Abstract
Two copper(II) complexes (Cu-L1, Cu-L2) derived from 2,3-substituted quinazolinone Schiff base ligands (L1, L2) were prepared to examine their anticancer activity. Compounds were characterized using various spectroscopic methods (FTIR, NMR, UV-vis) and quantum-chemical calculations. The biological effects of Cu(II) complexes bearing quinazolinone scaffolds were evaluated on two cancers’ cell lines (breast—MCF-7 and lung—A549), as well as on untransformed cells (keratinocytes—HaCaT). Copper complexes were highly cytotoxic, with IC50 in the low micromolar range, while the quinazoline ligands L1 and L2 remained inactive in inhibiting cell proliferation. Antioxidant activity was investigated in the model systems using DPPH and FRAP assays. The Cu-L1 and Cu-L2 complexes exhibited enhanced DPPH free radical scavenging efficiency compared to the L1 and L2 ligands, but their reducing ability was comparable to that of the free ligands. Evaluation of oxidative stress in vitro carried out by staining cells with various ROS-specific indicators showed reduced production of superoxide anion radical and hydrogen peroxide after treatment of cells with copper complexes. Such a negative impact on ROS formation in cells can lead to cellular redox imbalance and consequent cell death, among others, by inducing apoptosis and/or necrosis, depending on the copper complex used. We hypothesize that the high cytotoxic activity of the investigated copper complexes is apparently the result of multiple mechanisms of action, and the imbalance in the cellular antioxidant system partly contributes to the overall cytotoxic effect.
1. Introduction
Despite all recent advances in the treatment of various types of malignancies, cancer is still one of the leading causes of death in the world today [1]. The treatment options for cancer include mainly chemotherapy and radiotherapy. Although most anticancer drugs on the market are predominantly organic compounds, metal complexes have recently received considerable attention in anticancer drug research. The discovery of cis-platinum in the 1960s by Rosenberg et al. [2] was a major challenge that opened up new possibilities in the field of metal-based anticancer drugs. The platinum compounds are currently used in almost 50% of all cancer treatments. However, despite their widespread use, high toxicity and severe side effects are often observed [3,4,5]. Therefore, ongoing efforts are focused on metal-based compounds with lower toxicity and different mechanisms of action. The exceptional properties of Cu(II) complexes, including redox activity, multiple coordination modes, and reactivity, have initiated a number of studies on their therapeutic potential. A broad range of metal complexes have been investigated for their anticancer activity in order to develop new effective therapeutics with the ability to overcome the limitations of platinum complexes [6,7,8].
Among different transition metal complexes, copper complexes have attracted particular interest. Copper is an essential redox-active trace element that plays an important role in human physiological and pathological processes, and therefore, its homeostasis is tightly regulated. Copper can act as both an antioxidant and a pro-oxidant [9,10,11]. The copper-dependent metalloenzymes, such as Cu-Zn superoxide dismutase (SOD), reduce ROS production [12]. However, copper ions can also interact with molecular oxygen and generate ROS in undesirable Fenton-like reactions that can cause oxidative damage to DNA and other biomolecules [13,14,15]. Many metal complexes readily undergo redox reactions resulting in the generation of ROS as products of biotransformation. Increased intracellular ROS production can initiate selective cytotoxicity against different tumor cell lines, leading to their death. Therefore, the antiproliferative activity of several Cu(II) complexes is often linked to the redox chemistry of copper [16,17,18,19,20].
The special properties of copper allow it to be incorporated into a variety of organic and inorganic compounds. In the last two decades, a number of copper complexes with different ligands have been synthesized and characterized. It has been found that coordination with copper can cause significant changes in the biological properties of the ligands (e.g., improved specificity, greater control over drug release) [21,22,23].
Thus, among Cu(II) complexes, some serve as potent proteasome inhibitors and inducers of apoptosis, especially in cancer cells [24,25,26]. Multiple mechanistic studies demonstrated the anticancer effects of Cu(II) complexes bearing quinazolinone scaffolds. It was found that they are able to suppress proliferation of cancer cells mainly by generating ROS or inducing cell cycle arrest, leading to apoptosis. However, when redox activity is prevented, then the metal complex may act as a DNA intercalator [27,28,29,30,31]. Since the cytotoxicity of transition-metal complexes appears to be the result of multiple mechanisms, copper complexes with various heterocyclic Schiff base ligands have been studied. The study of copper–quinazolinone complexes is therefore of great interest in view of their capability to act as anticancer agents and diagnostic tools.
Considering the diverse applications of quinazolinones and their transition metal complexes in medicine, as well as our previous studies on different quinazolinone derivatives [32,33], this work is focused on the investigation of the anticancer activity of two new Cu(II) complexes based on quinazolinone Schiff base ligands. Both the ligands and the complexes were examined for their in vitro cytotoxic activity against the human breast (MCF-7) and lung (A549) cancer cell lines to determine their biological efficacy. In addition, the ligands and complexes were screened for their antioxidant behavior and radical scavenging activity in the model systems. The effect of Cu(II) complexes on intracellular ROS generation in cancer cells and the mechanism of cell death were investigated.
2. Results and Discussion
2.1. Synthesis and Characterization of Cu(II) Complexes
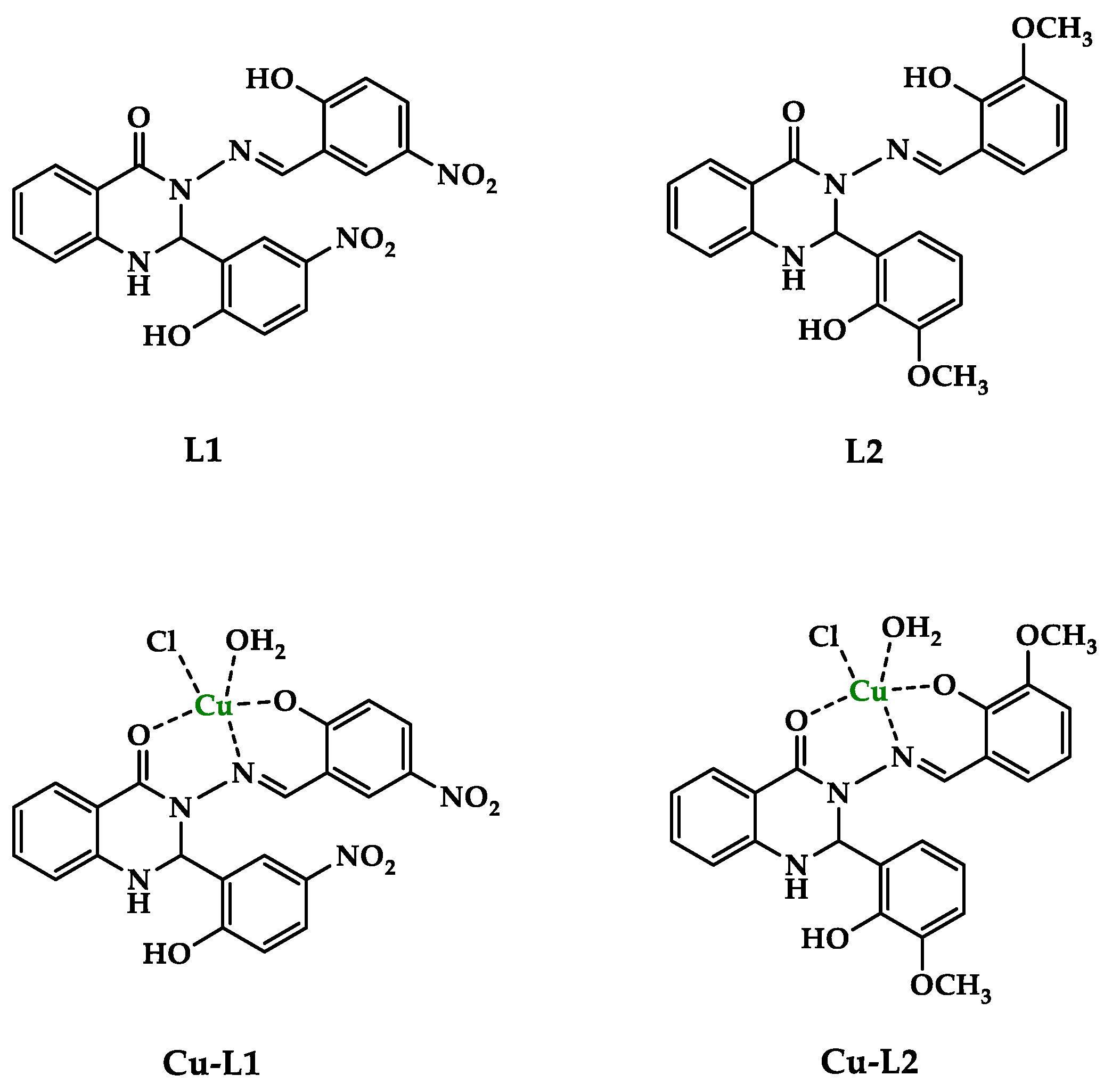
The 2,3-substituted quinazolinone Schiff base ligands (L1, L2) were prepared following the method described previously [32]. Analytical data for ligands L1 and L2 were in full agreement with those published [34,35]. The copper complexes Cu-L1 and Cu-L2 were synthesized via a reaction of the corresponding Schiff base ligand and copper chloride (1:1) in methanol under reflux conditions. Cu(II) complexes were obtained as colored solids in good yields (64–65%) and were characterized with analytical and spectroscopic methods. The structures of the ligands and Cu(II) complexes are in Figure 1.
