Combination of Direct Viable Count and Fluorescent In Situ Hybridization (DVC-FISH) as a Potential Method for Identifying Viable Vibrio parahaemolyticus in Oysters and Mussels
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. DVC Assays
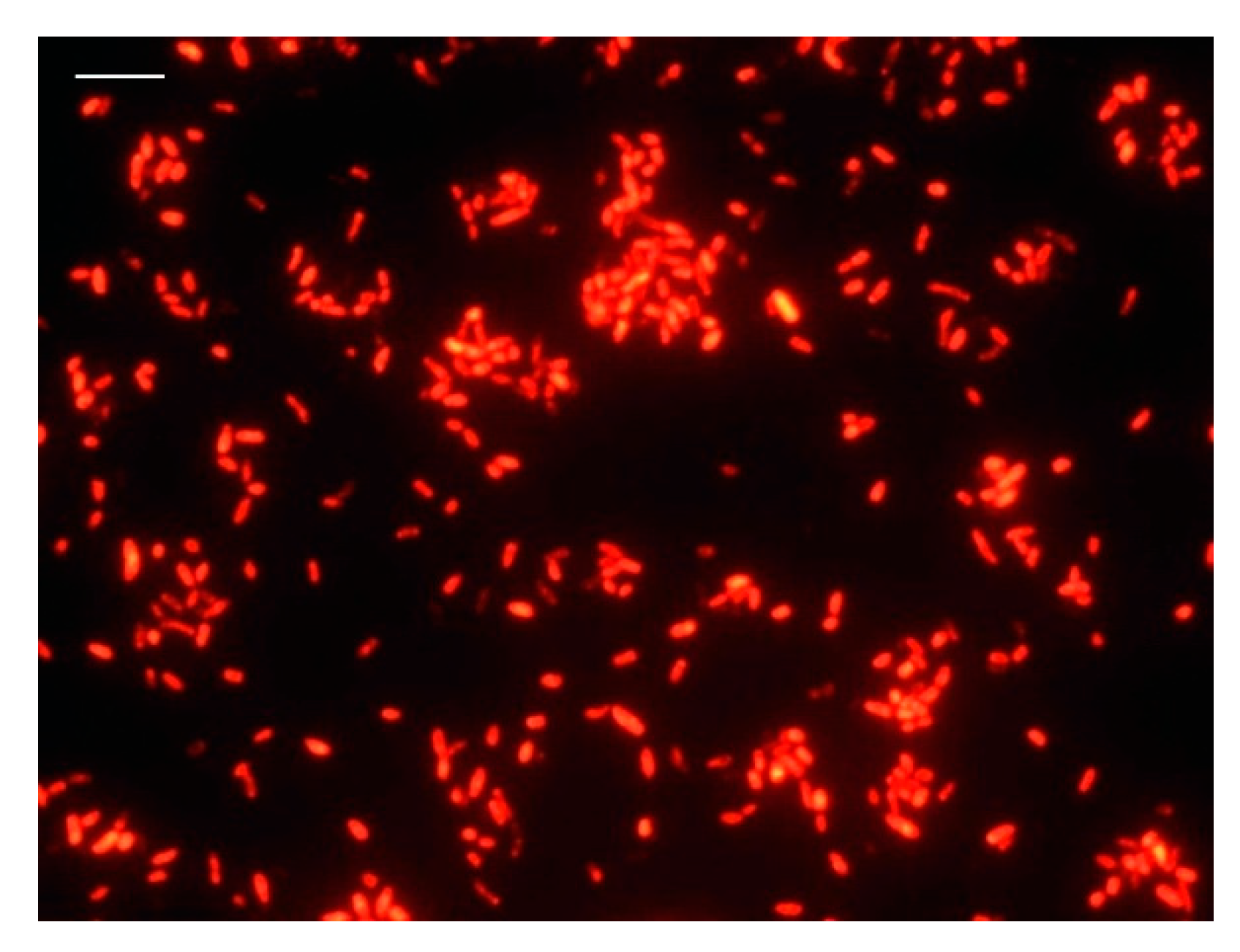
2.3. Fluorescent In Situ Hybridization (FISH) Analysis
2.4. DVC-FISH Sensitivity
2.5. Detection of Viable V. parahaemolyticus in Inoculated Samples
2.6. Viability Staining
2.7. Nucleic Acid Extraction
2.8. Detection and Testing of Nucleic Acid Stability of V. parahaemolyticus
3. Results
3.1. DVC-FISH Assay Optimization
3.2. Detection of Viable V. parahaemolyticus in Inoculated Samples
3.3. Detection and Testing of NA Stability of V. parahaemolyticus
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Romalde, J.; Dieguez, A.; Lasa, A.; Balboa, S. New Vibrio species associated to molluscan microbiota: A review. Front. Microbiol. 2014, 4, 413. [Google Scholar] [CrossRef]
- Oliver, J. The Biology of Vibrio vulnificus. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido-Maestu, A.; Lozano-Leon, A.; Rodríguez-Souto, R.R.; Vieites-Maneiro, R.; Chapela, M.J.; Ana, G.; Cabado, A.G. Presence of pathogenic Vibrio species in fresh mussels harvested in the southern Rias of Galicia (NW Spain). Food Control 2016, 59, 759–765. [Google Scholar] [CrossRef]
- Drake, S.L.; DePaola, A.; Jaykus, L.A. An overview of Vibrio vulnificus and Vibrio parahaemolyticus. Compr. Rev. Food Sci. Food Saf. 2007, 6, 120–144. [Google Scholar] [CrossRef]
- Kokashvili, T.; Whitehouse, C.A.; Tskhvediani, A.; Grim, C.J.; Elbakidze, T.; Mitaishvili, N.; Janelidze, N.; Janelidze, E.; Haley, E.J.; Lashkhi, E.; et al. Occurrence and Diversity of Clinically Important Vibrio Species in the Aquatic Environment of Georgia. Front. Public Health 2015, 3, 232. [Google Scholar] [CrossRef]
- Cantet, F.; Hervio-Heath, D.; Caro, A.; Le Mennec, C.; Monteil, C.; Quemere, C. Quantification of Vibrio parahaemolyticus, V. vulnificus and V. cholerae in French Mediterranean coastal lagoons. Res. Microbiol. 2013, 164, 867–874. [Google Scholar] [CrossRef] [Green Version]
- Johnson, C.N.; Flowers, A.R.; Noriea, N.F.; Zimmerman, A.M.; Bowers, J.C.; DePaola, A. Relationships between environmental factors and pathogenic Vibrios in the northern Gulf of Mexico. Appl. Environ. Microbiol. 2010, 76, 7076–7084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tall, A.; Teillon, A.; Boisset, C.; Delesmont, R.; Touron-Bodilis, A.; Hervio-Heath, D. Real-time PCR optimization to identify environmental Vibrio spp. strains. J. Appl. Microbiol. 2012, 113, 361–372. [Google Scholar] [CrossRef] [Green Version]
- European Commission. DG (SANTE) 2015-7486—MR—Final Report of an Audit Carried Out in the United States from 17 March 2015 to 27 March 2015 in Order to Evaluate the Control Systems in Place Governing the Production of Bivalve Molluscs and Fishery Products Derived Therefrom Intended for Export to the European Union; European Commission: Brussels, Belgium, 2015. [Google Scholar]
- Harwood, V.J.; Gandhi, J.P.; Wright, A.C. Methods for isolation and confirmation of Vibrio vulnificus from oysters and environmental sources: A review. J. Microbiol. Methods 2004, 59, 301–316. [Google Scholar] [CrossRef]
