mAb Production Modeling and Design Space Evaluation Including Glycosylation Process
Abstract
:1. Introduction
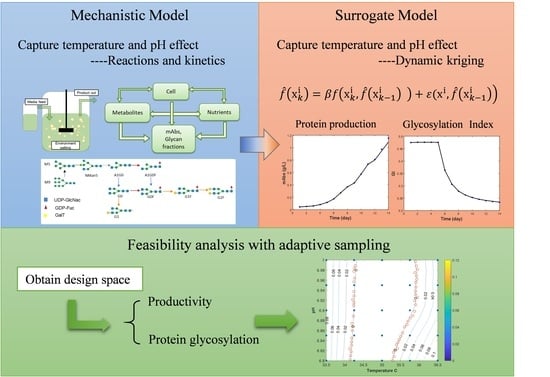
2. Methods
2.1. Kinetic Model Building
2.2. Design of Experiment
2.3. Kriging and Dynamic Kriging Model Building
2.4. Feasibility Analysis with Adaptive Sampling
3. Results and Discussion
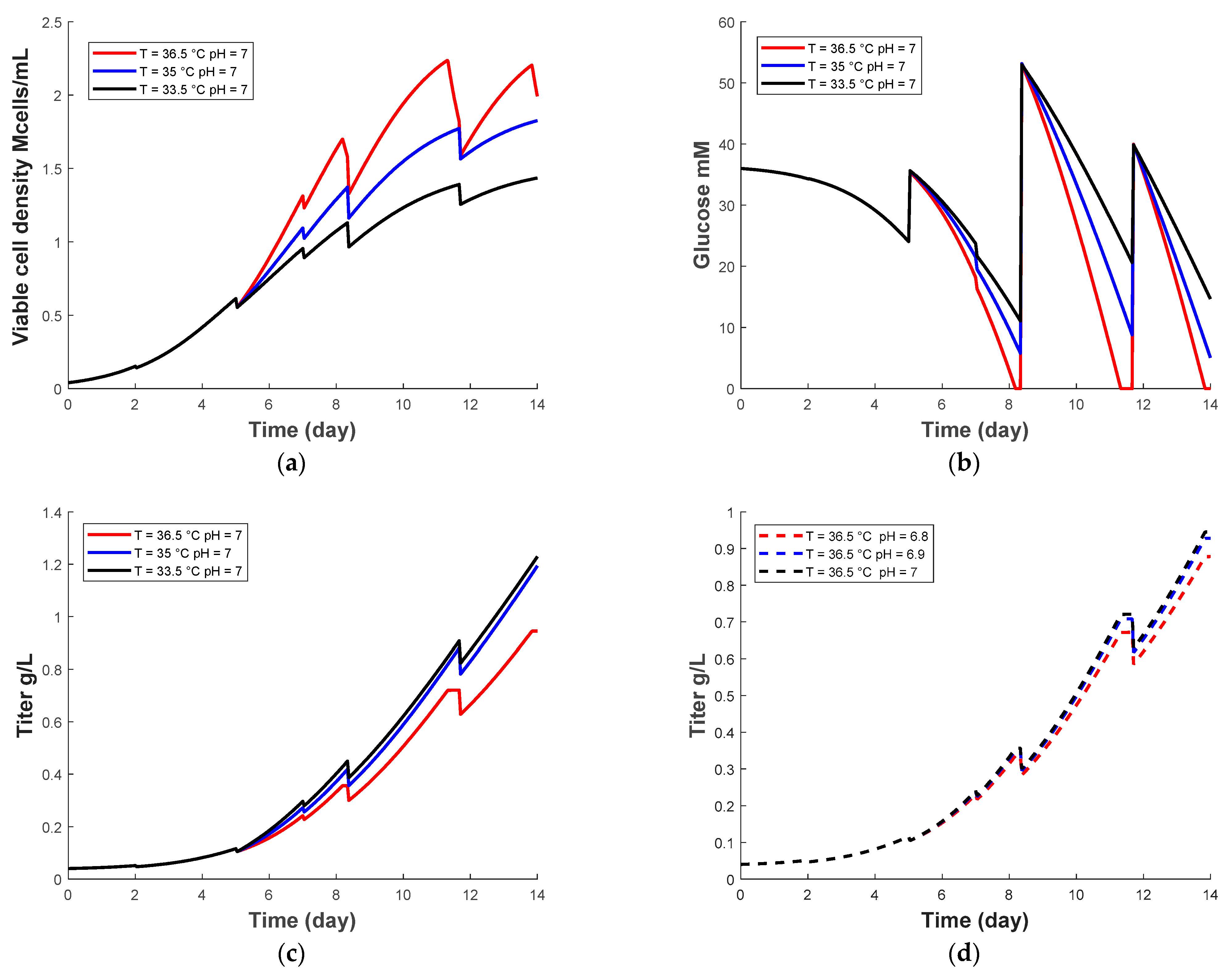
3.1. Prediction of Temperature and pH Effect Using the Kinetic Model
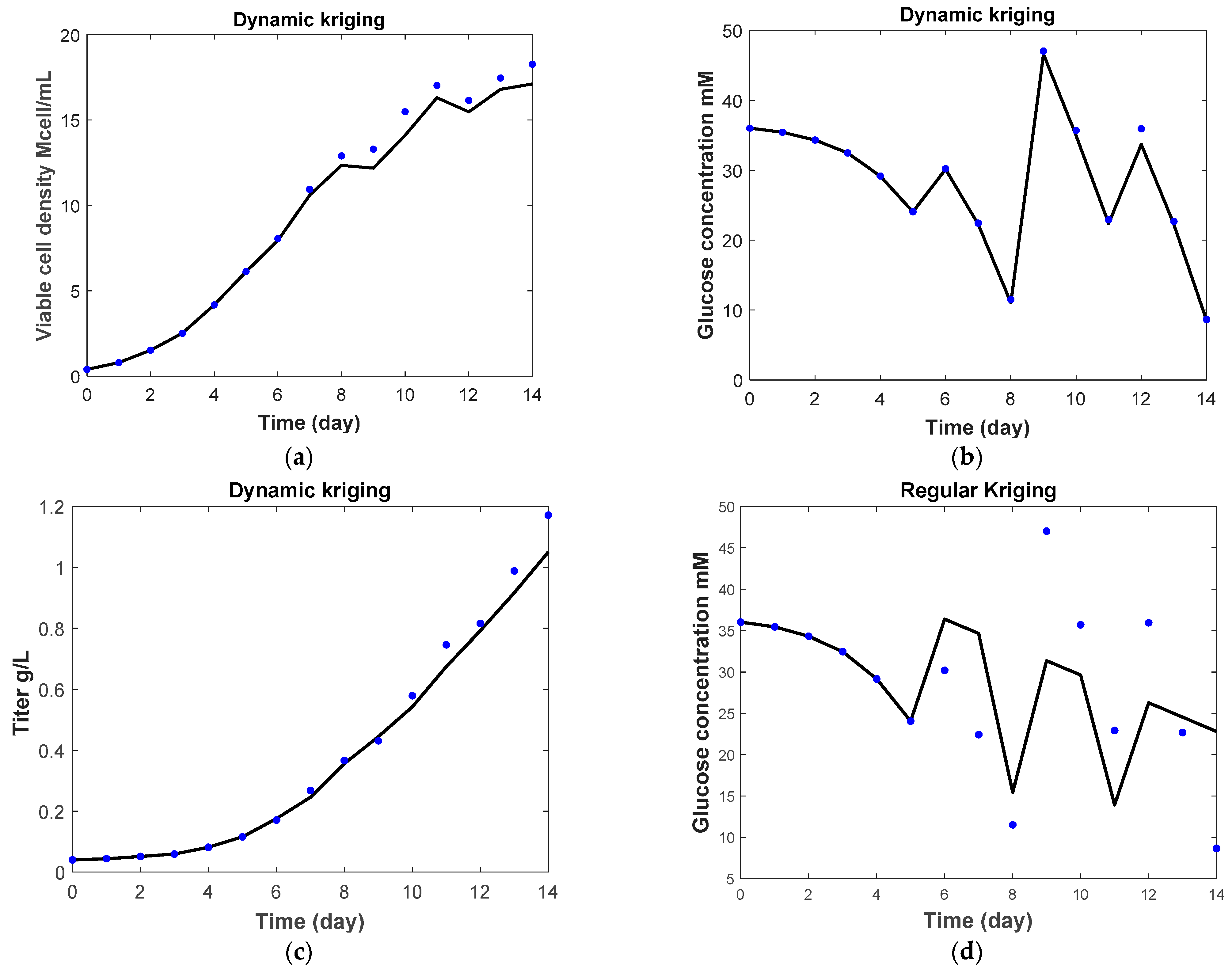
3.2. Kriging and Dynamic Kriging
3.2.1. Regular Kriging vs. Dynamic Kriging
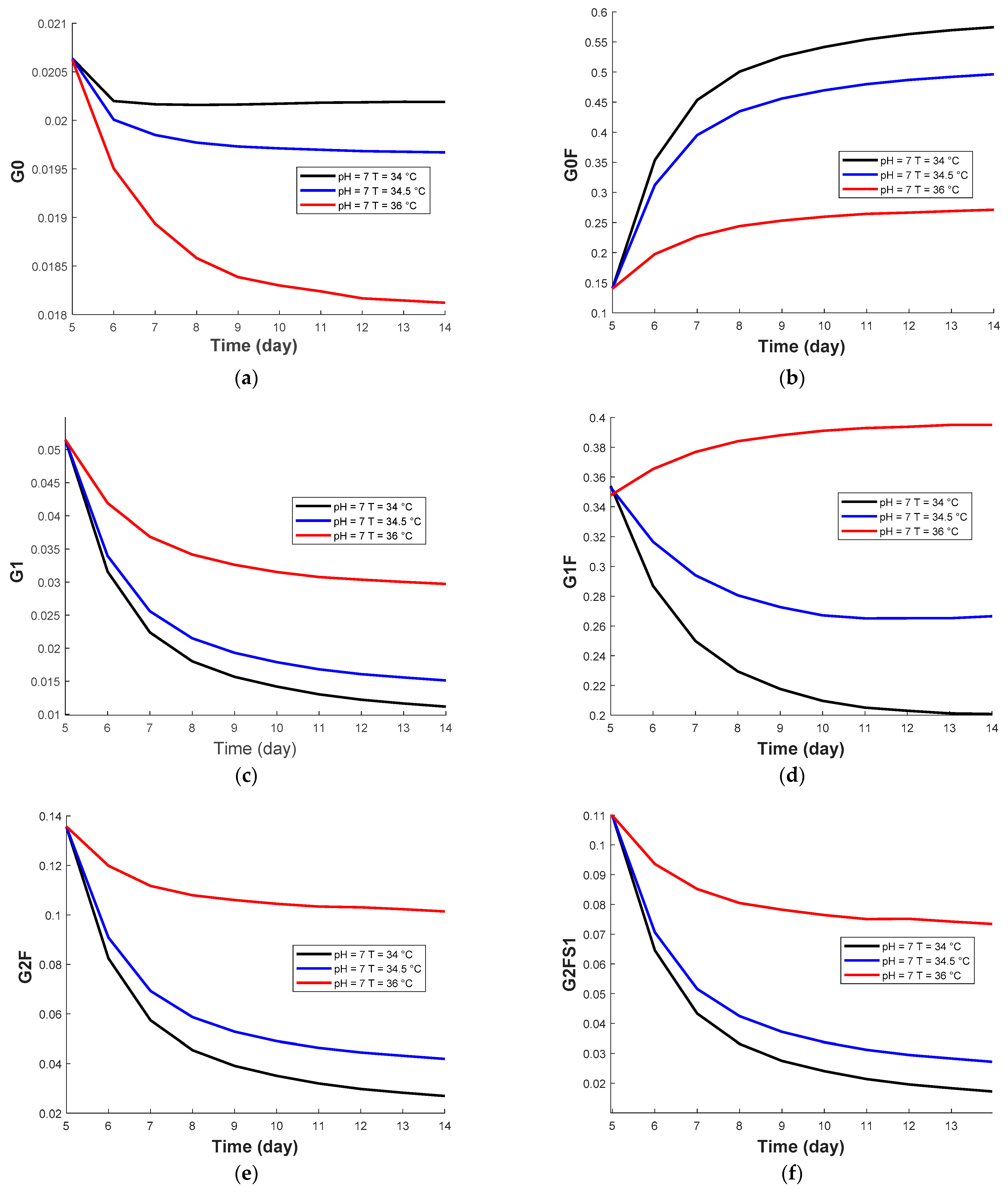
3.2.2. Prediction of Temperature Effect from Dynamic Kriging Model
3.3. Feasibility Analysis and Design Space
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tantibanchachai, C. Coronavirus (COVID-19) Update: FDA Authorizes Monoclonal Antibodies for Treatment of COVID-19. 2020. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-monoclonal-antibodies-treatment-covid-19 (accessed on 19 December 2020).