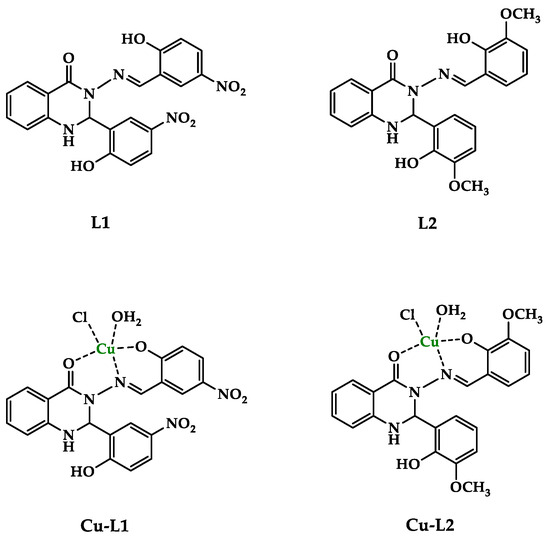
Figure 1.
The proposed structure of the studied compounds.
2.1.1. FTIR Spectroscopy
The infrared spectra of copper complexes Cu-L1 and Cu-L2 were analyzed and compared with the spectra of free ligands L1 and L2. The FTIR spectra of all studied compounds are shown in the supplementary material (Figures S1–S4). The significant decrease in the frequency of the bands corresponding to the azomethine ν(HC=N) and carbonyl group ν(C=O) in the Cu-L1 and Cu-L2 complexes compared to the ligands L1 and L2 indicated the coordination with the copper ion. Absorption bands at 3107 and 3081 cm−1 in the spectrum of free ligand L1 could be attributed to the ν(C-H) stretching and ν(N-H) aromatic vibrations of the quinazoline ring. Bands characteristic for the phenolic ν(O-H) group were observed at 3366 cm−1. A strong absorption peak of carbonyl group ν(C=O) appeared at 1650 cm−1. An absorption band at 1587 cm−1 was assigned to the stretching vibration of the azomethine ν(HC=N) group. The NO2 group stretch in L1 was confirmed by the occurrence of a band at 1334 cm−1. In the spectrum of the copper complex Cu-L1, these peaks were shifted to a lower frequency region. The IR band at 1587 cm−1 was shifted downfield (1575 cm−1), indicating a decrease in the nature of the C=N double bond due to coordination of the azomethine nitrogen with the copper ion. In the Cu-L1 complex, the band at 1650 cm−1 assigned to ν(C=O) shifted to the region with lower energy (1603 cm−1), which can be attributed to the weakening of the carbonyl group due to the coordination of oxygen with the metal center. Also, a negative shift for the (C-NO2) band at 1329 cm−1 was observed for the complex Cu-L1. In the spectrum of ligand L2, the strong absorption band at 1660 cm−1 was assigned to the ν(C=O) on the quinazolinone ring. In the spectrum of the Cu-L2 complex, this band was shifted downfield (1605 cm−1) by 55 cm−1, indicating the coordination of the carbonyl oxygen with copper ion. A strong band at 1608 cm−1 indicated the presence of the azomethine ν(HC=N) group in free L2, which shifted to a lower wave number in the corresponding copper complex (1582 cm−1) and supported the coordination of L2 through azomethine nitrogen. Phenolic hydroxyl groups of both ligands appeared as a broad band at 3584–3521 cm−1. In the spectra of the complexes, this band was not observed, suggesting the participation of corresponding OH groups in coordination via deprotonation, as described earlier for this compound by Gudasi et al. [35]. The tridentate behavior of the Schiff base ligands L1 and L2, coordinating through carbonyl oxygen, azomethine nitrogen, and phenolic oxygen, was proposed from their FTIR spectra.
2.1.2. UV-Vis Spectroscopy Was Proposed
The electronic absorption spectra of the ligands and complexes were recorded in 250–950 nm spectral range in DMSO. The UV-Vis spectra revealed considerable changes between the ligands and the corresponding copper complexes. The spectra of free ligands L1 and L2 and copper complexes Cu-L1 and Cu-L2 are presented in the supplementary material in Figures S5 and S6. UV-Vis of the ligand L1 (Figure S5, blue line) showed a broad absorption band in the region from 270 to 350 nm. This band corresponds to the π-π* transitions due to conjugation of the phenyl ring C with N–N=CH linkage [36]. The strong auxochromic effect of the NO2 group in the meta position causes a marked bathochromic shift towards the 430 nm region. In addition, the absorption band shift is also affected by the auxochromic effect of the ortho-positioned OH group. The absorption in this region can be assigned to n-π* transitions, which are typical of chromophores and auxochromic groups with heteroatoms linked by multiple bonds [37]. The spectrum of the Cu-L1 complex is shown in Figure S5 (orange line). Coordination of Cu(II) with the OH group on the phenyl ring led to decrease in the auxochromic effect due to this group. Consequently, the absorption band shifted to a shorter wavelength region, with a maximum at 383 nm. Moreover, the conjugation of Cu(II) with the heteroatoms of the N–N=CH linkage partially enhanced the hypsochromic shift. In addition, the Cu-L1 copper complex showed a single broad absorption band at 689 nm, which corresponds to the d-d Cu(II) transitions (see Figure S5, inset).
The UV-Vis spectrum of ligand L2 (Figure S6, blue line) was somewhat different compared to L1. Ligand L2 (bearing the OCH3 group) showed two close maxima in the region of about 300–310 nm. These bands can be assigned to π-π* transitions, as for the ligand L1. The second absorption maximum at 368 nm is due to n-π* transitions (also as seen for L1), showing a slight bathochromic shift because of the presence of the auxochromic OH group. In the case of the Cu-L2 complex (Figure S6 orange line), the broad absorption band at 418 nm is the result of changes in the N–N=C chromophore due to coordination of the central Cu(II) via non-covalent interactions with free electron pairs of the O=C–N–N=C(H) moiety. Furthermore, the OCH3 group (uncoordinated with Cu(II)) also caused a minor shift of the absorption maximum towards longer wavelengths [38]. Finally, the absorption band corresponding to the d-d transitions was identified at 670 nm (shoulder, see Figure S6, inset). The effect of Cu(II) was also manifested by the color change of the Cu-L1 and Cu-L2 solutions compared to the solutions of the free ligands L1 and L2.
2.1.3. NMR Spectroscopy
The high-resolution 1H and 13C NMR spectra of L1 in DMSO and its complex with Cu(II) are shown in Figure S7. All resonances of the non-complexed compound could be assigned and agreed with the published data [34]. As Cu(II) is a d9 metal ion with one unpaired electron, Cu(II) is consequently paramagnetic (also having a quadrupole moment), affecting chemical shifts and relaxation properties and having strong influence upon NMR spectra of Cu(II) complexes with organic ligands. NMR spectra have very wide lines, and many times they are even undetectable. The 1H NMR spectra of L1 and its complex with Cu(II) are shown in Figure S7. NMR spectra indicated that the line widths of the non-complexed compound signals increased (there was an 30% excess of the ligand in the NMR sample) and shifted. In addition, some signals with broad linewidths appeared: e.g., one OH signal shifted upfield (to 9.5 ppm); the other one was undetected (involved in the complex). There are some other signals with small intensities but with very broad linewidths which could not be assigned. It should also be noted that, due to the known wide range of 1H chemical shifts of resonances in Cu(II) complexes, the NMR spectra have also been measured with considerably broad spectral widths (up to 100 ppm) without detection of any signals of Cu-L1. Similar observations were also seen for compound Cu-L2 (Figure S8). The signals of the non-complexed ligand showed quite well-resolved signals; the signals of the Cu(II) complex were nearly undetectable—only very few resonances with extremely broad signals were detected.
2.1.4. DFT Calculations
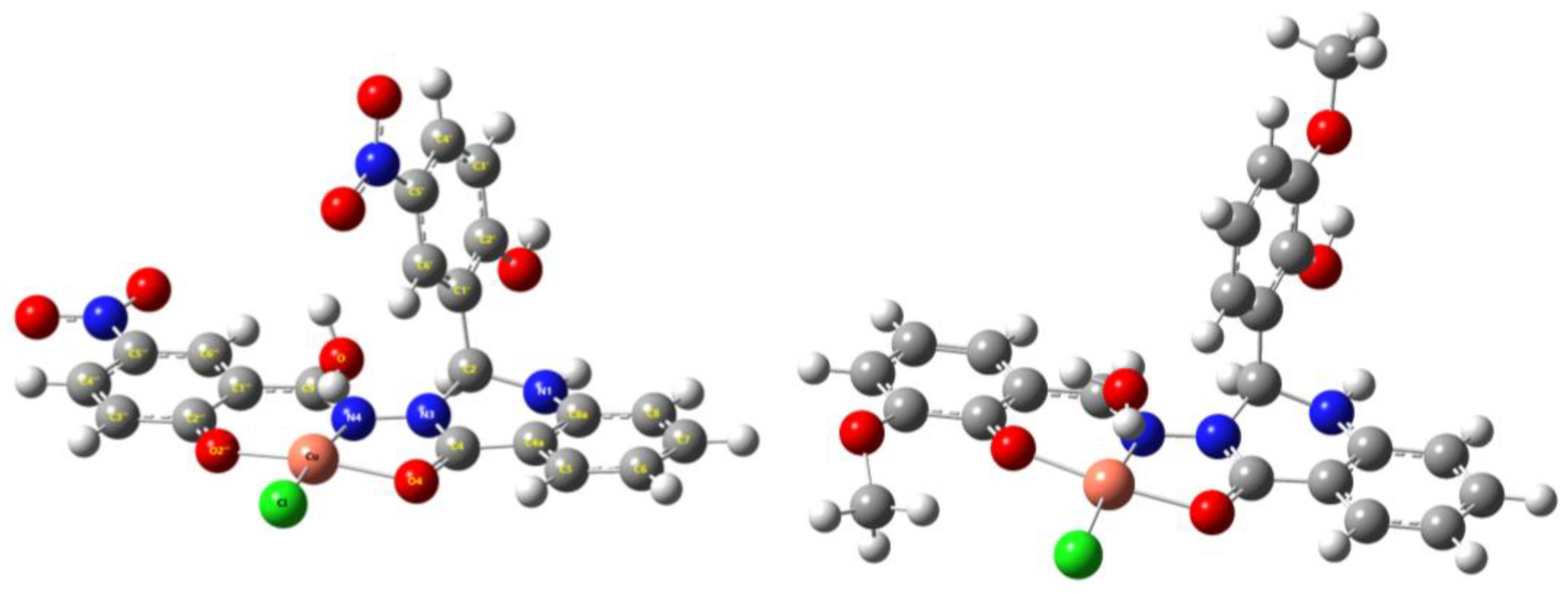
The analysis of the molecular structure of the Cu-L1 and Cu-L2 complexes was also performed with theoretical calculations using the DFT method. The APFD functional and the LanL2DZ basis set were used as a suitable approach for investigating the geometry and calculating structural parameters. DFT calculations showed that the copper atom is penta-coordinated, resulting in a tetragonal-pyramidal shape of the coordination sphere (Figure 2). Coordination of the copper with the ligand occurred in the conjugated region through the O=C=N–N=C(H) array and the OH group in the ortho position on the aromatic ring. From the geometries of both complexes, it can be seen that the copper atom is coordinated in the plane with three ligand atoms and one chlorine atom, with the water molecule in the axial position. It should be noted that the coordination sphere showed slight deviations from planarity, but in the case of both compounds, it was evident that the copper atom was coordinated at the same position. The selected structural parameters are summarized in Table S1.
