration (mmol/L); and
is the specific glucose consumption rate (mmol/cell/h).
Given that analytical solutions for different differential equations are not universally achievable, numerical methods were employed to integrate these equations with a step-size of 1 s. As the cell metabolism is relatively slow, process parameters are not expected to experience significant changes over such a short period of time; the system’s status can thus be assumed to be in quasi-steady state at each time step. This approach ensures that the system’s state is updated every second. It should be noted that if a larger time interval is used or for a fast-changing system, a higher-order scheme should be adopted to reduce numerical errors.
In order to optimize computing performance, all of the above-mentioned formulae were coded in C++. The C++ code was then compiled as a Python extension using Pybind11, thereby allowing for the execution of these C++ functions by Python applications for flexibility. A Python program was subsequently created to integrate these methods, enabling real-time data processing, realizing the implementation of the digital twin. Parameters were updated once sensor data or offline measurement were synchronized to the digital twin. The Levenberg–Marquardt algorithm included in the Python package Scipy was used to recalculate the kinetic parameters.
2.4. HMI Application
To effectively manage data, seamlessly integrate predictive and decision-making functionalities, and facilitate user interactions, a simple Human–Machine Interface (HMI) was created in Python. The application comprises four modules: data transmission, data storage (SQL database), the digital twin, and data visualization. The data transmission module communicated with the device based on the TCP/IP protocol. The module created a socket and connected to the device through a soft serial port created by the Arduino at the IP address 192.168.4.1 and the port number 9000 as required by the Wi-Fi chip. The data transmission happened on-demand whenever the status of the device or the digital twin changed. The data storage module is based on SQLite, recording the sensor data and the digital twin’s predictions. The digital twin module is based on the previously mentioned python extension named Pyrex. This module updates the predicted status in real time and iteratively corrects the model parameters when the device receives sensor data. Data visualization and user interface (UI) are based on the Python package bokeh (V2.3). The UI shows the data recorded by the device and predicted by the digital twin. Commands are sent to the digital twin or physical twin when the operator interacts with the HMI. Detailed usage of the HMI can be found in our published codes on GitHub.
2.5. Integration of the Digital-Twin-Based Cell Culture Framework
Through the integration of the aforementioned components, a digital-twin-based cell culture system was realized, as depicted in
Figure 3. This innovative framework seamlessly linked the hardware and software, as well as the virtual replica and physical device, in accordance with the BioDT paradigm. The bidirectional data flow between the system components facilitated the optimal utilization of data, thereby enhancing the accuracy and efficiency of the system.
4. Discussion
The present study details the development of a customizable and comprehensive digital-twin-based framework for cell culture, referred to as BioDT. This framework represents a standard paradigm for digital-twin-based cell culture and can serve as a reference for subsequent investigations in the field. To demonstrate the efficacy of the framework, we established a laboratory-scale CHO cell culture system using BioDT.
The design presented in this study consists of three key components: an innovative cell culture device, a digital twin, and a web HMI. The cell culture device is a miniature bioreactor that accommodates a T25 flask, featuring in-place monitoring of cell density and adjustable rocking speed and angle. A digital twin was constructed on a set of kinetic equations, which are used to calculate cell growth rate, glucose consumption, lactic acid production, and oxygen transfer. The HMI managed the exchange of data between the physical device and the digital twin, delivering real-time visualization of data, user interaction, and storage of data in a database. Through communication between the device and software, this system enabled dynamic and precise control over process parameters. The effectiveness of the proposed design was validated through a series of cell culture experiments. With the assistance of the digital twin in kinetics tracing and process predicting, a more comprehensive and deeper understanding of bioprocesses can be achieved, which would be considerably useful in optimizing the production process.
The current system consists of highly customizable components, and we have made all blueprints and codes open-source, thereby exemplifying a flexible approach for digital-twin-based systems. The hardware component of our design is tailored for suspension culture in a T25 flask. However, any culture device that is adapted to the communication protocol can be integrated into the system, and the model equations can be readily modified to suit other type of reactors. A major drawback of the current physical system is the lack of sensors, unlike commercial manufacturing facilities that are equipped with more advanced process analytics. It should be reasonable to expect that the digital twin can be significantly enhanced in the latter case, which would in turn accelerate the digitization of biomanufacturing. We also designed the bioprocess modelling library Pyrex in C++, which ensures high computing performance, and can be utilized by any Python application. The library currently provides modelling methods for cell behavior and mass transfer, albeit in a limited capacity. To handle the complicated phenomena in biomanufacturing, from upstream to downstream, methods that consider more parameters and their interactions need to be developed. Hartmann et al. [
27] comprehensively reviewed the modelling methods in bioprocesses and emphasized the necessity to develop multi-scale and multi-disciplinary models. Wang et al. [
28] proposed that models that integrate spatiotemporal multiscale cellular models and fluid dynamics would be an ideal tool for bioprocess design as well. It is also of critical importance to create a widely applicable computing library for bioprocess modelling that can help drive digitization in biomanufacturing, which would be the focus of our subsequent research. By making appropriate modifications to the HMI, users can develop new features to adapt to their own culture system. Incorporating more sophisticated concepts and technologies related to Process Analytical Technology (PAT) and Advanced Process Control (APC) into this system would hold great promise for the widespread implementation of digital twins in biomanufacturing. This could eventually lead to improvements in productivity and quality control, bolstering the overall efficiency of the biomanufacturing process.