Advanced Kinetic Modeling of Bio-co-polymer Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Production Using Fructose and Propionate as Carbon Sources
Abstract
:1. Introduction
2. Experimental Methods
2.1. Mircoorganism and Cultivation Conditions
2.2. Determination of Total Biomass
2.3. Enzyme Assay
High-Pressure Liquid Chromatography
3. Kinetic Modeling Approach
- A simple mass-action kinetic is assumed for the dynamics of the substrates;
- Propionate has an inhibiting effect and decelerates the growth of bacteria [33];
- The conversion of PHA into enzymatically active biomass (residual) is not affected by external propionic acid concentrations, as this is an internal process;
3.1. Numerical Solution
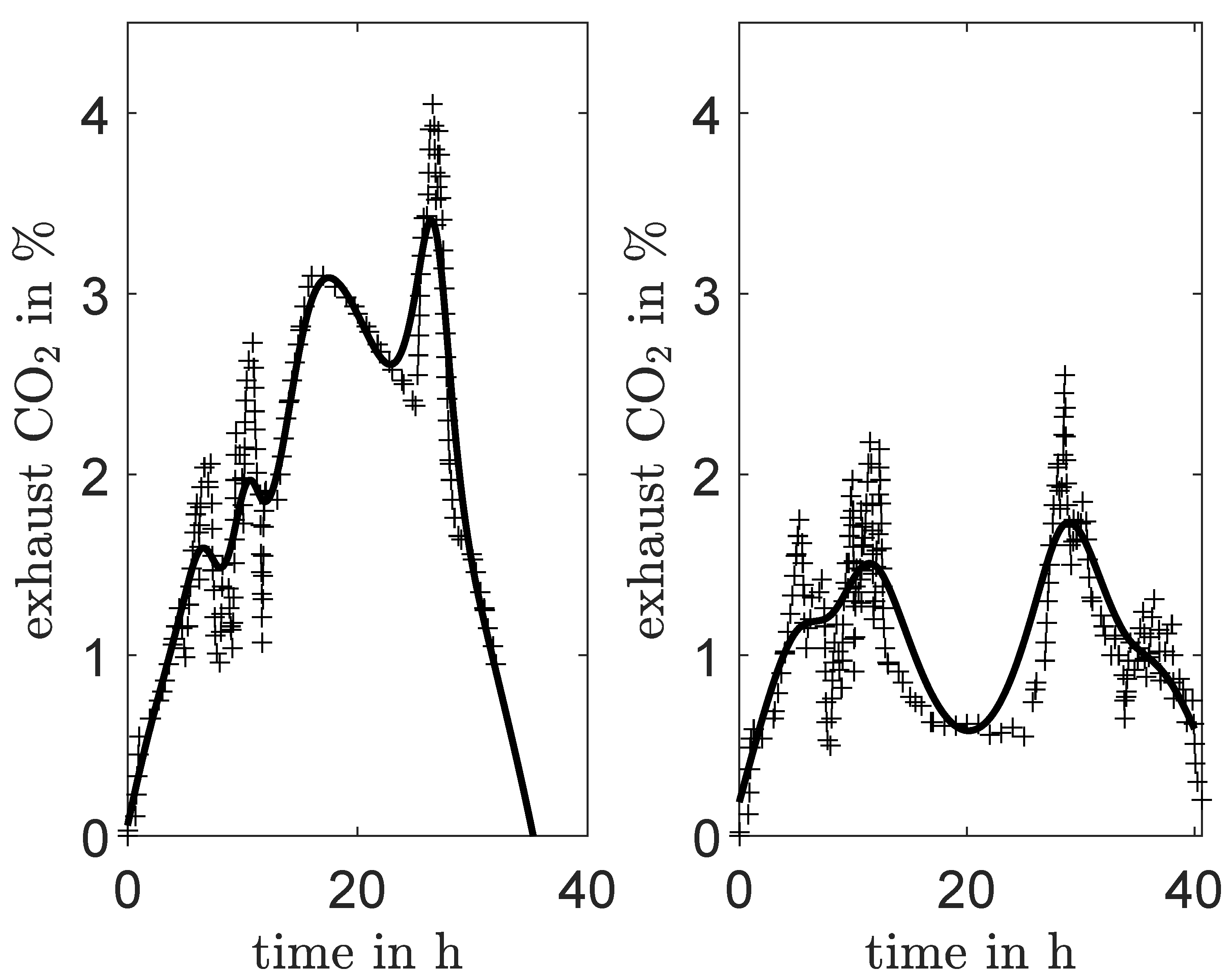
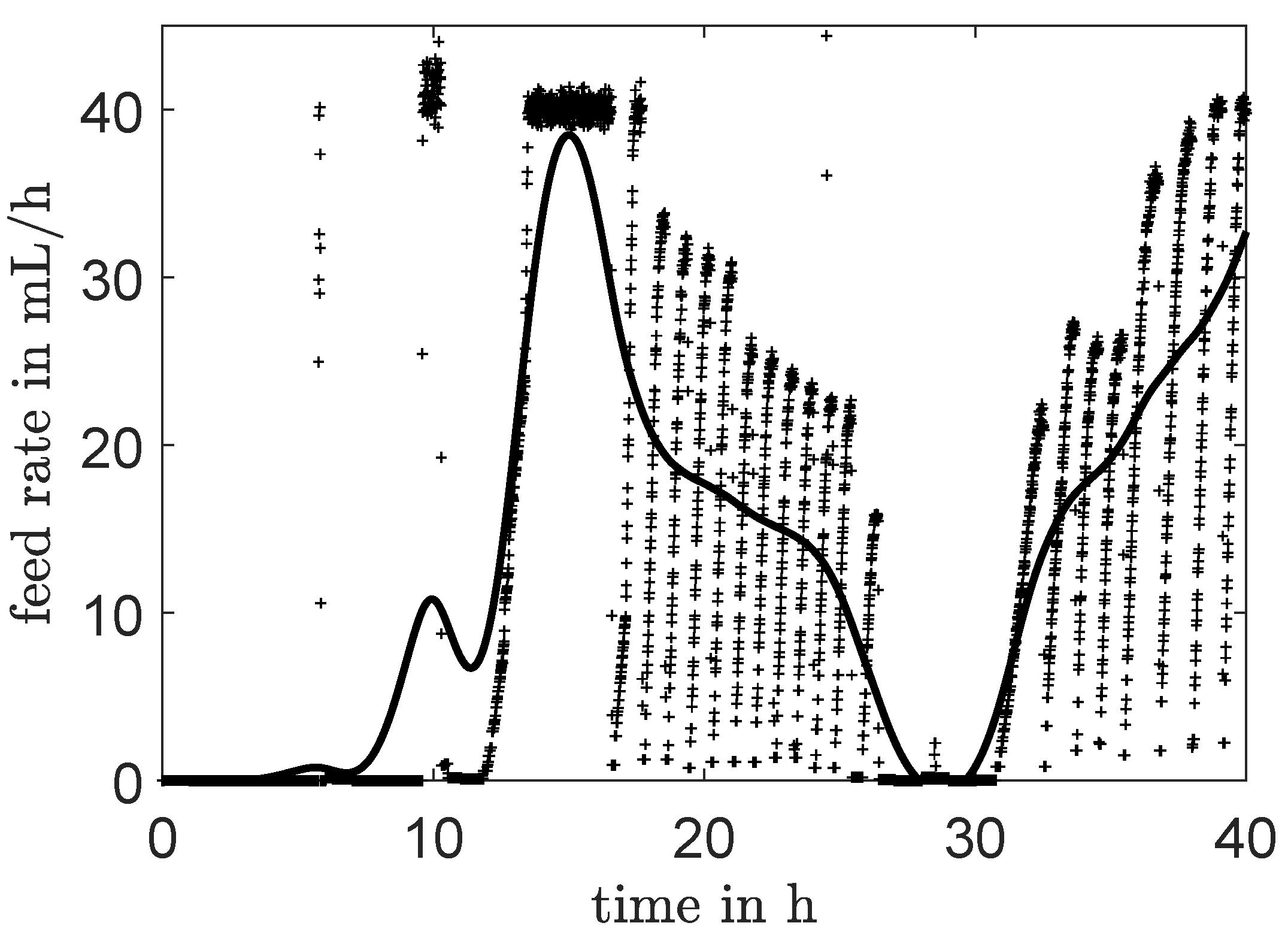
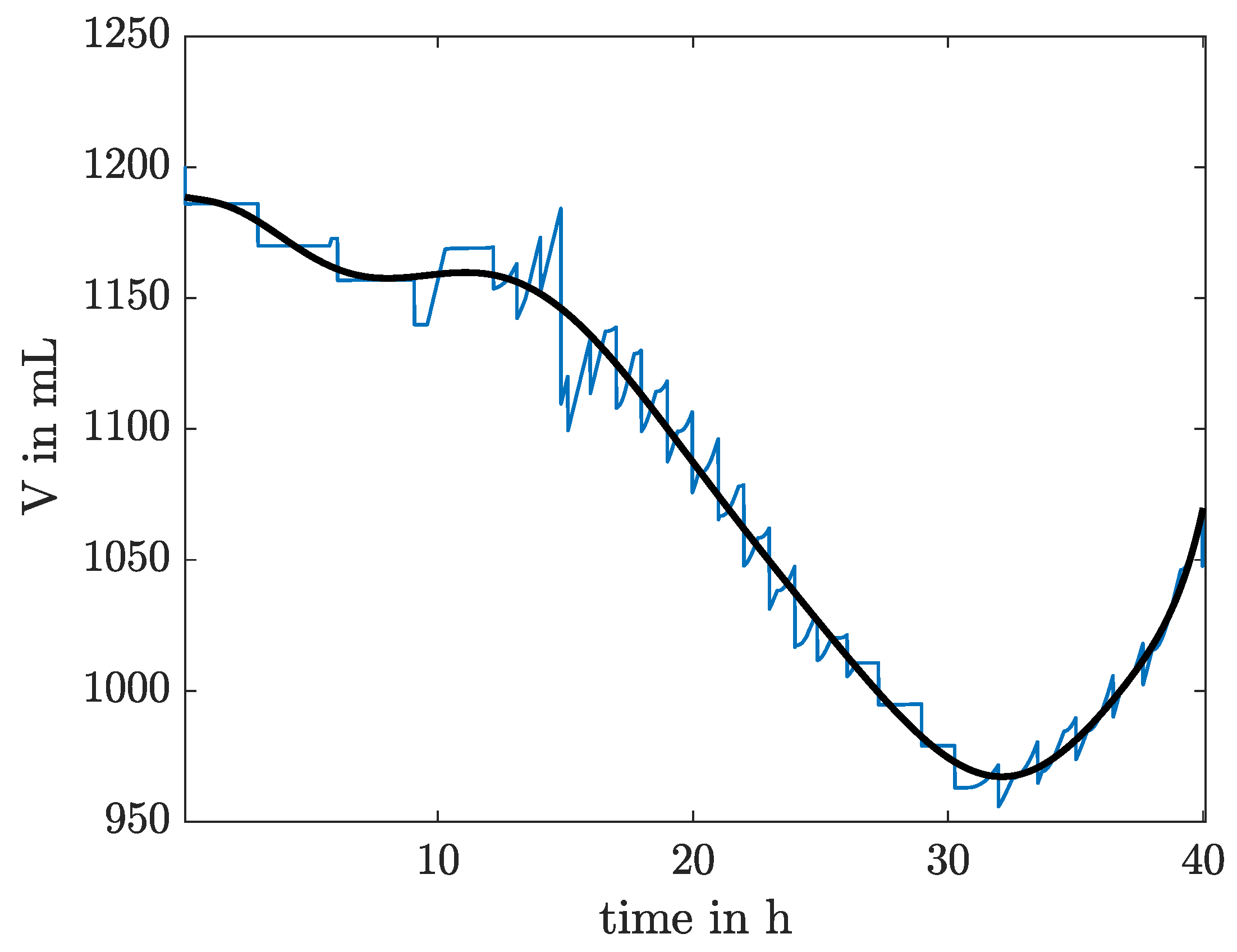
3.1.1. Interpolation for Volume, CO and Feed Rate
3.1.2. Parameter Identification
4. Results
4.1. Identification Using Different Data Sets
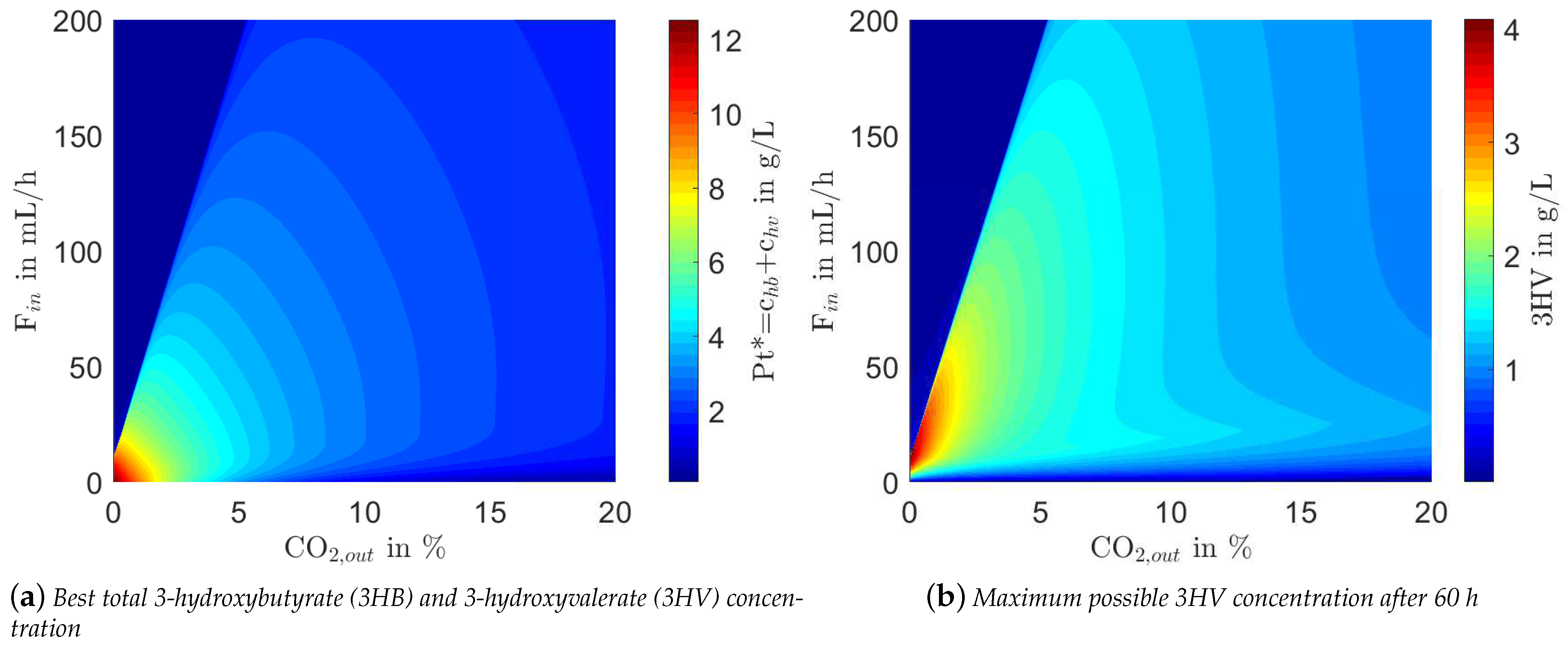
4.2. Computational Study
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Additional Figures—Profile Study


Appendix B. Parameter Identifiability
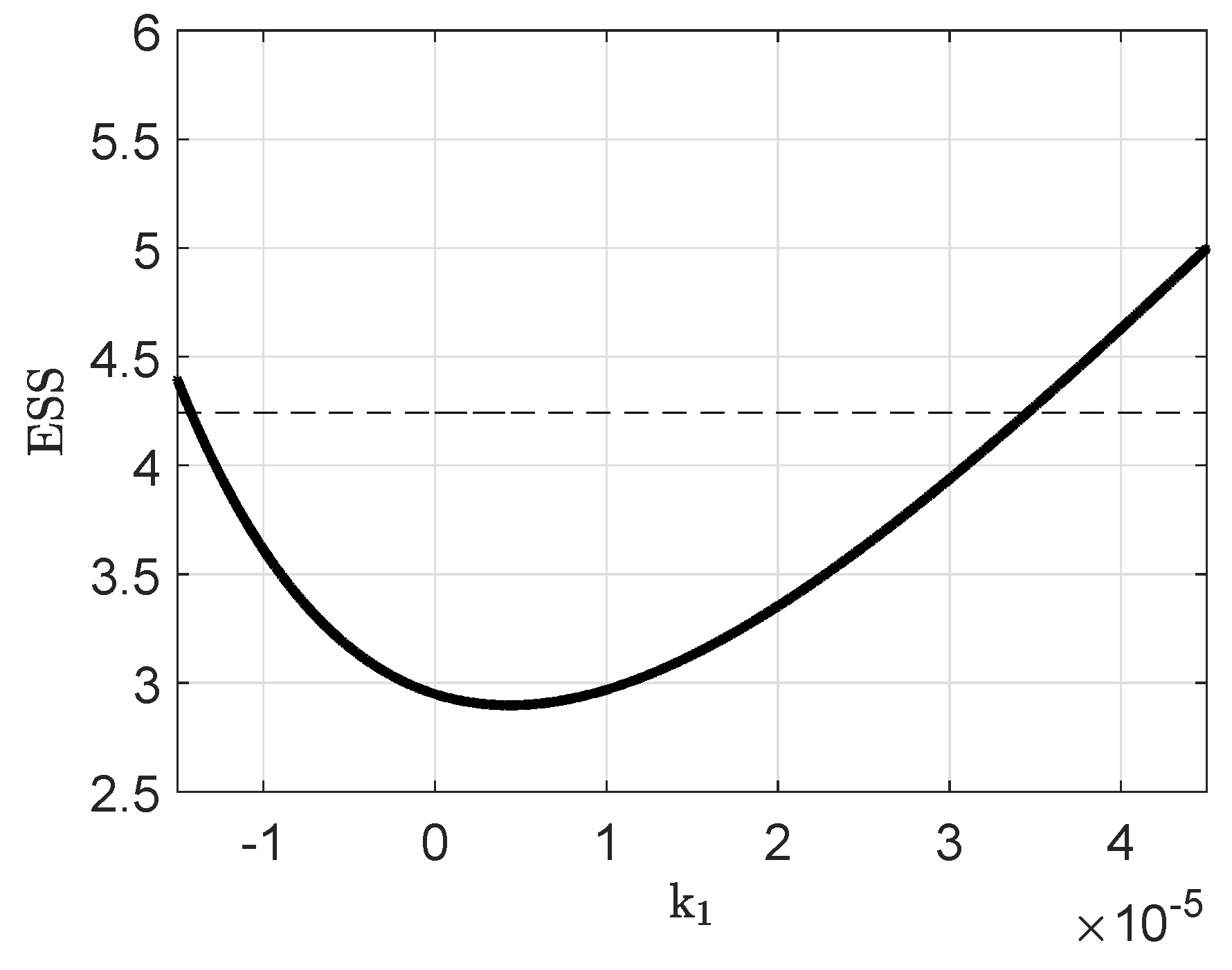

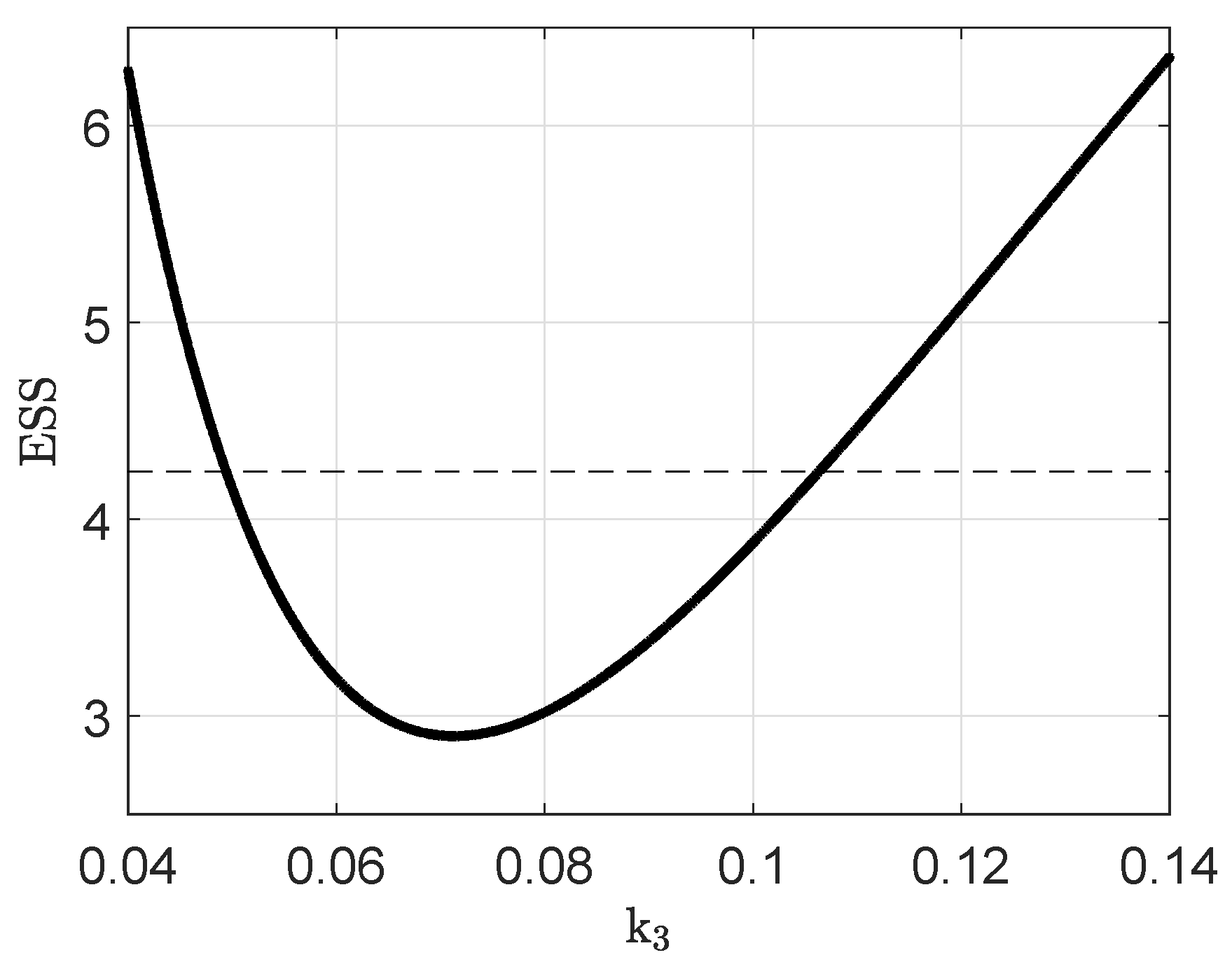

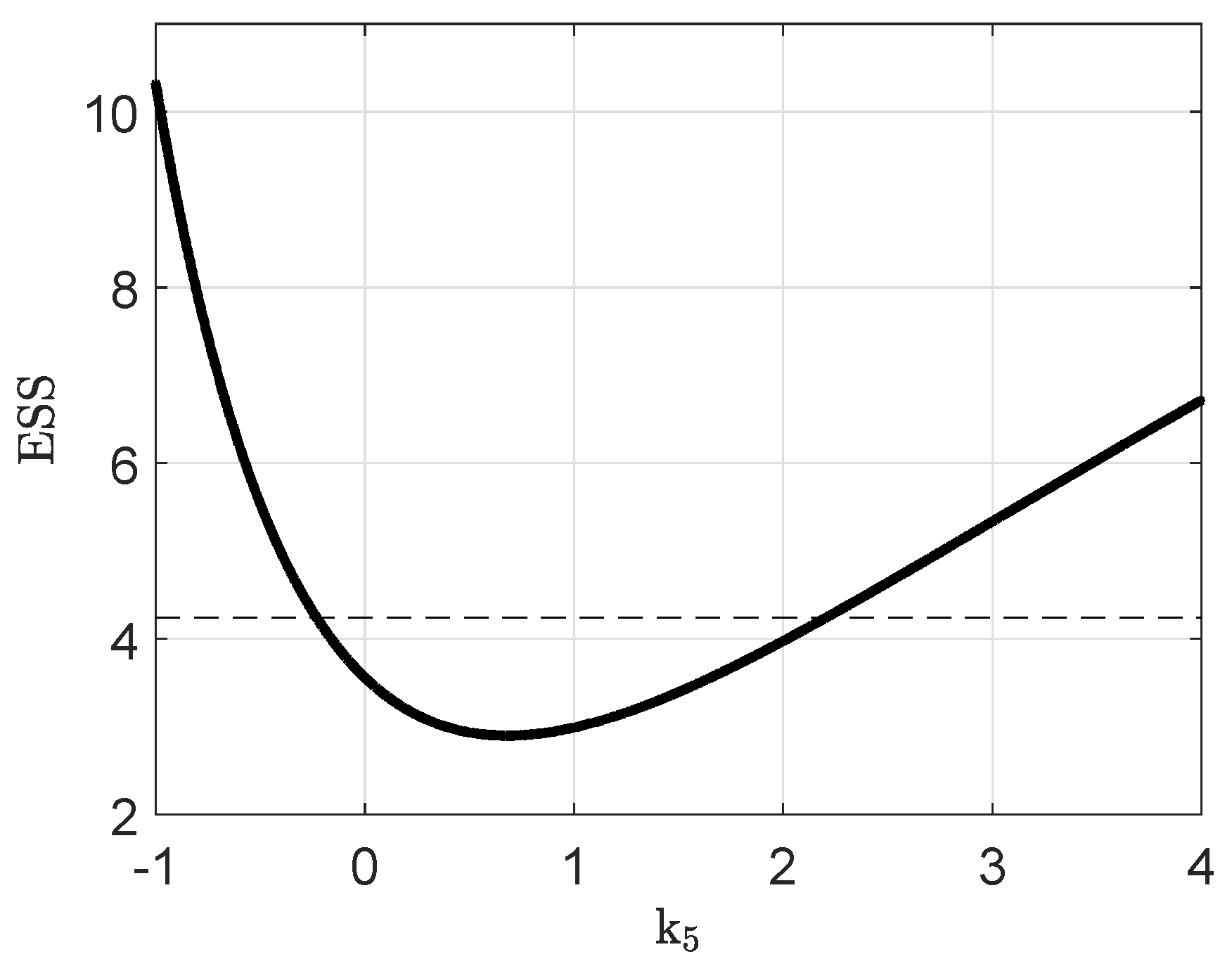

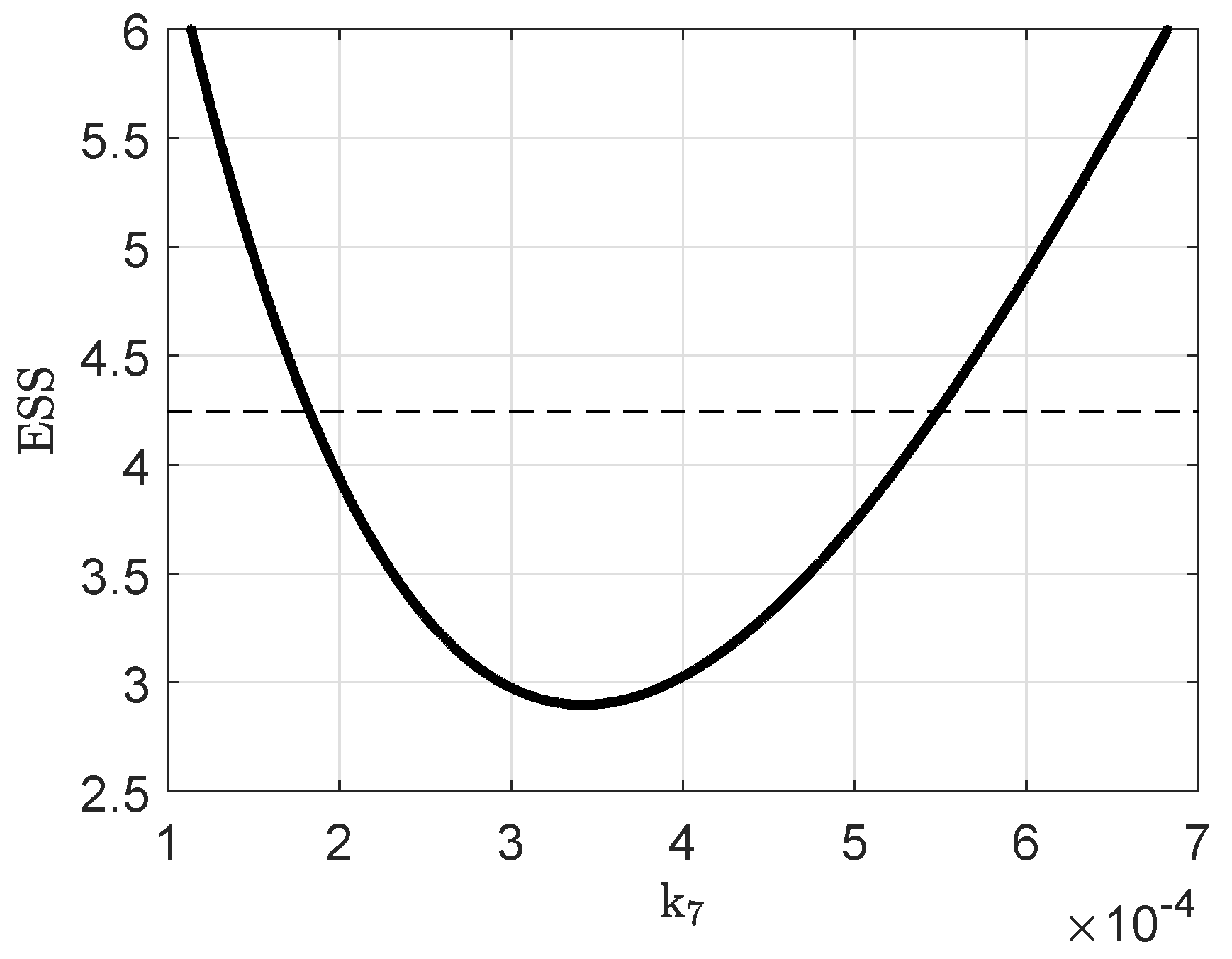

Appendix C. Parameter Values and Initial Conditions
| Parameter | Unit | Description | Value |
|---|---|---|---|
| fitted | |||
| k | (L/(gh)) | consumption of fructose | |
| and ammonium for growth | 4.34 × 10 | ||
| k | (L/(gh)) | consumption of propionic acid | |
| and ammonium for growth | 0.0048 | ||
| k | (L/(gh)) | HA consumption | 0.0713 |
| k | (L/(g h)) | consumption of fructose | |
| for 3HB accumulation | 0.0563 | ||