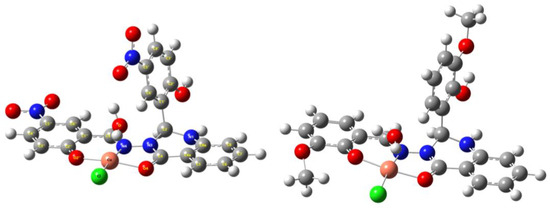
Figure 2.
The DFT-optimized structures of copper complexes Cu-L1 (left) and Cu-L2 (right) in the gas phase using the APFD/LanL2 approach.
From the above parameters, it can be seen that in the case of Cu-L1, the deviation from planarity is smaller but the water molecule in the axial position is significantly deviated (Table S1) due to strong hydrogen bonding with the NO2 group (O–H⋯O=N ≈ 1.92 Å). In the case of the Cu-L2 complex, the deviation from planarity is larger due to the influence of the OCH3 group, which slightly distorts the coordination sphere via hydrogen bonds between the oxygen atom in the 2” position and the chlorine atom coordinated with the copper atom (see Table S1). In the axial direction, there is no significant deviation, as the other OCH3 group is located on the opposite side and therefore has no effect on the coordination sphere.
2.2. Determination of Antioxidant Activity
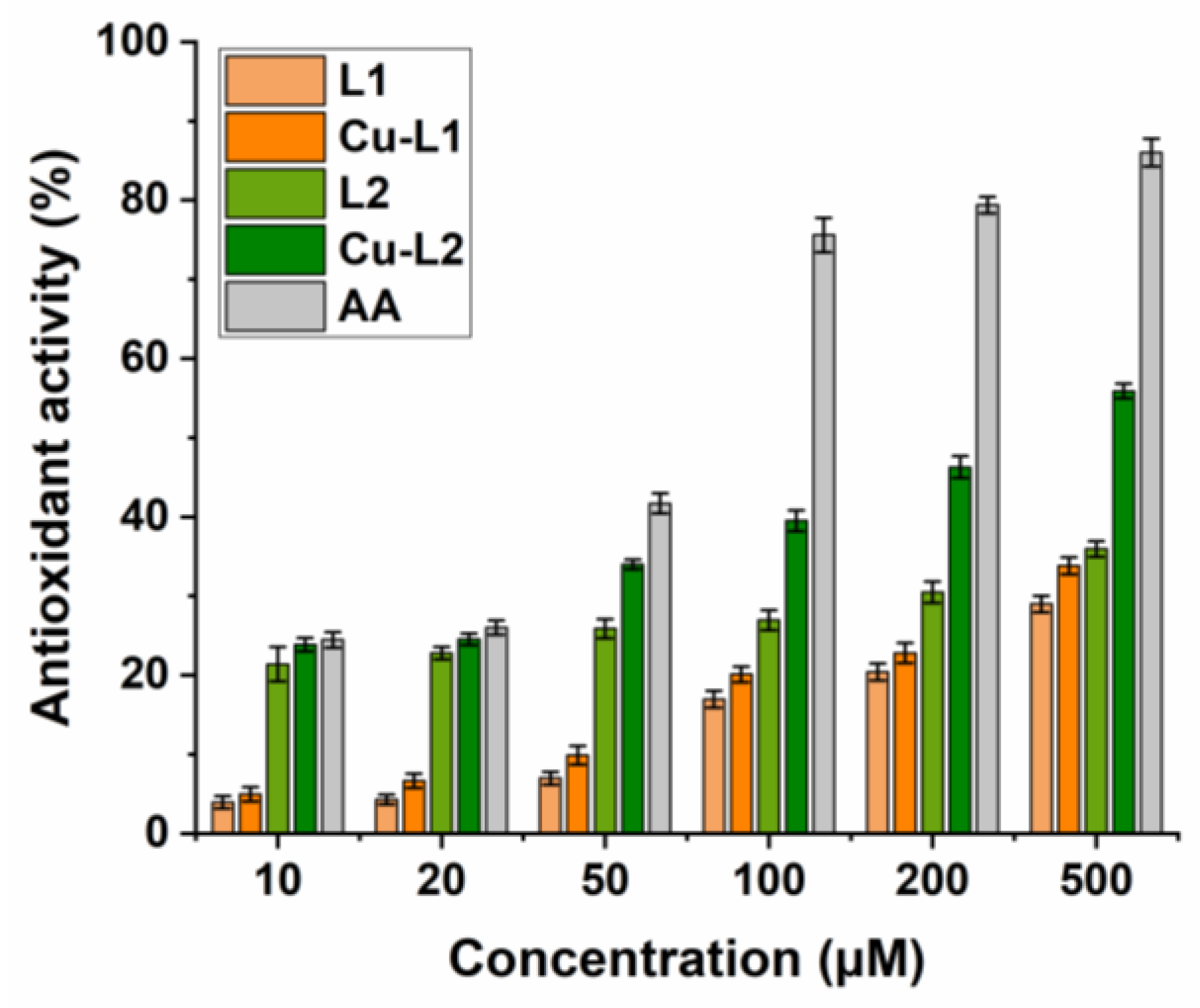
Antioxidant activity of substances is usually associated with their redox properties. Due to their oxidation reduction abilities, these compounds can scavenge free radicals by acting as reducing agents or hydrogen donors and can subsequently prevent undesirable reactions with other radicals, protecting biomolecules from oxidative damage. The antioxidant efficacy of the studied compounds is associated with the presence of electron-donating and/or electron-withdrawing groups in the phenyl rings in the 2 and 3 positions of the quinazolinone scaffold. The copper complexes Cu-L1 and Cu-L2 and Schiff base ligands L1 and L2 were screened for their radical scavenging activity via DPPH assay. The disappearance of the DPPH radical was followed spectrophotometrically at 517 nm. The higher the decrease in the absorbance of the DPPH solution, the stronger the antioxidant activity of the test compound (Figure 3).
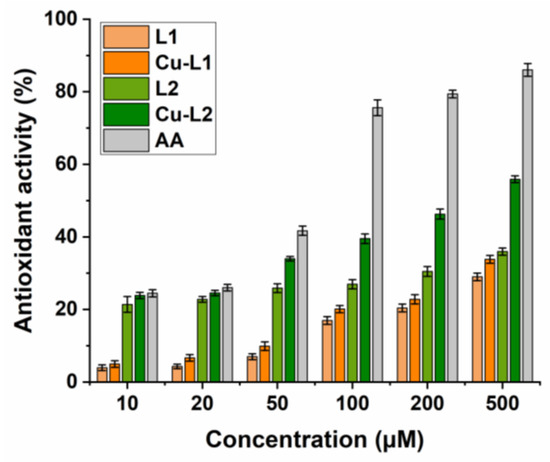
Figure 3.
DPPH assay. Determination of the radical scavenging activity of ligands L1 and L2 and copper complexes Cu-L1 and Cu-L2. Ascorbic acid (AA) positive control.
The results showed that the Cu-L1 complex displayed a concentration-dependent, low-to-moderate radical scavenging activity (5–34%) when compared to ascorbic acid (AA, 24–86%). AA was used as a positive control, with DPPH solution alone as a negative control. The DPPH radical scavenging effect of copper complex Cu-L1 was thus only slightly higher in comparison with the corresponding ligand L1 (4–29%). In this case, the presence of an electron-withdrawing nitro group resulted in a decreased radical scavenging ability. In contrast, the Cu-L2 complex and ligand L2 (bearing electron-donating methoxy/hydroxy substituents on the phenyl rings in the 2 and 3 positions of the quinazolinone ring) displayed significantly better radical scavenging activity (24–57% and 21–36%, respectively). Compared to the standard antioxidant AA, Cu-L2 complex’s efficacy is good, especially at the higher concentrations tested.
The enhanced radical scavenging effect is assigned to the presence of electron-donating groups in the quinazolinone derivatives that can provide hydrogen atoms, an important factor in their capability to scavenge DPPH radicals. After hydrogen atom donation, the compounds exist in the form of a radical, and the effect of electron conjugation in the quinazolinone structure stabilizes this radical, which has a beneficial effect on the course of the reaction. In addition, complexes of transition metal with Schiff bases possess scavenging activity due to the presence of an azomethine group, which can release a proton during the complexation, or due to the reducing ability of metal complexes.