- Kiyoshi, M.; Caaveiro, J.M.; Tada, M.; Tamura, H.; Tanaka, T.; Terao, Y.; Morante, K.; Harazono, A.; Hashii, N.; Shibata, H.; et al. Assessing the Heterogeneity of the Fc-Glycan of a Therapeutic Antibody Using an engineered FcγReceptor IIIa-Immobilized Column. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Del Val, I.J.; Kontoravdi, C.; Nagy, J.M. Towards the implementation of quality by design to the production of therapeutic monoclonal antibodies with desired glycosylation patterns. Biotechnol. Prog. 2010, 26, 1505–1527. [Google Scholar] [CrossRef]
- Ehret, J.; Zimmermann, M.; Eichhorn, T.; Zimmer, A. Impact of cell culture media additives on IgG glycosylation produced in Chinese hamster ovary cells. Biotech-Nol. Bioeng. 2019, 116, 816–830. [Google Scholar]
- Lodish, H.; Berk, A.; Zipursky, S.L.; Matsudaira, P.; Baltimore, D.; Darnell, J. Protein Glycosylation in the ER and Golgi Complex. In Molecular Cell Biology; W.H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Higel, F.; Seidl, A.; Sörgel, F.; Friess, W. N-glycosylation heterogeneity and the influence on structure, function and pharmacokinetics of monoclonal antibodies and Fc fusion proteins. Eur. J. Pharm. Biopharm. 2016, 100, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Aghamohseni, H.; Spearman, M.; Ohadi, K.; Braasch, K.; Moo-Young, M.; Butler, M.; Budman, H.M. A semi-empirical glycosylation model of a camelid monoclonal antibody under hypothermia cell culture conditions. J. Ind. Microbiol. Biotechnol. 2017, 44, 1005–1020. [Google Scholar] [CrossRef]
- Kim, S.-M.; Chang, K.-H.; Oh, D.J. Effect of Environmental Parameters on Glycosylation of Recombinant Immunoglobu-lin G Produced from Recombinant CHO Cells. Biotechnol. Bioprocess Eng. 2018, 23, 456–464. [Google Scholar] [CrossRef]
- Sumit, M.; Dolatshahi, S.; Chu, A.-H.A.; Cote, K.; Scarcelli, J.J.; Marshall, J.K.; Cornell, R.J.; Weiss, R.; Lauffenburger, U.A.; Mulukutla, B.C.; et al. Dissecting N-Glycosylation Dynamics in Chinese Hamster Ovary Cells Fed-batch Cultures using Time Course Omics Analyses. iScience 2019, 12, 102–120. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.C.F.; Wong, K.T.K.; Goh, L.T.; Heng, C.K.; Yap, M.G.S. Impact of dynamic online fed-batch strategies on metabolism, productivity and N-glycosylation quality in CHO cell cultures. Biotechnol. Bioeng. 2004, 89, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Raju, T.S.; Jordan, R.E. Galactosylation variations in marketed therapeutic antibodies. mAbs 2012, 4, 385–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Surve, T.; Gadgil, M. Manganese increases high mannose glycoform on monoclonal antibody expressed in CHO when glucose is absent or limiting: Implications for use of alternate sugars. Biotechnol. Prog. 2015, 31, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, D.; Robinson, A.S.; Ogunnaike, B.A. Controlling the Glycosylation Profile in mAbs Using Time-Dependent Media Supplementation. Antibodies 2017, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Kotidis, P.; Jedrzejewski, P.; Sou, S.N.; Sellick, C.; Polizzi, K.M.; Del Val, I.J.; Kontoravdi, C. Model-based optimization of antibody galactosylation in CHO cell culture. Biotechnol. Bioeng. 2019, 116, 1612–1626. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Lovelett, R.J.; Price, J.V.; Radhakrishnan, D.; Barnthouse, K.; Hu, P.; Schaefer, E.; Cunningham, J.; Lee, K.H.; Shivappa, R.B.; et al. Modeling the Effect of Amino Acids and Copper on Monoclonal Antibody Productivity and Glycosylation: A Modular Approach. Biotechnol. J. 2020, 16, 2000261. [Google Scholar] [CrossRef]