| k | (L/(g h)) | consumption of propionic acid | |
| for 3HB accumulation | 0.6803 | ||
| k | (L/(g h)) | consumption of propionic acid | |
| for 3HV accumulation | 2.0208 | ||
| k | (L/(gh)) | fructose consumption | |
| for maintenance | 3.4184 × 10 | ||
| k | (L/(gh)) | propionic acid consumption | |
| for maintenance | 0.0125 | ||
| fixed | |||
| c | (g/L) | inhibitory propionic acid concentration | 1.5 [33] |
| c | (g/L) | Michalis–Menten rate for ammonium | 0.2 |
| c | (g/L) | propionic acid concentration in the feed | 20 |
| c(0) | (g/L) | initial fructose concentration | |
| data set 1/data set 2 | 21.96/21.75 | ||
| c(0) | (g/L) | initial propionic acid concentration | |
| data set 1/data set 2 | 0/0.48 | ||
| c(0) | (g/L) | initial ammonium concentration | |
| data set 1/data set 2 | 1.74/1.40 | ||
| c(0) | (g/L) | initial residual biomass concentration | |
| data set 1/data set 2 | 1.47/1.16 | ||
| c(0) | (g/L) | initial 3HB concentration | |
| data set 1/data set 2 | 0.03/0.03 | ||
| c(0) | (g/L) | initial 3HV concentration | |
| data set 1/data set 2 | 0/0.01 | ||
| P | (g/L) | HA (3HB+3HV) concentration | time dependent |
| CO | (%) | exhaust CO | measurement |
| CO | (%) | inlet CO | measurement |
| (-) | CO dependent metabolic activity | time dependent | |
| D | (-) | dilution rate | time dependent |
| F | (%) | feed rate for propionic acid | pH controlled [33] |
| P | (g/L) | maximum concentration of PHA | 0.89 [37] |
References
- Tan, G.Y.A.; Chen, C.L.; Li, L.; Ge, L.; Wang, L.; Razaad, I.M.N.; Li, Y.; Zhao, L.; Mo, Y.; Wang, J.Y. Start a research on biopolymer polyhydroxyalkanoate (PHA): A review. Polymers 2014, 6, 706–754. [Google Scholar] [CrossRef] [Green Version]