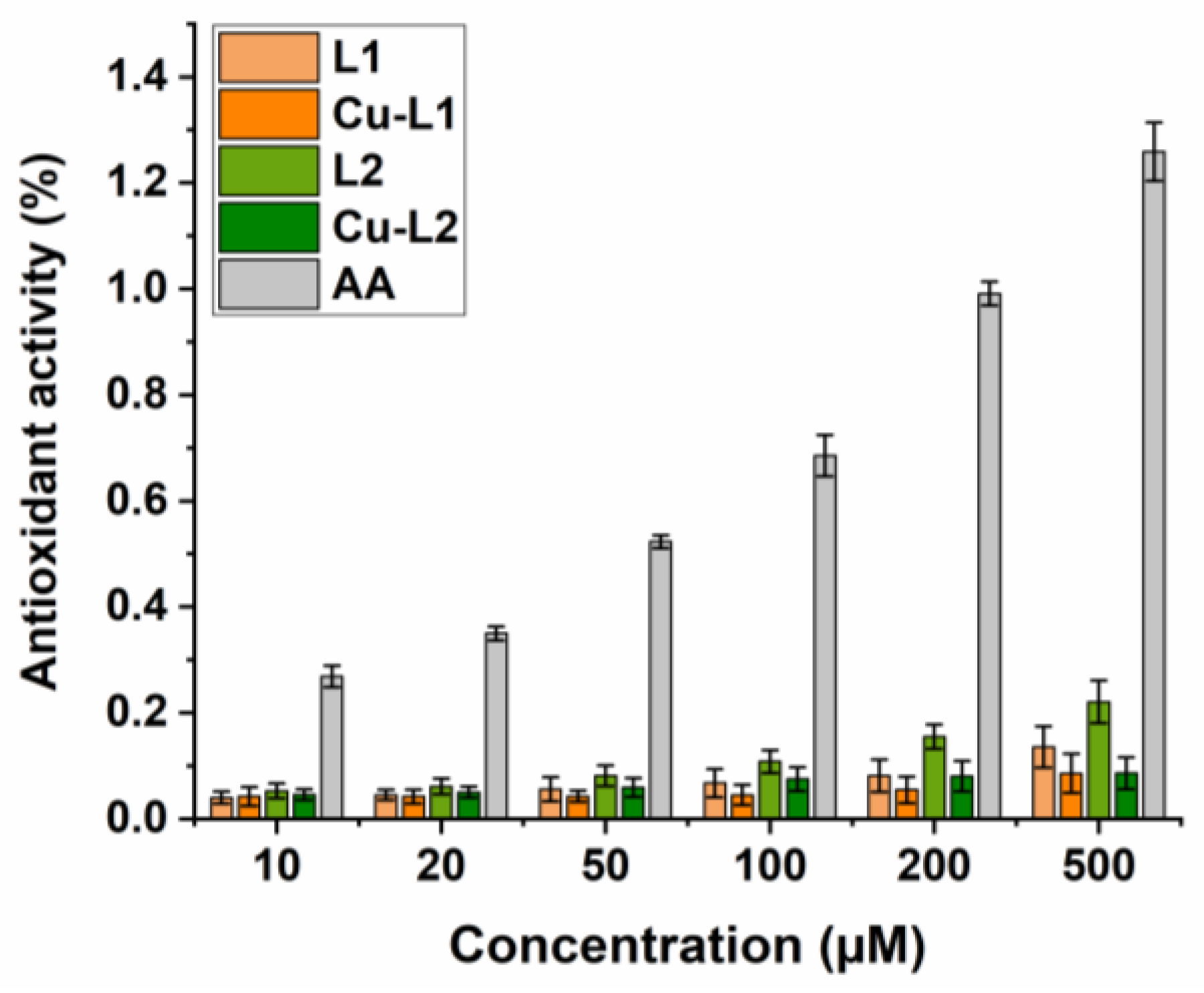
Results from the FRAP test, in which Fe3+ ions are reduced to Fe2+, i.e., electron donation by an antioxidant compound, showed that both the ligands and Cu(II) complexes exhibited weak reducing ability. Their activity was markedly lower compared to the standard antioxidant, and differences in their activity were observable only at the higher concentrations tested (Figure 4). Several other Cu(II) complexes with various N-heterocyclic ligands displayed similar properties, which have been described in the literature [39,40,41].
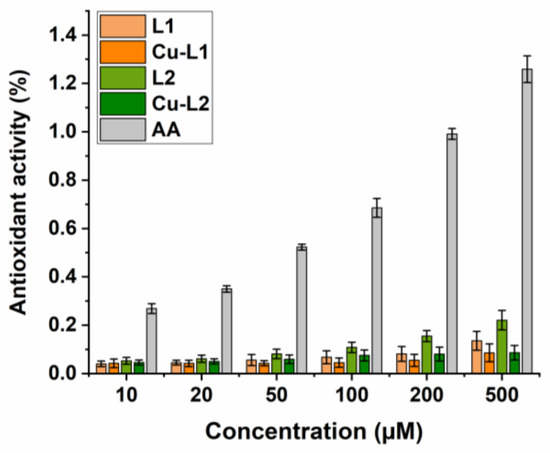
Figure 4.
FRAP assay. Determination of Ferric Reducing Antioxidant Power of ligands L1, L2 and copper complexes Cu-L1, Cu-L2. Ascorbic acid (AA) positive control.
It can be concluded that the antioxidant activity of the studied compounds is concentration-dependent, and the activity of the ligands and their complexes with Cu(II) ions is on a similar level. This suggests that the ligand is a key player in scavenging of DPPH radicals or reduction in iron(III) complexes. The antioxidant activity of Cu(II) complexes has been previously reported [42], and model studies have assessed that it cannot be directly linked to cytotoxic activity determined in vitro (vide infra), as in this study.
2.3. Cytotoxicity
The cytotoxic effect of the studied compounds was tested on two cancer cell lines: human estrogen receptor-positive breast cancer (MCF-7) and human lung adenocarcinoma (A549). Cell viability was measured using the MTT assay, which is based on the ability of the mitochondrial enzyme dehydrogenase to reduce the yellow, water-soluble tetrazolium salt (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, MTT) to an insoluble dark blue formazan. The absorption of the dissolved formazan is proportional to the oxidative activity of the cell’s mitochondria and, under experimental conditions, to the number of metabolically active (living) cells [43]. Additionally, the resazurin assay was used, in which the cell-permeable, nontoxic, and weakly fluorescent blue indicator dye resazurin is used. This dye is converted to resorufin with strong pink fluorescence via oxidoreductive processes. The fluorescence intensity of resorufin is proportional to the number of living cells. Thus, resazurin is a direct indicator of cell health and, on this basis, allows us to measure cell viability and cytotoxicity. The resazurin test is known to be slightly more sensitive compared to MTT, as it can detect cell densities as low as 200 cells/well [44]. As shown in Table 1, copper complexes Cu-L1 and Cu-L2 have IC50 in a low micromolar range, making them very cytotoxic. Under similar conditions, IC50 for cisplatin was ca. 54 and 126 μM for MCF-7 and A549, respectively [45,46]. By contrast, L1 and L2 ligands did not exhibit cytotoxicity below 32 μM (a higher concentration was not checked due to the ligands’ poor solubility).

Table 1.
IC50 against A549 and MCF-7 cell lines. Measured using MTT and Resazurin assays after 24 h treatment.
Cu(II) complexes with coordinated quinazolinone Schiff base derivatives often possess higher anticancer activity than their organic ligands [27,30,47]. In other studies, it was shown that L1 had IC50 of ca. 23 μM [34] against MCF-7 cells. It is not possible to directly compare this data with those obtained by us, since the experimental conditions used in the MTT test (cells’ density, passage number, DMSO concentration) were different. However, it is possible to state that L1 has rather low cytotoxic activity compared to the reference used, doxorubicin (IC50 was ca. 4 μM). This was further supported by in vivo data showing the lack of acute toxicity in mice [34]. Furthermore, the cytotoxic effect was assessed for untransformed human keratinocytes (HaCaT) cells by applying the resazurin test. The tested complexes showed cytotoxicity at a level similar to that of cancer cells, with IC50 values of 1.11 ± 0.01 and 0.64 ± 0.07 μM for Cu-L1 and Cu-L2, respectively. Thus, the complexes do not show specificity to cancer cells, so a targeted delivery system needs to be used to avoid side effects.