- Villiger, T.K.; Scibona, E.; Stettler, M.; Broly, H.; Morbidelli, M.; Soos, M. Controlling the time evolution of mAb N-linked glycosylation—Part II: Model-based predictions. Biotechnol. Prog. 2016, 32, 1135–1148. [Google Scholar] [CrossRef]
- Sou, S.N.; Jedrzejewski, P.M.; Lee, K.; Sellick, C.; Polizzi, K.M.; Kontoravdi, C. Model-based investigation of intracellular processes determining antibody Fc-glycosylation under mild hy-pothermia. Biotechnol. Bioeng. 2017, 114, 1570–1582. [Google Scholar] [CrossRef]
- Narayanan, H.; Luna, M.F.; Von Stosch, M.; Bournazou, M.N.C.; Polotti, G.; Morbidelli, M.; Butté, A.; Sokolov, M. Bioprocessing in the Digital Age: The Role of Process Models. Biotechnol. J. 2020, 15, e1900172. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, M.; Morbidelli, M.; Butté, A.; Souquet, J.; Broly, H. Sequential Multivariate Cell Culture Modeling at Multiple Scales Supports Systematic Shaping of a Mono-clonal Antibody Toward a Quality Target. Biotechnol. J. 2018, 13, 1700461. [Google Scholar] [CrossRef] [PubMed]
- Zürcher, P.; Sokolov, M.; Brühlmann, D.; Ducommun, R.; Stettler, M.; Souquet, J.; Jordan, M.; Broly, H.; Morbidelli, M.; Butté, A. Cell culture process metabolomics together with multivariate data analysis tools opens new routes for bio-process development and glycosylation prediction. Biotechnol. Prog. 2020, 36, e3012. [Google Scholar] [CrossRef]
- Del Val, I.J.; Fan, Y.; Weilguny, D. Dynamics of immature mAb glycoform secretion during CHO cell culture: An integrated modelling framework. Biotechnol. J. 2016, 11, 610–623. [Google Scholar] [CrossRef] [Green Version]
- Sou, S.N.; Sellick, C.; Lee, K.; Mason, A.; Kyriakopoulos, S.; Polizzi, K.M.; Kontoravdi, C. How does mild hypothermia affect monoclonal antibody glycosylation? Biotechnol. Bioeng. 2015, 112, 1165–1176. [Google Scholar] [CrossRef] [Green Version]
- Jimenez del Val, I.; Nagy, J.M.; Kontoravdi, C. A dynamic mathematical model for monoclonal antibody N-linked glyco-sylation and nucleotide sugar donor transport within a maturing Golgi apparatus. Biotechnol. Prog. 2011, 27, 1730–1743. [Google Scholar] [CrossRef]
- Fan, Y.; Del Val, I.J.; Müller, C.; Lund, A.M.; Sen, J.W.; Rasmussen, S.K.; Kontoravdi, C.; Baycin-Hizal, D.; Betenbaugh, M.J.; Weilguny, D.; et al. A multi-pronged investigation into the effect of glucose starvation and culture duration on fed-batch CHO cell culture. Biotechnol. Bioeng. 2015, 112, 2172–2184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, E.; Ierapetritou, M. A kriging method for the solution of nonlinear programs with black-box functions. AIChE J. 2007, 53, 2001–2012. [Google Scholar] [CrossRef]