- Koller, M. A review on established and emerging fermentation schemes for microbial production of polyhydroxyalkanoate (PHA) biopolyesters. Fermentation 2018, 4, 30. [Google Scholar] [CrossRef] [Green Version]
- Bugnicourt, E.; Cinelli, P.; Lazzeri, A.; Alvarez, V. Polyhydroxyalkanoate (PHA): Review of synthesis, characteristics, processing and potential applications in packaging. Express Polym. Lett. 2014, 8, 791–808. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, C.; Rahman, A.; Rehman, A.U.; Walsh, M.K.; Miller, C.D. Minireview Food waste conversion to microbial polyhydroxyalkanoates. Microb. Biotechnol. 2017, 10, 1338–1352. [Google Scholar] [CrossRef] [PubMed]
- Koller, M.; Atli, A.; Dias, M.; Reiterer, A.; Braunegg, G. Plastics from Bacteria—Microbial PHA Production from Waste Raw Materials; Springer: Berlin/Heidelberg, Germany, 2010; Volume 14, pp. 85–119. [Google Scholar] [CrossRef]
- Riedel, S.L.; Jahns, S.; Koenig, S.; Bock, M.C.E.; Brigham, C.J.; Bader, J.; Stahl, U. Polyhydroxyalkanoates production with Ralstonia eutropha from low quality waste animal fats. J. Biotechnol. 2015, 214, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Brigham, C.J.; Riedel, S.L. The potential of polyhydroxyalkanoate production from food wastes. Appl. Food Biotechnol. 2019, 6, 7–18. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, L.; De Wever, H. Acetic acid as an indirect sink of CO2 for the synthesis of polyhydroxyalkanoates (PHA): Comparison with PHA production processes directly using CO2 as feedstock. Appl. Sci. 2018, 8, 1416. [Google Scholar] [CrossRef] [Green Version]
- Jia, Q.; Xiong, H.; Wang, H.; Shi, H.; Sheng, X.; Sun, R.; Chen, G. Production of polyhydroxyalkanoates (PHA) by bacterial consortium from excess sludge fermentation liquid at laboratory and pilot scales. Bioresour. Technol. 2014, 171, 159–167. [Google Scholar] [CrossRef]