2.4. In Vitro Antioxidant Activity
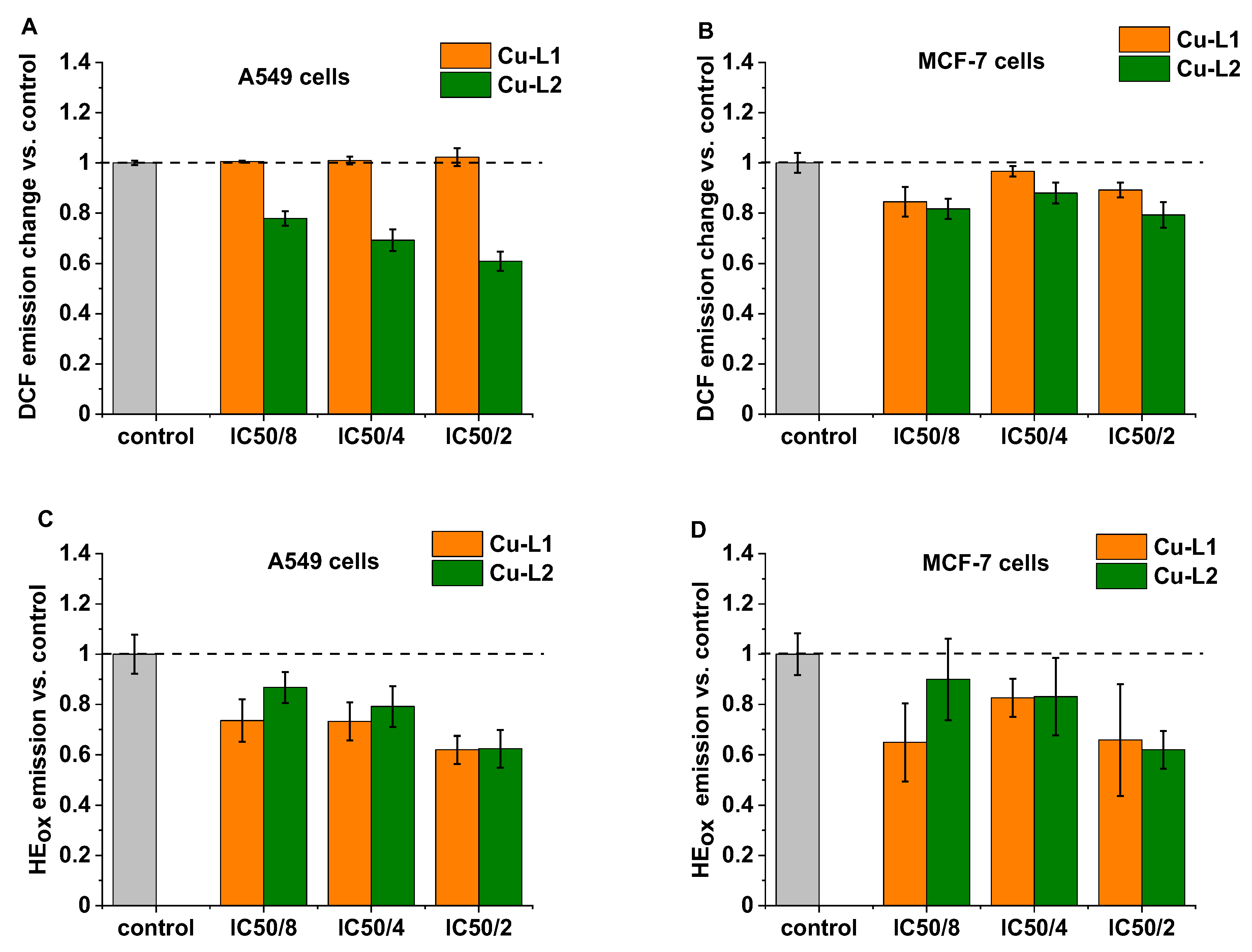
Often, copper complexes induce cell death via increased reactive oxygen production (ROS) in cancer cells [48,49,50]. Therefore, to evaluate the effect of the synthesized Cu complexes on ROS production, a 2,7-dichlorodihydrofluorescein diacetate (H2DCF-DA) probe was used. H2DCF-DA is cell-permeable and can easily diffuse into cells, where it can be converted into 2,7-dichlorodihydrofluorescein (DCFH) with the use of cellular esterases. DCFH is rapidly oxidized to fluorescent 2,7-dichlorofluorescein (DCF) in the presence of ROS. It must be noted that this probe exhibits high sensitivity towards H2O2; however, other types of ROS, such as hydroxyl radical, hydroperoxides, or peroxynitrite, can also oxidize this dye. MCF-7 and A549 cells were incubated for 24 h with a subtoxic dose of both copper complexes at concentrations of IC50/8, IC50/4, and IC50/2, and then the H2DCF-DA probe was applied. As shown in Figure 5, the complex Cu-L2 is quite active in reducing oxidative stress, particularly in A549 cells, as demonstrated by the decreasing DCF emission. No effect of the Cu-L1 complex was observed on DCF emission in A549 cells, while a slight decrease was observed in MCF-7 cells. Next, the effect of the studied copper complexes on the inducing superoxide anion radical (O2•–) formation in cells was investigated by applying hydroethidium (HE), which is oxidized by O2•– (HEox). As shown in Figure 5C,D, a clear decrease in the formation of O2•– was observed for both studied copper complexes on both cell lines. For the highest tested concentration, IC50/2, which corresponds to 1 and 0.5 μM for Cu-L1 and Cu-L2, respectively, superoxide anion radical production was diminished by ca. 40%. Such a huge effect induced by such a small concentration of applied copper complexes suggests interference with signaling pathways and/or the cellular antioxidant system rather than just radical scavenging activity. The role of ROS in promoting tumor development through impact on growth signals and selected gene expression is well established [51]. Therefore, a reduction in ROS formation, particularly in O2•– and H2O2 by cells, can lead to cell death through an antioxidant effect, as observed for other compounds [52]. This hypothesis is further supported by the fact that no effect of copper complexes on hydroxyl radical formation, a molecule that is not considered a signaling molecule, was observed in cells. Additionally, the antioxidant activity observed in model studies indicates that radical scavenging properties for Cu-L1 are much lower than for Cu-L2, while the difference in the cytotoxicity of both metal complexes is rather small. Instead, the L1 and L2 ligands show no significant cytotoxicity in vitro, even though the L2 ligand exhibits radical scavenging activity in model studies.
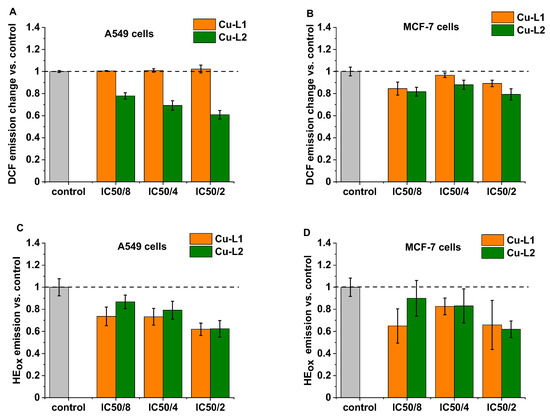
Figure 5.
The level of oxidative stress in A549 and MCF-7 cells measured with selective fluorescent probes (A,B). H2O2 measured via formation of 2,7-dichlorofluorescein (DCF) probe (C,D). O2•– measured via conversion of hydroethidium (HE) probe to oxidized form HEox after 24 h treatment with Cu-L1 (orange) and Cu-L2 (green).
2.5. Mechanism of Cell Death
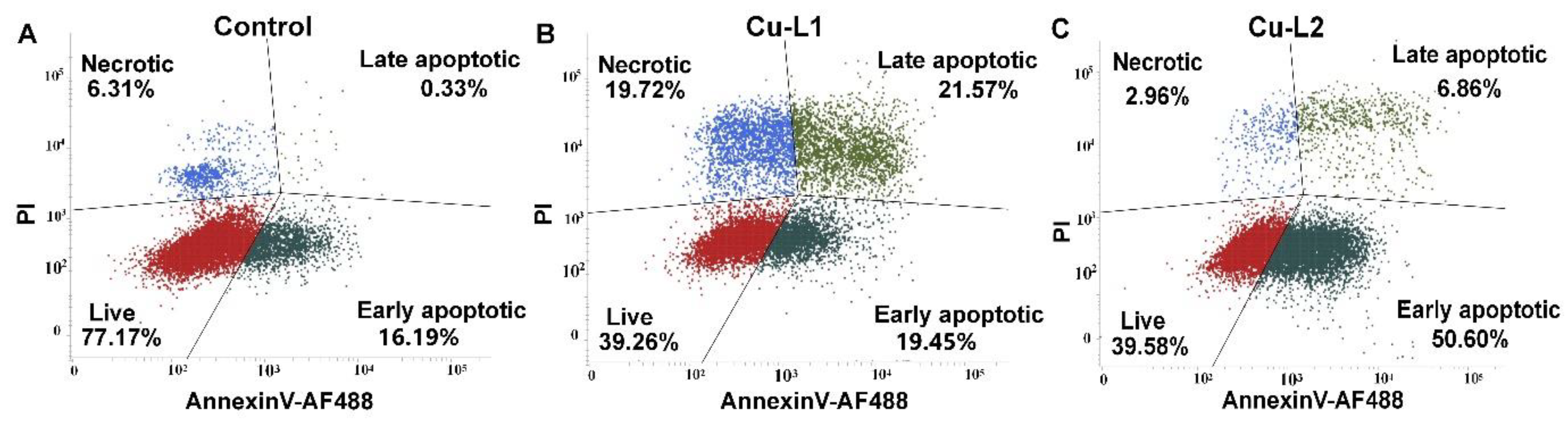
The mechanism of cellular death induced by synthesized Cu complexes was investigated using flow cytometry. In this assay, the percentage of apoptotic cells was estimated by evaluating the amount of cells stained with fluorescently labeled Annexin V, while necrotic cells were measured from the incorporation of propidium iodide in the damaged cells’ nuclei. A 24 h treatment with toxic dosages of Cu complexes (2 × IC50), which corresponds to 4 and 2 μM of Cu-L1 and Cu-L2, respectively, resulted in profound cell death in MCF-7 cells (Figure 6). Despite the similarity in chemical structure of the studied complexes, the induced mechanism of cellular death differed significantly. Although Cu-L1 induced cell death mainly through the late apoptotic and necrotic pathway, Cu-L2 caused early-stage cellular apoptosis. The difference in the observed mechanism can be explained by the change in the antioxidant properties of the compounds studied and/or the variations in the cellular accumulation. In our previous studies, the apoptotic mechanism of cellular death induced in MCF-7 cells by L1 was confirmed with several morphological and biochemical assays [34]. Therefore, the complexation of L1 with copper not only amplified the ligand’s cytotoxic activity but also changed the mechanism of cellular death.
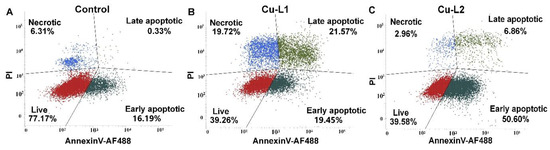
Figure 6.
The representative dot plots of MCF-7 untreated cells (A) incubated with 2 × IC50 of Cu-L1 (B) or Cu-L2 (C) for 24 h and later labeled with Annexin V- AF488 and PI.