- Zhao, F.; Zhao, D.; Cao, H.; Ma, L.; Jiang, Y. Distribution, serological and molecular characterization of Vibrio parahaemolyticus from shellfish in the eastern coast in China. Food Control 2011, 22, 1095–1100. [Google Scholar] [CrossRef]
- Abdullah Sani, N.; Ariyawansa, S.; Salam Babji, A.; Khair Hashim, J. The risk assessment of Vibrio parahaemolyticus in cooked black tiger shrimps (Penaeus monodon) in Malaysia. Food Control 2013, 31, 546–552. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Stockley, L.; Rangdale, R.; Martinez-Urtaza, J. Environmental occurrence and clinical impact of Vibrio vulnificus and Vibrio parahaemolyticus: A European perspective. Environ. Microbiol. Rep. 2010, 2, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; Trinanes, J.A.; Taylor, N.G.H.; Hartnell, R.; Siitonen, A.; Martinez-Urtaza, J. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat. Clim. Chang. 2012, 3, 73–77. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Trinanes, J.; Gonzalez-Escalona, N.; Martinez-Urtaza, J. Noncholera vibrios: The microbial barometer of climate change. Trends Microbiol. 2016, 25, 76–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Urtaza, J.; Baker-Austin, C.; Jones, J.; Newton, A.; Gonzalez-Aviles, G.; DePaola, A. Spread of Pacific Northwest Vibrio parahaemolyticus Strain. N. Engl. J. Med. 2013, 369, 1573–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scallan, E.; Hoekstra, R.; Angulo, F.; Tauxe, R.; Widdowson, M.; Roy, S.; Jones, J.; Griffin, P. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Hartnell, R.E.; Stockley, L.; Keay, W.; Rosec, J.P.; Hervio-Heath, D.; Van den Berg, H.; Leoni, F.; Ottaviani, D.; Henigman, U.; Denayer, S.; et al. A pan-European ring trial to validate an International Standard for detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus in seafoods. Int. J. Food Microbiol. 2019, 288, 58–65. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Roberts, N.; Singleton, F.L.; Attwell, R.W.; Grimes, D.J.; Colwell, R.R. Survival and viability of non culturable Escherichia coli and Vibrio cholerae in the estuarine and marine environment. Microb. Ecol. 1982, 8, 313–323. [Google Scholar] [CrossRef]
- Li, L.; Mendis, N.; Trigui, H.; Oliver, J.D.; Faucher, S.P. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 2014, 5, 258. [Google Scholar] [CrossRef] [Green Version]
- Meng, L.; Alter, T.; Aho, T.; Huehn, S. Gene expression profiles of Vibrio parahaemolyticus in viable but non-culturable state. FEMS Microbiol. Ecol. 2015, 91, 91. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef] [Green Version]
- Datta, P.P.; Bhadra, R.K. Cold shock response and major cold shock proteins of Vibrio cholerae. Appl. Environ. Microbiol. 2003, 69, 6361–6369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Limthammahisorn, S.; Brady, Y.; Arias, C. In vivo gene expression of cold shock and other stress-related genes in Vibrio vulnificus during shellstock temperature control conditions in oysters. J. Appl. Microbiol. 2009, 106, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Nowakowska, J.; Oliver, J.D. Resistance to environmental stresses by Vibrio vulnificus in the viable but nonculturable state. FEMS Microbiol. Ecol. 2013, 84, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Colwell, R.; Grimes, D. Nonculturable Microrganisms in the Environment; American Society for Microbiology Press: Washington, DC, USA, 2000; pp. 325–342. [Google Scholar]