- Bengtsson, S.; Karlsson, A.; Alexandersson, T.; Quadri, L.; Hjort, M.; Johansson, P.; Morgan-Sagastume, F.; Anterrieu, S.; Arcos-Hernandez, M.; Karabegovic, L.; et al. A process for polyhydroxyalkanoate (PHA) production from municipal wastewater treatment with biological carbon and nitrogen removal demonstrated at pilot-scale. New Biotechnol. 2017, 35, 42–53. [Google Scholar] [CrossRef]
- Sabapathy, P.C.; Devaraj, S.; Meixner, K.; Anburajan, P.; Kathirvel, P.; Ravikumar, Y.; Zabed, H.M.; Qi, X. Recent developments in Polyhydroxyalkanoates (PHAs) production—A review. Bioresour. Technol. 2020, 306, 123–132. [Google Scholar] [CrossRef]
- Meereboer, K.W.; Misra, M.; Mohanty, A.K. Review of recent advances in the biodegradability of polyhydroxyalkanoate (PHA) bioplastics and their composites. Green Chem. 2020, 22, 5519–5558. [Google Scholar] [CrossRef]
- Gholami, A.; Mohkam, M.; Rasoul-Amini, S.; Younes, G. Industrial production of polyhydroxyalkanoates by bacteria: Opportunities and challenges. Minerva Biotecnol. 2016, 28, 59–74. [Google Scholar]
- Atli, A.; Koller, M.; Scherzer, D. Continuous production of poly ([R]-3-hydroxybutyrate) by Cupriavidus necator in a multistage bioreactor cascade. Biotechnol. Prod. Process. Eng. 2011, 91, 295–304. [Google Scholar] [CrossRef]
- Penloglou, G.; Vasileiadou, A.; Chatzidoukas, C.; Kiparissides, C. Model-based intensification of a fed-batch microbial process for the maximization of polyhydroxybutyrate (PHB) production rate. Bioprocess Biosyst. Eng. 2017, 40, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Carius, L.; Pohlodek, J.; Morabito, B.; Franz, A.; Mangold, M.; Findeisen, R.; Kienle, A. Model-based State Estimation Based on Hybrid Cybernetic Models. IFAC-PapersOnLine 2018, 51, 197–202. [Google Scholar] [CrossRef]