3. Materials and Methods
3.1. General Methods
All chemicals were of analytical grade and were used without further purification. A multimode microwave reactor CEM Discover, with an operator-selectable power of 0–300 W and a microwave frequency source of 2.45 GHz, was used for the synthesis of compounds. Thin-layer chromatography (TLC) on aluminum sheets pre-coated with Silica Gel 60 F254 (Merck, Darmstadt, Germany) was used to follow the conversions to products. High-resolution NMR spectra were recorded on a Bruker Avance III HD spectrometer (Bruker, Rheinstetten, Germany) at 25 °C in deuterated dimethyl sulfoxide (DMSO-d6) (Sigma-Aldrich, Steinheim, Germany). The proton and carbon chemical shifts were referenced with tetramethylsilane (TMS). Chemical shifts (δ) are quoted in ppm. One-dimensional 600 MHz 1H and 150 MHz 13C NMR spectra and two-dimensional (COSY, HSQC, HMBC) spectroscopic techniques were used for the determination of the chemical shifts. The electronic absorption spectra were recorded in DMSO Uvasol (Merck) using quartz cuvettes with a 1 cm path length, at 25 °C, on a Shimazu UV-3600 (Thermo Fisher Scientific, Waltham, Massachusetts, United States) UV-Vis spectrometer from 250 to 950 nm. Elemental analyses were performed on a Flash 2000 CHNS/O Elemental Analyzer (Thermo Fisher Scientific). FT-IR (Fourier transform infrared spectra) measurements were performed on a Nicolet 6700 spectrometer (Thermo Fisher Scientific) with a DTGS detector and OMNIC 8.0 software. The spectra were measured in the 4000–400 cm−1 region with a spectral resolution of 4 cm−1. The number of accumulated scans was 128. The calculations were performed using Gaussian 16 software [53] utilizing an APFD functional [54] and LanL2DZ basis set [55]. The convergence criteria were set to default, using an ultrafine integration grid.
3.2. Synthesis of Cu(II) Complexes (Cu-L1 and Cu-L2)
Two quinazolinone Schiff base ligands (L1, L2) were synthesized using a microwave-assisted protocol with a phosphomolybdic acid catalyst, as described in our previous report [32]. The analytical and spectroscopic data of the studied ligands were in full agreement with those published recently [32,34,35].
The copper complexes Cu-L1 and Cu-L2 were prepared by adding the methanolic solution of the corresponding Schiff base ligand (1 mmol, L1 0.045 g; L2 0.042 g) in absolute methanol (15 mL) to the methanolic solution of copper chloride (CuCl2·2 H2O, 1 mmol, 0.170 g in 10 mL) with continuous stirring, and then they were refluxed at 80 °C for 4 h. After cooling to r.t., the volume of the reaction mixture was subsequently reduced to one-third via evaporation. After 1–2 days, tiny crystals of the Cu(II) complex were collected, washed with cold methanol, a mixture of diethyl ether and methanol, and dried.
- Complex Cu-L1: Green solid; yield 64%; FT-IR (ATR, diamond): νmax 3065 (N–H), 1603 (C=O), 1575 (HC=N), 1329 (C-NO2) cm−1; UV-Vis (DMSO): λmax/nm 296, 383. Elemental analysis calculated for C21H15N5O7CuCl (%): C 46.00; H 2.76; N 12.77. Found: C 45.51; H 2.36; N 12.68; 1H NMR (600 MHz, DMSO-d6). δ: 9.48 (broad s, 1H, OH″), 8.36 (m, 2H, H2″ H4″).
- Complex Cu-L2: Brown solid; yield 65%; FT-IR (ATR, diamond): νmax 3364 (N–H), 1605 (C=O), 1582 (HC=N), cm−1; UV-Vis (DMSO): λmax/nm 298, 311, 418. Elemental analysis calculated for C23H21N3O5CuCl (%): C 53.29; H 4.08; N 8.11. Found: C 52.99; H 3.98; N 8.02; 1H NMR (600 MHz, DMSO-d6). δ: 12.74 (s, 1H, OH′), 8.14 (s, 1H, =CH), 7.25 (m, 2H, H4″, 5″).
3.3. Determination of Antioxidant Activity
3.3.1. DPPH Assay
The radical-scavenging ability of ligands and Cu(II) complexes was evaluated through DPPH assay [56] with some modifications. DPPH assay is a simple method for screening radical scavenging activity based on the reduction in semipersistent free radical 2,2-diphenyl-1-picrylhydrazyl (DPPH•) via antioxidants, which is followed by the decolorization of the DPPH radical solution (purple) to form light yellow 2,2-diphenyl-1-picrylhydrazine (DPPH-H). The compounds (L1, L2, Cu-L1, Cu-L2) at different concentrations (10, 20, 50, 100, 200, 500 µM) dissolved in DMSO were mixed with DPPH solution. A reaction mixture consisting of the test derivative (10 µL) and DPPH solution (190 µL) was added to a 96-well plate and incubated in the dark. After 30 min, absorbance was measured at 517 nm on an xMark™ Microplate Spectrophotometer (Bio-Rad Laboratories, Hercules, CA, USA). The reaction was conducted at laboratory temperature, and all experiments were repeated three times. Ascorbic acid was used as a positive control, and DPPH solution was used as a negative control. The percentage of antioxidant activity was evaluated using the following equation:
where Acontrol is the absorbance of negative control; Asample is the absorbance of the tested compound.
Scavenging of DPPH radicals (%) = (Acontrol − Asample/Acontrol) × 100
3.3.2. FRAP Assay
All compounds L1, L2, Cu-L1, and Cu-L2 were screened for their reducing ability via colorimetric Fe(III)-reducing power assay [57]. The principle of this method is that substances with a high reduction potential cause the conversion of the Fe3+/ferricyanide complex to Fe2+/ferrocyanide forms, which is accompanied by a color change. The test protocol is as follows: solution of the test compound (200 µL) in methanol (concentration 0.1–1 mM) was mixed with a phosphate buffer (500 µL, 0.2 M, pH 6.6) and a solution of potassium ferricyanide K3[Fe(CN)6] (1%, 500 µL). The mixture was incubated at 50 °C for 20 min. Subsequently, a solution of trichloroacetic acid (10%, 500 µL) was added, and the mixture was centrifuged (3000 rpm for 10 min). The supernatant upper layer (500 µL) was combined with distilled water (500 µL) and an aqueous solution of FeCl3 (0.1%, 100 µL). The resulting mixture was allowed to stand (10 min), and the absorbance at 700 nm was measured on an xMarkTM Microplate Spectrophotometer. An increase in the absorbance of the reaction mixture was indicative of an increase in the reducing power of the compound. The experiments were run in triplicate.
3.4. Cell Culture Conditions
The in vitro studies were conducted using human estrogen receptor-positive breast cancer MCF-7 (Sigma-Aldrich), human lung adenocarcinoma A549 (Sigma-Aldrich), and human keratinocytes HaCaT (CLS Cell Lines Service GmbH) cell lines. A549 and HaCaT cells were cultured in DMEM, while MCF-7 cells were cultured in EMEM medium. For all cell lines, the medium was supplemented with 10% fetal bovine serum (FBS) (v/v) and 1% penicillin–streptomycin (100 units/mL–100µg/mL) (v/v). Furthermore, the medium for MCF-7 cells was enriched with 2 mM glutamine and 1% non-essential amino acids (NEAA) (v/v). Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 (v/v).
3.5. Cytotoxicity
Cell viability upon treatment with Cu complexes and ligands was determined using the resazurin and MTT assays reported by us previously [58]. Cells were seeded into 96-well plates with the density of 3 × 104 cells per cm2 in a complete medium and cultured for 24 h. Subsequently, cells were incubated with various concentrations of the studied compounds for 24 h. Stock solutions of the Cu(II) complexes and ligands were prepared in DMSO. The final concentration of DMSO in cell culture was fixed at 0.1% (v/v). After 24 h of incubation, the cells were washed with PBS and incubated in resazurin or MTT solutions for 3 h at 37 °C. Subsequently, for the resazurin assay, the fluorescence was measured using a Tecan Infinite 200 microplate reader at 605 nm using 560 nm excitation light. For the MTT assay, the MTT solution was removed after 3 h of incubation, and the formed violet formazan crystals were dissolved in 100 µL of DMSO–methanol (1:1) mixture. The absorbance was measured using a Tecan Infinite 200 microplate reader(Tecan, Männedorf, canton of Zürich, Switzerland) at 565 nm with 700 nm as a reference wavelength. The experiments were performed in triplicate and repeated three times. Results are presented as the mean value and the standard error of the mean. IC50 parameters were determined using the Hill equation (OriginPro 2020).
3.6. Oxidative Stress Evaluation
Cells were seeded into a 96-well plate with the density of 3 × 104 cells per cm2 in a complete medium and cultured for 24 h. Then, the medium was removed, and various concentrations of the studied complexes were added for 24 h incubation. Subsequently, cells were washed with PBS, and 100 µL of ROS probes 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA, 20 µM), aminophenyl fluorescein (APF, 5 µM), and hydroethidium (HE, 10 µM) were added to each well for 30 min. Then, the ROS indicator solutions were removed, the cells were washed with PBS, and the fluorescence of the cells was quantified using a Tecan Infinite 200 plate reader (λex = 535 nm and λem = 485 nm for DCFDA and APF and λex = 520 nm and λem = 605 nm for HE). Experiments were performed in triplicate and repeated twice.
3.7. Mechanism of Cell Death
The mechanism of cellular death was determined by staining cells with the Annexin V (Annexin V—Alexa Fluor 488 Life technologies) and propidium iodide (PI, BioRad Laboratories, Hercules, California, United States). MCF-7 cells were seeded in a 6-well plate with a density of 3 × 104 cells per cm2 in a complete medium and cultured for 24 h. Then the medium was removed, and the studied complexes at the concentration of 2 × IC50 were added for another 24 h incubation. Subsequently, the incubated cells were washed with PBS. The cells were stained with Annexin V—Alexa Fluor 488 for 30 min in the dark and then with PI for 10 min. Cells were analyzed using a flow cytometer BD Facs Versa (λext/em = 488/527 ± 16 nm, or λext/em = 488/586 ± 21 nm). As a positive control, H2O2 (600 µM, 6 h incubation) was used.