- Baffone, W.; Citterio, B.; Vittoria, E.; Casaroli, A.; Campana, R.; Falzano, L.; Donelli, G. Retention of virulence in viable but non-culturable halophilic Vibrio spp. Int. J. Food Microbiol. 2003, 89, 31–39. [Google Scholar] [CrossRef]
- Falcioni, T.; Papa, S.; Campana, R.; Manti, A.; Battistelli, M.; Baffone, W. State transitions of Vibrio parahaemolyticus VBNC cells evaluated by flow cytometry. Cytom. B Clin. Cytom. 2008, 74, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Okoh, A.I.; Odjadjare, E.E.; Igbinosa, E.O.; Osode, A.N. Wastewater treatment plants as a source of microbial pathogens in receiving watersheds. Afr. J. Biotechnol. 2007, 6, 2932–2944. [Google Scholar]
- Moreno, Y.; Ballesteros, L.; García-Hernández, J.; Santiago, P.; González, A.; Ferrús, M. Specific detection of viable Listeria monocytogenes in Spanish wastewater treatment plants by Fluorescent in situ Hybridization and PCR. Water Res. 2011, 45, 4634–4640. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Ueno, D.; Inoue, K.; Someya, T. Direct Viable Count Combined with Fluorescence in situ Hybridization (DVC-FISH) for Specific Enumeration of Viable Escherichia coli in Cow Manure. Microbes Environ. 2009, 24, 33–38. [Google Scholar] [CrossRef] [Green Version]
- Frickmann, H.; Zautner, A.; Moter, A.; Kikhney, J.; Hagen, R.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef]
- Kogure, K.; Simidu, U.; Taga, N. A tentative direct microscopic method for counting living marine bacteria. Canad. J. Microb. 1979, 25, 415–420. [Google Scholar] [CrossRef]
- Servais, P.; Prats, J.; Passerat, J.; Garcia-Armisen, T. Abundance of culturable versus viable Escherichia coli in freshwater. Canad. J. Microb. 2009, 55, 905–909. [Google Scholar] [CrossRef]
- Duodu, S.; Colquhoun, D. Monitoring the survival of fish-pathogenic Francisella in water microcosms. FEMS Microbiol. Ecol. 2010, 74, 534–541. [Google Scholar] [CrossRef] [Green Version]
- Piqueres, P.; Moreno, Y.; Alonso, J.; Ferrús, M. A combination of direct viable count and fluorescent in situ hybridization for estimating Helicobacter pylori cell viability. Res. Microbiol. 2006, 157, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Kalmbach, S.; Manz, W.; Szewzyk, U. Isolation of new bacterial species from drinking water biofilms and proof of their in situ dominance with highly specific 16S rRNA probes. Appl. Environ. Microbiol. 1997, 63, 4164–4170. [Google Scholar] [CrossRef] [Green Version]
- Armisen, T.G.; Servais, P. Combining direct viable count (DVC) and fluorescent in situ hybridisation (FISH) to enumerate viable E-coli in rivers and wastewaters. Water Sci. Technol. 2004, 50, 271–275. [Google Scholar] [CrossRef]