- Morabito, B.; Kienle, A.; Findeisen, R.; Carius, L. Multi-mode Model Predictive Control and Estimation for Uncertain Biotechnological Processes. IFAC-PapersOnLine 2019, 52, 709–714. [Google Scholar] [CrossRef]
- Novak, M.; Koller, M.; Braunegg, G.; Horvat, P. Mathematical Modelling as a Tool for Optimized PHA Production. Chem. Biochem. Eng. 2015, 29, 183–220. [Google Scholar] [CrossRef]
- Špoljaric’, I.V.; Lopar, M.; Koller, M.; Muhr, A.; Salerno, A.; Reiterer, A.; Malli, K.; Angerer, H.; Strohmeier, K.; Schober, S.; et al. Mathematical modeling of poly [(R)-3-hydroxyalkanoate] synthesis by Cupriavidus necator DSM 545 on substrates stemming from biodiesel production. Bioresour. Technol. 2013, 133, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.M.; Serafim, L.S.; Lemos, P.C.; Reis, M.A.; Oliveira, R. Mathematical modelling of a mixed culture cultivation process for the production of polyhydroxybutyrate. Biotechnol. Bioeng. 2005, 92, 209–222. [Google Scholar] [CrossRef]
- Dias, J.M.; Oehmen, A.; Serafim, L.S.; Lemos, P.C.; Reis, M.A.; Oliveira, R. Metabolic modelling of polyhydroxyalkanoate copolymers production by mixed microbial cultures. BMC Syst. Biol. 2008, 2, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Duvigneau, S.; Dürr, R.; Carius, L.; Kienle, A. Hybrid Cybernetic Modeling of Polyhydroxyalkanoate Production in Cupriavidus necator using Fructose and Acetate as Substrates. IFAC-PapersOnLine 2020, 53, 16872–16877. [Google Scholar] [CrossRef]
- Franz, A.; Song, H.S.; Ramkrishna, D.; Kienle, A. Experimental and theoretical analysis of poly(beta-hydroxybutyrate) formation and consumption in Ralstonia eutropha. Biochem. Eng. J. 2011, 55, 49–58. [Google Scholar] [CrossRef]