4. Conclusions
Copper(II) complexes of 2,3-substituted quinazolinone Schiff bases were prepared, and their structures were analyzed via physicochemical and spectroscopic methods. Quantum-chemical DFT calculations showed that the copper atom was penta-coordinated, resulting in a tetragonal-pyramidal shape of the coordination sphere. Coordination of the ligands with copper ions resulted in a pronounced enhancement of their cytotoxic activity. Cu(II) complexes are one to two orders of magnitude more effective than cisplatin, a well-known cytotoxic drug. Therefore, they can be considered highly cytotoxic compounds operating at low micromolar concentrations. The antioxidant activity of copper compounds was demonstrated in both model cell-free and in vitro studies. The difference between the two copper complexes manifests itself not only in radical scavenging properties observed in model studies, but also in the mechanism of inducing cellular death. The results suggest that under the experimental conditions used in this study, these Cu(II) complexes behave as antioxidants rather than as ROS inducers as would be expected. However, in addition to inducing ROS balance impairment, cytotoxic activity may also arise from targeting other biomacromolecules. To clarify specific targets at the molecular level, more advanced cellular studies are needed. However, outcomes of this study could be useful as a starting point for the development of new copper-based anticancer agents.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/inorganics11100391/s1, Figure S1: The FT-IR spectrum of the free ligand L1; Figure S2. The FT-IR spectrum of the Cu-L1 complex; Figure S3. The FT-IR spectrum of the free ligand L2; Figure S4. The FT-IR spectrum of the Cu-L2 complex; Figure S5. The electronic absorption spectra of the free ligand L1 (blue line) and the Cu-L1 complex (orange line) measured in DMSO at 0.1 mM concentration. The inset shows absorption band of Cu-L1 complex obtained at higher concentration (1 mM) in order to register the d-d transitions; Figure S6. The electronic absorption spectra of the ligand L2 (blue line) and the Cu-L2 complex (orange line) measured in DMSO at 0.1 mM concentration. The inset shows absorption band of Cu-L2 complex obtained at higher concentration (1 mM) in order to register the d-d transitions; Figure S7. 1H NMR and 13C spectra of the ligand L1 (A and C, respectively) and its copper complex (with 30% excess of the ligand L1) Cu-L1 (B and D); Figure S8. 1H NMR and 13C spectra of the ligand L2 (A and C, respectively) and its copper complex (with 30% excess of the ligand L2) Cu-L2 (B and D); Table S1. The selected theoretical bond lengths (in Å) data of the studied Cu-L1 and Cu-L2 complexes obtained from gas-phase DFT calculations using APFD/LanL2DZ approach; Table S2. The selected theoretical bond angles (in degrees) data of the studied Cu-L1 and Cu-L2 complexes obtained from gas-phase DFT calculations using APFD/LanL2DZ approach; Table S3. The selected theoretical dihedral angles (in degrees) data of the studied Cu-L1 and Cu-L2 complexes obtained from gas-phase DFT calculations in the using APFD/LanL2DZ approach.
Author Contributions
Conceptualization, Z.H. and M.B.; methodology, formal analysis, and visualization, I.G., J.H., O.M., Z.H. and M.B.; investigation, I.G., J.H., O.M. and Z.H.; validation, I.G., J.H., O.M., Z.H. and M.B; writing—original draft preparation, I.G., J.H., O.M., Z.H. and M.B.; writing—review and editing, Z.H. and M.B.; supervision, Z.H. and M.B.; funding acquisition, Z.H. and M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Faculty of Chemistry of the Jagiellonian University, Kraków, Poland, and by the Slovak Grant Agency VEGA, grant No 2/0071/22. Calculations were performed at the Computing Centre of the SAS using the supercomputing infrastructure acquired in projects ITMS 26230120002 and 26210120002, both supported by the Research and Development Operational Program funded by the ERDF.
Data Availability Statement
The data presented in this study are available in the main text.
Acknowledgments
The authors thank Viera Dujnic for the FTIR spectra and Michal Hricovíni for DFT calculations and helpful discussion.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.; VanCamp, L.; Trosko, J. Platinum Compounds: A New Class of Potent Antitumour Agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Disc. 2005, 4, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Luo, Q.; Zhang, Y.; Jia, F.; Zhao, Y.; Wang, F. Advances in Toxicological Research of the Anticancer Drug Cisplatin. Chem. Res. Toxicol. 2019, 32, 1469–1486. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, C.; Gao, X.; Yao, Q. Platinum-based drugs for cancer therapy and anti-tumor strategies. Theranostics 2022, 12, 2115–2132. [Google Scholar] [CrossRef]
- Bergamo, A.; Sava, G. Linking the future of anticancer metal-complexes to the therapy of tumour metastases. Chem. Soc. Rev. 2015, 44, 8818–8835. [Google Scholar] [CrossRef]
- Medici, S.; Peana, M.; Nurchi, V.M.; Lachowicz, J.I.; Crisponi, G.; Zoroddu, M.A. Noble metals in medicine: Latest advances. Coord. Chem. Rev. 2015, 284, 329–350. [Google Scholar] [CrossRef]
- Zhang, P.; Sadler, P.J. Redox-Active Metal Complexes for Anticancer Therapy. Eur. J. Inorg. Chem. 2017, 2017, 1541–1548. [Google Scholar] [CrossRef]
- Tapiero, H.; Townsend, D.M.; Tew, K.D. Trace elements in human physiology and pathology. Copper. Biomed. Pharmacother. 2003, 57, 386–398. [Google Scholar] [CrossRef]
- Kodama, H.; Fujisawa, C.; Bhadhprasit, W. Inherited Copper Transport Disorders: Biochemical Mechanisms, Diagnosis, and Treatment. Curr. Drug Metabol. 2012, 13, 237–250. [Google Scholar] [CrossRef]
- Mustafa, S.; Al Sharif, M. Copper (Cu) an Essential Redox-Active Transition Metal in Living System—A Review Article. Am. J. Anal. Chem. 2018, 9, 15–26. [Google Scholar] [CrossRef]
- Wang, Y.; Branicky, R.; Noë, A.; Hekimi, S. Superoxide dismutases: Dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018, 217, 1915–1928. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: New York, NY, USA, 2007. [Google Scholar]
- Thomas, C.; Mackey, M.M.; Diaz, A.A.; Cox, D.P. Hydroxyl radical is produced via the Fenton reaction in submitochondrial particles under oxidative stress: Implications for diseases associated with iron accumulation. Redox Rep. 2009, 14, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, A.; Kozlov, A. Biological Activities of Reactive Oxygen and Nitrogen Species: Oxidative Stress versus Signal Transduction. Biomolecules 2015, 5, 472–484. [Google Scholar] [CrossRef]
- Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Copper in diseases and treatments, and copper-based anticancer strategies. Med. Res. Rev. 2010, 30, 708–749. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in copper complexes as anticancer agents. Chem. Rev. 2013, 114, 815–862. [Google Scholar] [CrossRef]
- Akladios, F.N.; Andrew, S.D.; Parkinson, C.J. Increased generation of intracellular reactive oxygen species initiates selective cytotoxicity against the MCF-7 cell line resultant from redox active combination therapy using copper–thiosemicarbazone complexes. J. Biol. Inorg. Chem. 2016, 21, 407–419. [Google Scholar] [CrossRef]
- Rogolino, D.; Cavazzoni, A.; Gatti, A.; Tegoni, M.; Pelosi, G.; Verdolino, V.; Fumarola, C.; Cretella, D.; Petronini, P.G.; Carcelli, M. Anti-proliferative effects of copper(II) complexes with hydroxyquinoline-thiosemicarbazone ligands. Eur. J. Med. Chem. 2017, 128, 140–153. [Google Scholar] [CrossRef]
- Uddin, M.N.; Ahmed, S.S.; Rahatul Alam, S.M. Review: Biomedical applications of Schiff base metal complexes. J. Coord. Chem. 2020, 73, 3109–3149. [Google Scholar] [CrossRef]
- Chen, D.; Cui, Q.C.; Yang, H.; Dou, Q.P. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006, 66, 10425–10433. [Google Scholar] [CrossRef]
- Guo, A.J.; Xu, X.S.; Hu, Y.H.; Wang, M.Z.; Tan, X. Effects of ternary complexes of copper with salicylaldehyde-aminoacid Schiff base coordination compounds on the proliferation of BGC823 cells. Chin. J. Cancer 2010, 29, 277–282. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Szymański, P.; Frączek, T.; Markowicz, M.; Mikiciuk-Olasik, E. Development of copper based drugs, radiopharmaceuticals and medical materials. BioMetals 2012, 25, 1089–1112. [Google Scholar] [CrossRef] [PubMed]