- Baudart, J.; Coallier, J.; Laurent, P.; Prevost, M. Rapid and Sensitive Enumeration of Viable Diluted Cells of Members of the Family Enterobacteriaceae in Freshwater and Drinking Water. Appl. Environ. Microbiol. 2002, 68, 5057–5063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Hernández, J.; Moreno, Y.; Amorocho, C.M.; Hernández, M. A combination of direct viable count and fluorescence in situ hybridization for specific enumeration of viable Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus. Lett. Appl. Microbiol. 2011, 54, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Mesonero, L.; Moreno, Y.; Alonso, J.L.; Ferrús, M.A. DVC-FISH and PMA-qPCR techniques to assess the survival of Helicobacter pylori inside Acanthamoeba castellanii. Res. Microbiol. 2016, 167, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Barcina, I.; Arana, I.; Santoram, P.; Iriberri, J.; Egca, L. Direct viable count of Gram positive and Gram negative bacteria using ciprofloxacin as inhibitor of cellular division. J. Microbiol. Met. 1995, 22, 139–150. [Google Scholar] [CrossRef]
- Anupama, K.P.; Deeksh, A.K.; Deeksha, A.; Karunasagar, I.; Karunasagar, I.; Maiti, B. Comparative performance of TCBS and TSA for the enumeration of trh+ Vibrio parahaemolyticus by direct colony hybridization. J. Microbiol. Methods 2019, 157, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Sawabe, T.; Yoshizawa, A.; Kawanishi, Y.; Komatsu-Takeda, E.; Nakagawa, S.; Sawabe, T.; Ootubo, M.; Satomi, M.; Yano, Y.; Yamazaki, K. Multi-Probe-Fluorescence in situ Hybridization for the Rapid Enumeration of Viable Vibrio parahaemolyticus. Microbes Environ. 2009, 24, 259–264. [Google Scholar] [CrossRef] [Green Version]
- Bisha, B.; Simonson, J.; Janes, M.; Bauman, K.; Goodridge, L. A review of the current status of cultural and rapid detection of Vibrio parahaemolyticus. Int. J. Food Sci. Technol. 2012, 47, 885–899. [Google Scholar] [CrossRef]
- Amann, R.I.; ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Mol. Biol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [Green Version]
- Besnard, V.; Federighi, M.; Cappelier, J.M. Development of a direct viable count procedure for the investigation of VBNC state in Listeria monocytogenes. Lett. Appl. Microbiol. 2000, 31, 77–81. [Google Scholar] [CrossRef]
- Yoshimitsu, M.; Wai, S.N.; Ishikawa, T.; Takade, A.; Yoshida, S.-I. Resuscitation of viable but nonculturable cells of Vibrio parahaemolyticus induced at low temperature under starvation. FEMS Microbiol. Lett. 2000, 186, 115–120. [Google Scholar]
- Bej, A.K.; Patterson, D.P.; Brasher, C.W.; Vickery, M.; Jones, D.D.; Kaysner, C.A. Detection of total and haemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 1999, 36, 215–225. [Google Scholar] [CrossRef]
- Rohde, A.; Hammerl, J.A.; Appel, B.; Dieckmann, R.; Al Dahouk, S. Differential detection of pathogenic Yersinia spp. by fluorescence in situ hybridization. Food Microbiol. 2017, 62, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Joux, F.; Lebaron, P. Ecological implications of an improved direct viable count method for aquatic bacteria. Appl. Environ. Microbiol. 1997, 63, 3643–3647. [Google Scholar] [CrossRef] [Green Version]
- Kogure, K.; Simidu, U.; Taga, N. An improved direct viable count method for aquatic bacteria. Arch. Hydrobiol. 1984, 102, 117–122. [Google Scholar]
- Bakermans, C.; Madsen, E.L. Use of substrate responsive-direct viable counts to visualize naphthalene degrading bacteria in a coal tar-contaminated groundwater microbial community. J. Microbiol. Methods 2000, 43, 81–90. [Google Scholar] [CrossRef]
- Garcia-Armisen, T.; Servais, P. Enumeration of viable E. coli in rivers and wastewaters by fluorescent in situ hybridization. J. Microbiol. Methods 2004, 58, 269–279. [Google Scholar] [CrossRef]