- Katoh, T.; Yuguchi, D.; Yoshii, H.; Shi, H.D.; Shimizu, K. Dynamics and modeling on fermentative production of poly(β-hydroxybutyric acid) from sugars via lactate by a mixed culture of Lactobacillus delbrueckii and Alcaligenes eutrophus. J. Biotechnol. 1999, 67, 113–134. [Google Scholar] [CrossRef]
- Duvigneau, S.; Dürr, R.; Kranert, L.; Wilisch-Neumann, A.; Carius, L.; Findeisen, R.; Kienle, A. Hybrid Cybernetic Modeling of the Microbial Production of Polyhydroxyalkanoates Using Two Carbon Sources. Comput. Aided Chem. Eng. 2021, 50, 1969–1974. [Google Scholar] [CrossRef]
- Franz, A.; Dürr, R.; Kienle, A. Population balance modeling of biopolymer production in cellular systems. IFAC Proc. Vol. 2014, 47, 1705–1710. [Google Scholar] [CrossRef] [Green Version]
- Dürr, R.; Franz, A.; Kienle, A. Combination of limited measurement information and multidimensional population balance models. IFAC-PapersOnLine 2015, 48, 261–266. [Google Scholar] [CrossRef]
- Strong, P.; Laycock, B.; Mahamud, S.; Jensen, P.; Lant, P.; Tyson, G.; Pratt, S. The Opportunity for High-Performance Biomaterials from Methane. Microorganisms 2016, 4, 11. [Google Scholar] [CrossRef]
- Jiang, Y.; Hebly, M.; Kleerebezem, R.; Muyzer, G.; van Loosdrecht, M.C. Metabolic modeling of mixed substrate uptake for polyhydroxyalkanoate (PHA) production. Water Res. 2011, 45, 1309–1321. [Google Scholar] [CrossRef]
- García, C.; Alcaraz, W.; Acosta-Cárdenas, A.; Ochoa, S. Application of process system engineering tools to the fed-batch production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from a vinasses–molasses Mixture. Bioprocess Biosyst. Eng. 2019, 42, 1023–1037. [Google Scholar] [CrossRef]