- Boukouvala, F.; Muzzio, F.J.; Ierapetritou, M.G. Dynamic Data-Driven Modeling of Pharmaceutical Processes. Ind. Eng. Chem. Res. 2011, 50, 6743–6754. [Google Scholar] [CrossRef]
- Sacks, J.; Welch, W.J.; Mitchell, T.J.; Wynn, H.P. Design and Analysis of Computer Experiments. Stat. Sci. 1989, 4, 409–423. [Google Scholar] [CrossRef]
- Bhosekar, A.; Ierapetritou, M. Advances in surrogate based modeling, feasibility analysis, and optimization: A review. Comput. Chem. Eng. 2018, 108, 250–267. [Google Scholar] [CrossRef]
- Lophaven, S.N.; Nielsen, H.B.; Søndergaard, J. DACE: A Matlab Kriging Toolbox; Technical Report IMM-TR-2002-12; Technical University of Denmark: Lyngby, Denmark, August 2002; Volume 2. [Google Scholar]
- Hernandez, A.F.; Grover, M.A. Stochastic dynamic predictions using Gaussian process models for nanoparticle synthesis. Comput. Chem. Eng. 2010, 34, 1953–1961. [Google Scholar] [CrossRef]
- Boukouvala, F.; Ierapetritou, M.G. Derivative-free optimization for expensive constrained problems using a novel ex-pected improvement objective function. AIChE J. 2014, 60, 2462–2474. [Google Scholar] [CrossRef]
- Vergara, M.; Becerra, S.; Berrios, J.; Osses, N.; Reyes, J.G.; Rodríguez-Moyá, M.; Gonzalez, R.; Altamirano, C. Differential Effect of Culture Temperature and Specific Growth Rate on CHO Cell Behavior in Chemostat Culture. PLoS ONE 2014, 9, e93865. [Google Scholar] [CrossRef] [PubMed]
- Trummer, E.; Fauland, K.; Seidinger, S.; Schriebl, K.; Lattenmayer, C.; Kunert, R.; Vorauer-Uhl, K.; Weik, R.; Borth, N.; Katinger, H.; et al. Process parameter shifting: Part II. Biphasic cultivation—A tool for enhancing the volumetric productivity of batch processes using Epo-Fc expressing CHO cells. Biotechnol. Bioeng. 2006, 94, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Tang, P.; Yongky, A.; Drew, B.; Borys, M.C.; Liu, S.; Li, Z.J. Systematic development of temperature shift strategies for Chinese hamster ovary cells based on short duration cultures and kinetic modeling. MAbs 2019, 11, 191–204. [Google Scholar] [CrossRef] [Green Version]
- Villiger, T.K.; Roulet, A.; Périlleux, A.; Stettler, M.; Broly, H.; Morbidelli, M.; Soos, M. Controlling the time evolution of mAb N-linked glycosylation, Part I: Microbioreactor experiments. Biotechnol. Prog. 2016, 32, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Chen, H.; Xu, S. pH excursions impact CHO cell culture performance and antibody N-linked glycosylation. Bioprocess Biosyst. Eng. 2018, 41, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Agarabi, C.D.; Schiel, J.E.; Lute, S.C.; Chavez, B.K.; Boyne, M.T., 2nd; Brorson, K.A.; Khan, M.; Read, E.K. 2nd; Brorson, K.A.; Khan, M.; Read, E.K. Bioreactor process parameter screening utilizing a Plackett-Burman design for a model monoclonal an-tibody. J. Pharm. Sci. 2015, 104, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Ažman, K.; Kocijan, J. Application of Gaussian processes for black-box modelling of biosystems. ISA Trans. 2007, 46, 443–457. [Google Scholar] [CrossRef]
- FDA. FDA Briefing Document Oncologic Drugs Advisory Committee Meeting. 2017. Available online: https://www.fda.gov/media/106566/download (accessed on 22 December 2020).