- Kenzaka, T.; Yamaguchi, N.; Prapagdee, B.; Mikami, E.; Nasu, M. Bacterial community composition and activity in urban rivers in Thailand and Malaysia. J. Health Sci. 2001, 47, 353–361. [Google Scholar] [CrossRef] [Green Version]
- Bjergbaek, L.; Roslev, P. Formation of nonculturable Escherichia coli in drinking water. J. Appl. Microbiol. 2005, 99, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Guyard, S.; Mary, P.; Defives, C.; Hornez, J.P. Enumeration and characterization of bacteria in mineral water by improved direct viable count method. J. Appl. Microbiol. 1999, 86, 841–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, Y.; Sanchez-Contreras, J.; Montes, R.M.; García-Hernández, J.; Ballesteros, L.; Ferrús, M.A. Detection and enumeration of viable Listeria monocytogenes cells from ready-to-eat and processed vegetable foods by culture and DVC-FISH. Food Control 2012, 27, 374–379. [Google Scholar] [CrossRef]
- García Hernández, J.; Moreno Trigos, Y.; Hernández Pérez, M. In Vivo Study of the Survival of Lactobacillus delbruecki subsp. bulgaricus CECT 4005T and Streptococcus thermophilus CECT 801 by DVC-FISH after Consumption of Fermented Milk. J. Food Sci. 2012, 77, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Laupland, K.; Valiquette, L. The changing culture of the microbiology laboratory. Can. J. Infect. Dis Med. Microbiol. 2013, 24, 125–128. [Google Scholar] [CrossRef]
- Regnault, B.; Martin-Delautre, S.; Lejay-Collin, M.; Lefèvre, M.; Grimont, P.A.D. Oligonucleotide probe for the visualization of Escherichia coli/Escherichia fergusonii cells by in situ hybridation: Specificity and potential applications. Res. Microbiol. 2000, 151, 521–533. [Google Scholar] [CrossRef]
- Moreno, Y.; Ferrús, M.A. Specific detection of cultivable Helicobacter pylori cells from wastewater treatment plants. Helicobacter 2012, 17, 327–332. [Google Scholar] [CrossRef]
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, Y.; Hamano, K.; Satomi, M.; Tsutsui, I.; Aue-umneoy, D. Diversity and characterization of oxytetracycline-resistant bacteria associated with non-native species, white-leg shrimp (Litopenaeus vannamei), and native species, black tiger shrimp (Penaeus monodon), intensively cultured in Thailand. J. Appl. Microbiol. 2014, 110, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Akinbowale, O.L.; Peng, H.; Barton, M.D. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 2006, 100, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Baker-Austin, C.; McArthur, J.V.; Lindell, A.H.; Wright, M.F.; Tuckfield, R.C.; Gooch, J.; Warner, L.; Oliver, J.; Stepanauskas, R. Multi-site Analysis Reveals Widespread Antibiotic Resistance in the Marine Pathogen Vibrio vulnificus. Microb. Ecol. 2009, 57, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Walker, R.D.; Janes, M.E.; Prinyawiwatkul, W.; Ge, B. Antimicrobial Susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus Isolates from Louisiana Gulf and Retail Raw Oysters. Appl. Environ. Microbiol. 2007, 73, 7096–7098. [Google Scholar] [CrossRef] [Green Version]
- Huber, D.; von Voithenberg, V.L.; Kaigala, G.V. Fluorescence in situ hybridization (FISH): History, limitations and what to expect from micro-scale FISH? Micro Nano Eng. 2018, 1, 15–24. [Google Scholar] [CrossRef]