- Ochoa, S.; García, C.; Alcaraz, W. Real-time optimization and control for polyhydroxybutyrate fed-batch production at pilot plant scale. J. Chem. Technol. Biotechnol. 2020, 95, 3221–3231. [Google Scholar] [CrossRef]
- Dürr, R.; Duvigneau, S.; Kienle, A. Microbial Production of Polyhydroxyalkanoates—Modeling of Chain Length Distribution. Comput. Aided Chem. Eng. 2021, 50, 1975–1981. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, B.G.; Choi, C.Y. Effect of propionic acid on poly (β-hydroxybutyric-co-β-hydroxyvaleric) acid production by Alcaligenes eutrophus. Biotechnol. Lett. 1992, 14, 903–906. [Google Scholar] [CrossRef]
- Duvigneau, S.; Kettner, A.; Carius, L.; Griehl, C.; Findeisen, R.; Kienle, A. Fast, inexpensive, and reliable HPLC method to determine monomer fractions in poly (3-hydroxybutyrate-co-3-hydroxyvalerate). Appl. Microbiol. Biotechnol. 2021, 105, 4743–4749. [Google Scholar] [CrossRef] [PubMed]
- Satoh, H.; Sakamoto, T.; Kuroki, Y.; Kudo, Y.; Mino, T. Application of the Alkaline-Digestion-HPLC Method to the Rapid Determination of Polyhydroxyalkanoate in Activated Sludge. J. Water Environ. Technol. 2016, 14, 411–421. [Google Scholar] [CrossRef] [Green Version]
- Koller, M.; Horvat, P.; Hesse, P.; Bona, R.; Kutschera, C.; Atlić, A.; Braunegg, G. Assessment of formal and low structured kinetic modeling of polyhydroxyalkanoate synthesis from complex substrates. Bioprocess Biosyst. Eng. 2006, 29, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Mohidin Batcha, A.F.; Prasad, D.M.; Khan, M.R.; Abdullah, H. Biosynthesis of poly(3-hydroxybutyrate) (PHB) by Cupriavidus necator H16 from jatropha oil as carbon source. Bioprocess Biosyst. Eng. 2014, 37, 943–951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storn, R.; Price, K. Differential Evolution—A Simple and Efficient Heuristic for global Optimization over Continuous Spaces. J. Glob. Optim. 1997, 11, 341–359. [Google Scholar] [CrossRef]