- Iakovidis, I.; Delimaris, I.; Piperakis, S.M. Copper and Its Complexes in Medicine: A Biochemical Approach. Mol. Biol. Int. 2011, 2011, 594529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, H.; Yan, M.; Wang, H.; Zhang, C. Novel copper complexes as potential proteasome inhibitors for cancer treatment. Mol. Med. Rep. 2016, 15, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Shagufta; Ahmad, I. Transition metal complexes as proteasome inhibitors for cancer treatment. Inorg. Chim. Acta 2020, 506, 119521. [Google Scholar] [CrossRef]
- Ashok, U.P.; Kollur, S.P.; Arun, B.P.; Sanjay, C.; Suresh, K.S.; Anil, N.; Baburao, H.V.; Markad, D.; Castro, J.O.; Frau, J.; et al. In vitro anticancer activity of 4(3H)-quinazolinone derived Schiff base and its Cu(II), Zn(II) and Cd(II) complexes: Preparation, X-ray structural, spectral characterization and theoretical investigations. Inorg. Chim. Acta 2020, 511, 119846. [Google Scholar] [CrossRef]
- Li, S.X.; Luo, P.; Jiang, Y.M. Copper complexes with 4(3H)-quinazolinone: Thermal gravimetric analysis and anticancer activity of [Cu(L)2(H2O)2(NO3)2], [Cu(L–)(NO3)]n, and [Cu(L)2(H2O)2(Cl)2]. Russ. J. Coord. Chem. 2017, 43, 238–243. [Google Scholar] [CrossRef]
- Lazou, M.; Tarushi, A.; Gritzapis, P.; Psomas, G. Transition metal complexes with a novel guanine-based (E)-2-(2-(pyridin-2-ylmethylene)hydrazinyl)quinazolin-4(3H)-one: Synthesis, characterization, interaction with DNA and albumins and antioxidant activity. J. Inorg. Biochem. 2020, 206, 111019. [Google Scholar] [CrossRef]
- Ubale, P.; Mokale, S.; More, S.; Waghamare, S.; More, V.; Munirathinam, N.; Dilipkumar, S.; Das, R.K.; Reja, S.; Helavi, V.B.; et al. Evaluation of in vitro anticancer, antimicrobial and antioxidant activities of new Cu(II) complexes derived from 4(3H)-quinazolinone: Synthesis, crystal structure and molecular docking studies. J. Mol. Struct. 2022, 1251, 131984. [Google Scholar] [CrossRef]
- Brissos, R.F.; Caubet, A.; Gamez, P. Possible DNA-Interacting Pathways for Metal-Based Compounds Exemplified with Copper Coordination Compounds. Eur. J. Inorg. Chem. 2015, 2015, 2633–2645. [Google Scholar] [CrossRef]
- Hricovíniová, Z.; Hricovíni, M.; Kozics, K. New series of quinazolinone derived Schiff’s bases: Synthesis, spectroscopic properties and evaluation of their antioxidant and cytotoxic activity. Chem. Pap. 2018, 72, 1041–1053. [Google Scholar] [CrossRef]
- Hricovíniová, J.; Hricovíniová, Z.; Kozics, K. Antioxidant, Cytotoxic, Genotoxic, and DNA-Protective Potential of 2,3-Substituted Quinazolinones: Structure-Activity Relationship Study. Int. J. Mol. Sci. 2021, 22, 610. [Google Scholar] [CrossRef] [PubMed]
- Zahedifard, M.; Faraj, F.L.; Paydar, M.; Looi, C.Y.; Hajrezaei, M.; Hasanpourghadi, M.; Kamalidehghan, B.; Majid, N.A.; Ali, H.M.; Abdulla, M.A. Synthesis, characterization and apoptotic activity of quinazolinone Schiff base derivatives toward MCF-7 cells via intrinsic and extrinsic apoptosis pathways. Sci. Rep. 2015, 5, 11544. [Google Scholar] [CrossRef] [PubMed]
- Gudasi, K.B.; Patil, S.A.; Vadavi, R.S.; Shenoy, R.V.; Nethaji, M. Crystal structure of 2-[2-hydroxy-3-methoxyphenyl]-3-[2-hydroxy-3-methoxybenzylamino]-1,2-dihydroquinazolin-4(3H)-one and the synthesis, spectral and thermal investigation of its transition metal complexes. Trans. Metal Chem. 2006, 31, 586–592. [Google Scholar] [CrossRef]
- Tapabashi, N.O.; Taha, N.I.; El-Subeyhi, M. Design, Microwave Assisted Synthesis of Some Schiff Bases Derivatives of Congo Red and Conventional Preparation of Their Structurally Reversed Analogous Compounds. Int. J. Org. Chem. 2021, 11, 35–45. [Google Scholar] [CrossRef]
- Nielsen, I.B.; Petersen, M.Å.; Lammich, L.; Nielsen, M.B.; Andersen, L.H. Absorption Studies of Neutral Retinal Schiff Base Chromo-phores. J. Phys. Chem. A 2006, 110, 12592–12596. [Google Scholar] [CrossRef]
- Crompton, E.M.; Lewis, F.D. Positional effects of the hydroxy substituent on the photochemical and photophysical behavior of 3- and 4-hydroxystilbene. Photochem. Photobiol. Sci. 2004, 3, 660–668. [Google Scholar] [CrossRef]
- Salga, M.; Sada, M.; Mustapha, I.A. Influence of Steric Hindrance on The Antioxidant Activity of Some Schiff Base Ligands And Their Copper(II) Complexes. Orient. J. Chem. 2014, 30, 1529–1534. [Google Scholar] [CrossRef]
- Turan, N.; Adigüzel, R.; Buldurun, K.; Bursal, E. Spectroscopic, Thermal and Antioxidant Properties of Novel Mixed Ligand-Metal Complexes Obtained from Saccharinate Complexes and Azo Dye Ligand (mnppa). Int. J. Pharmacol. 2016, 12, 92–100. [Google Scholar] [CrossRef]
- Buldurun, K.; Turan, N.; Aras, A.; Mantarci, A.; Turkan, F.; Bursal, E. Spectroscopic and Structural Characterization, Enzyme Inhibitions, and Antioxidant Effects of New Ru(II) and Ni(II) Complexes of Schiff Base. Chem. Biodivers. 2019, 16, e1900243. [Google Scholar] [CrossRef]
- Pinheiro, A.C.; Nunes, I.J.; Ferreira, W.V.; Tomasini, P.P.; Trindade, C.; Martins, C.C.; Wilhelm, E.A.; Oliboni, R.d.S.; Netz, P.A.; Stieler, R.; et al. Antioxidant and Anticancer Potential of the New Cu(II) Complexes Bearing Imine-Phenolate Ligands with Pendant Amine N-Donor Groups. Pharmaceuticals 2023, 15, 376. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, P.J.; Irwin, W.; Diaz, E.D.; Howard-Cofield, E.E.; Krejsa, C.M.; Slaughter, M.R.; Gao, E.B.; Kaludercic, N.; Angeline, E.A.; Bernardi, E.P.; et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch. Toxicol. 2006, 80, 580–604. [Google Scholar] [CrossRef] [PubMed]
- Mazuryk, O.; Suzenet, F.; Kieda, C.; Brindell, M. Biological effect of nitroimidazole derivative of polypyridyl ruthenium complex on cancer and endothelial cells. Metallomics 2015, 7, 553–566. [Google Scholar] [CrossRef]
- Gurgul, I.; Janczy-Cempa, E.; Mazuryk, O.; Lekka, M.; Łomzik, M.; Suzenet, F.; Gros, P.G.; Brindell, M. Inhibition of Metastasis by Polypyridyl Ru(II) Complexes through Modification of Cancer Cell Adhesion–In Vitro Functional and Molecular Studies. J. Med. Chem. 2022, 65, 10459–10470. [Google Scholar] [CrossRef]
- Kesavan, M.P.; Vinoth Kumar, G.G.; Dhaveethu Raja, J.; Anitha, K.; Karthikeyan, S.; Rajesh, J. DNA interaction, antimicrobial, antioxidant and anticancer studies on Cu(II) complexes of Luotonin A. J. Photochem. Photobiol. B Biol. 2017, 167, 20–28. [Google Scholar] [CrossRef]
- Peña, Q.; Lorenzo, J.; Sciortino, G.; Rodríguez-Calado, S.; Maréchal, J.-D.; Bayóna, P.; Simaan, A.J.; Iranzo, O.; Capdevila, M.; Palaciosa, Ò. Studying the reactivity of “old” Cu(II) complexes for “novel” anticancer purposes. J. Inorg. Biochem. 2019, 195, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Sawant, A.V.; Prassanawar, S.S.; Panda, D. Copper-Plumbagin Complex Produces Potent Anticancer Effects by Depolymerizing Microtubules and Inducing Reactive Oxygen Species and DNA Damage. ACS Omega 2023, 8, 3221–3235. [Google Scholar] [CrossRef]
- Sîrbu, A.; Palamarciuc, O.; Babak, M.V.; Lim, J.M.; Ohui, K.; Enyedy, E.A.; Shova, S.; Darvasiová, D.; Rapta, P.; Ang, W.H.; et al. Copper(II) thiosemicarbazone complexes induce marked ROS accumulation and promote nrf2-mediated antioxidant response in highly resistant breast cancer cells. Dalton Trans. 2017, 46, 3833–3847. [Google Scholar] [CrossRef]
- Schumacker, P.T. Reactive oxygen species in cancer: A dance with the devil. Cancer Cell 2015, 27, 156–157. [Google Scholar] [CrossRef]
- Kang, K.A.; Piao, M.J.; Ryu, Y.S.; Hyun, Y.J.; Park, J.E.; Shilnikova, K.; Zhen, A.X.; Kang, H.K.; Koh, Y.S.; Jeong, Y.J.; et al. Luteolin induces apoptotic cell death via antioxidant activity in human colon cancer cells. Int. J. Oncol. 2017, 51, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16, Rev. B.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Austin, A.; Petersson, G.A.; Frisch, M.J.; Dobek, F.J.; Scalmani, G.; Throssell, K. A density functional with spherical atom dispersion terms. J. Chem. Theor. Comput. 2012, 8, 4989–5007. [Google Scholar] [CrossRef] [PubMed]
- Dunning, T.H.; Hay, P.J. Gaussian Basis Sets for Molecular Calculations. In Methods of Electronic Structure Theory, Modern Theoretical Chemistry; Schaefer, H.F., Ed.; Springer: Boston, MA, USA, 1977; Volume 3, pp. 1–28. [Google Scholar]
- Locatelli, M.; Gindro, R.; Travaglia, F.; Coïsson, J.D.; Rinaldi, M.; Arlorio, M. Development of a free software for the correct interpretation of data. Food Chem. 2009, 114, 889–897. [Google Scholar] [CrossRef]
- Gupta, D. Methods for determination of antioxidant capacity: A review. Int. J. Pharm. Sci. Res. 2015, 6, 546–566. [Google Scholar] [CrossRef]
- Gurgul, I.; Mazuryk, O.; Łomzik, M.; Gros, P.C.; Rutkowska-Zbik, D.; Brindell, M. Unexplored features of Ru(II) polypyridyl complexes-towards combined cytotoxic and antimetastatic activity. Metallomics 2020, 12, 784–793. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).