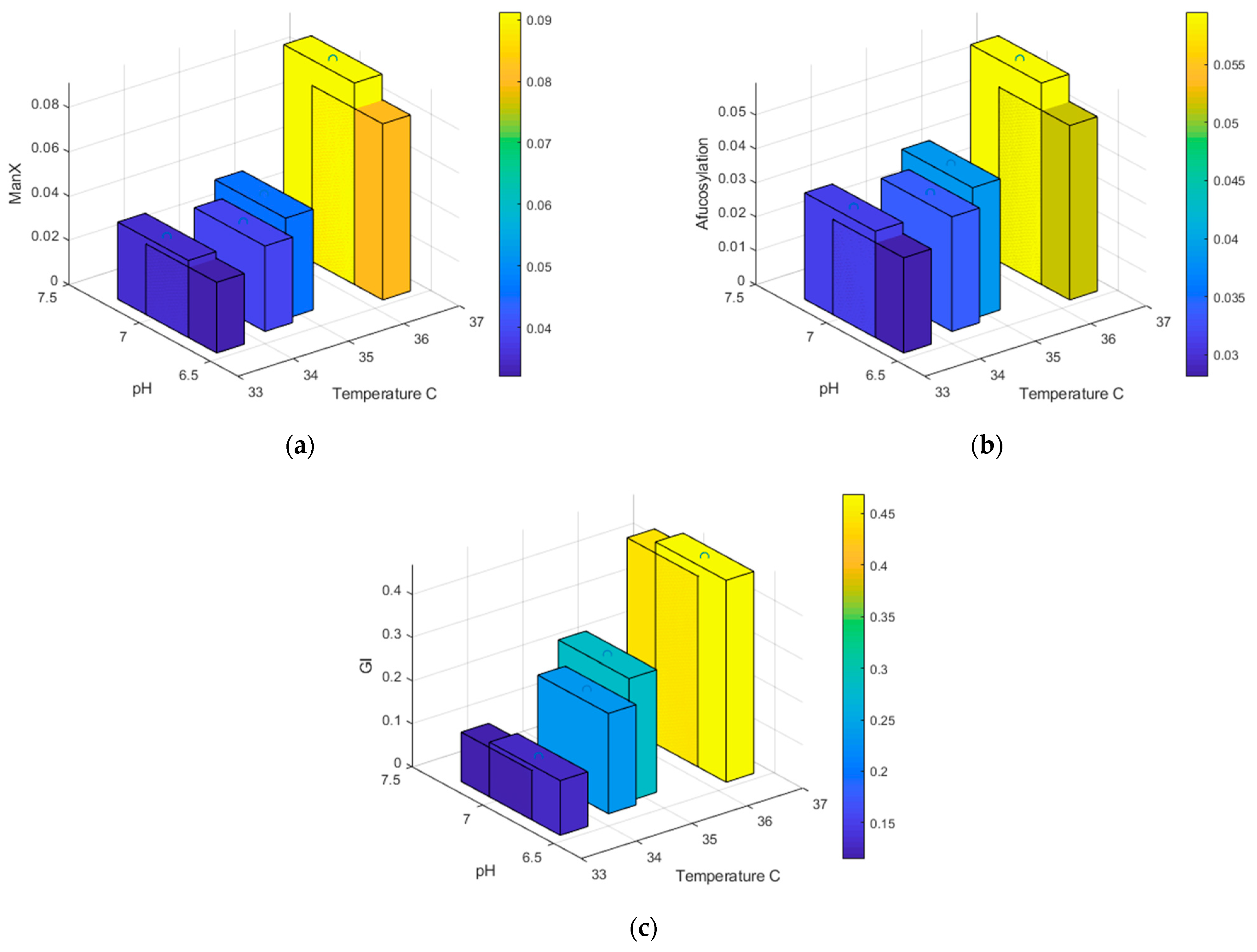
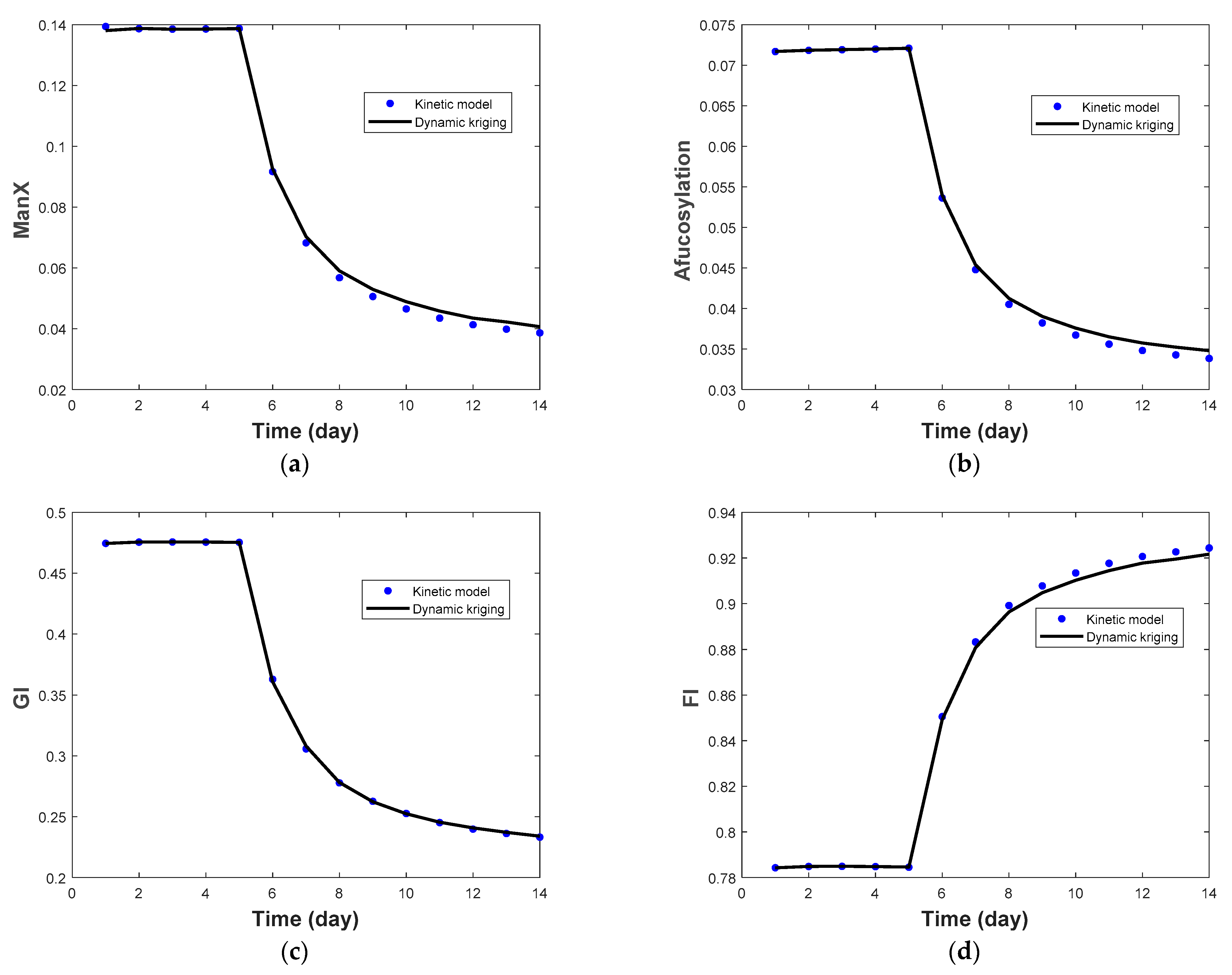

| Protein | pH Range | qp | Glycan Fraction | Ref |
|---|---|---|---|---|
| mAb | pH 7.15–6.70. | Reduced: G0F, G0, Man5 Increased: G1F, G2F, G2FS1 | [16] | |
| mAb | pH 7.2–6.9 | - | Reduced: G0 | [37] |
| mAb | pH 6.9–6.7 | Increased: G1F+G2F, Man5, galactosylation Reduced: Sialylation | [36] | |
| pH 6.9–7.3 |
| Conditions | Titer | G0 | G0F | G1/G2 | G1F/G2F | G2F1S |
|---|---|---|---|---|---|---|
| pH shifted down | ||||||
| Temperature shifted down |
| Glycan Fractions | Man5 | G0 | G1 | G1F | G2F | G2FS1 |
|---|---|---|---|---|---|---|
| Day 5 | 0.029 | 0.004 | 0.027 | 0.088 | 0.073 | 0.088 |
| Day 1 | 0.018 | 0.004 | 0.017 | 0.072 | 0.044 | 0.0510 |
| Glycan Index | Afucoyslation | ManX | GI | FI |
|---|---|---|---|---|
| MRSE | 0.0121 | 0.0186 | 0.0025 | 0.0026 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, O.; Ierapetritou, M. mAb Production Modeling and Design Space Evaluation Including Glycosylation Process. Processes 2021, 9, 324. https://doi.org/10.3390/pr9020324
Yang O, Ierapetritou M. mAb Production Modeling and Design Space Evaluation Including Glycosylation Process. Processes. 2021; 9(2):324. https://doi.org/10.3390/pr9020324
Chicago/Turabian StyleYang, Ou, and Marianthi Ierapetritou. 2021. "mAb Production Modeling and Design Space Evaluation Including Glycosylation Process" Processes 9, no. 2: 324. https://doi.org/10.3390/pr9020324
APA StyleYang, O., & Ierapetritou, M. (2021). mAb Production Modeling and Design Space Evaluation Including Glycosylation Process. Processes, 9(2), 324. https://doi.org/10.3390/pr9020324