- Raue, A.; Kreutz, C.; Maiwald, T.; Bachmann, J.; Schilling, M.; Klingmüller, U.; Timmer, J. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics 2009, 25, 1923–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, L.; Jiang, M.; Ryu, C.H.; Chang, H.N.; Cho, S.H.; Lee, J.W. Inhibitory effect of carbon dioxide on the fed-batch culture of Ralstonia eutropha: Evaluation by CO2 pulse injection and autogenous CO2 methods. Biotechnol. Bioeng. 2003, 83, 312–320. [Google Scholar] [CrossRef]
- Park, J.M.; Kim, T.Y.; Lee, S.Y. Genome-scale reconstruction and in silico analysis of the Ralstonia eutropha H16 for polyhydroxyalkanoate synthesis, lithoautotrophic growth, and 2-methyl citric acid production. BMC Syst. Biol. 2011, 5, 101. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duvigneau, S.; Dürr, R.; Behrens, J.; Kienle, A. Advanced Kinetic Modeling of Bio-co-polymer Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Production Using Fructose and Propionate as Carbon Sources. Processes 2021, 9, 1260. https://doi.org/10.3390/pr9081260
Duvigneau S, Dürr R, Behrens J, Kienle A. Advanced Kinetic Modeling of Bio-co-polymer Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Production Using Fructose and Propionate as Carbon Sources. Processes. 2021; 9(8):1260. https://doi.org/10.3390/pr9081260
Chicago/Turabian StyleDuvigneau, Stefanie, Robert Dürr, Jessica Behrens, and Achim Kienle. 2021. "Advanced Kinetic Modeling of Bio-co-polymer Poly(3-hydroxybutyrate-co-3-hydroxyvalerate) Production Using Fructose and Propionate as Carbon Sources" Processes 9, no. 8: 1260. https://doi.org/10.3390/pr9081260