- Rocha, R.; Almeida, C.; Azevedo, N.F. Influence of the fixation/permeabilization step on peptide nucleic acid fluorescence in situ hybridization (PNA-FISH) for the detection of bacteria. PLoS ONE 2018, 13, e0196522. [Google Scholar]
- Locke, A.; Fitzgerald, S.; Mahadevan-Jansen, A. Advances in Optical Detection of Human-Associated Pathogenic Bacteria. Molecules 2020, 25, 5256. [Google Scholar] [CrossRef]
- García, E.; Hernández, A.; Herrero, J.A.; Gómez, J. Cholera and other infections of the Vibrio genus. Medicine 2014, 11, 3317–3321. [Google Scholar]
- Qadri, F.; Chowdhury, N.R.; Takeda, Y.; Nair, G.B. Vibrio parahaemolyticus—seafood safety and associations with higher organisms. In Oceans and Health: Pathogens in the Marine Environment; Springer: Boston, MA, USA, 2005; pp. 277–295. [Google Scholar]
- Letchumanan, V.; Chan, K.G.; Lee, L.H. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 2014, 5, 705. [Google Scholar] [CrossRef] [Green Version]
- Love, D.; Lane, R.; Davis, B.; Clancy, K.; Fry, J.; Harding, J.; Hudson, B. Performance of Cold Chains for Chesapeake Bay Farmed Oysters and Modeled Growth of Vibrio parahaemolyticus. J. Food Prot. 2019, 82, 168–178. [Google Scholar] [CrossRef] [PubMed]
- López-Joven, C.; de Blas, I.; Roque, A. Temperature effects on the growth and survival of tdh positive Vibrio parahaemolyticus in tissues of postharvest Manila clam (Ruditapes philippinarum). Food Microbiol. 2017, 75, 61–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, J.L.; Lydon, K.A.; Kinsey, T.P.; Friedman, B.; Curtis, R.; Schuster, R.; Bowers, J.C. Vibrio parahaemolyticus abundances in oysters. Int. J. Food Microbiol. 2017, 253, 54–58. [Google Scholar] [CrossRef]
- Ndraha, N.; Wong, H.; Hsiao, H. Managing the risk of Vibrio parahaemolyticus infections associated with oyster consumption: A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1187–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W. The Identification and Detection Technology of Research in Microorganisms Including Living or Dead Bacteria. In Functional Nucleic Acids Detection in Food Safety; Springer: Singapore, 2016; pp. 343–364. [Google Scholar]
- Ju, W.; Moyne, A.; Marco, M.L. RNA-based detection does not accurately enumerate living Escherichia coli O157:H7 cells on plants. Front. Microbiol. 2016, 7, 1–9. [Google Scholar] [CrossRef] [Green Version]



| Time | Viable (SYTO) (cells/mL) | Not Viable (PI) (cells/mL) | mTSA Plate Count (cfu/mL) | FISH (cells/mL) |
|---|---|---|---|---|
| Initial culture (0 h) | 9.6 × 106 | 3.4 × 106 | 1.4 × 107 | 1.5 × 107 |
| DVC (5 h) | 2.4 × 107 | 3.3 × 107 | 1.7 × 107 | 3.2 × 107 |
| Control (5 h) | 6.6 × 107 | 2.6 × 107 | 7.3 × 107 | 1.3 × 108 |
| Time (Minutes) | mTSA Plate Count (cfu/mL) | FISH (cells/mL) | DVC-FISH (cells/mL) | |||
|---|---|---|---|---|---|---|
| Oysters | Mussels | Oysters | Mussels | Oysters | Mussels | |
| 0 | 1.70 × 107 | 1.30 × 106 | 1.80 × 107 | 9.70 × 106 | 1.70 × 107 | 4.90 × 106 |
| 10 | 7.00 × 106 | 6.90 × 106 | 8.70 × 106 | 2.10 × 107 | 8.10 × 106 | 9.70 × 106 |
| 20 | 8.80 × 106 | 9.70 × 106 | 1.40 × 107 | 4.00 × 107 | 9.40 × 106 | 2.80 × 107 |
| Time (Hours) | mTSA Plate Count (cfu/mL) | FISH (cells/mL) | DVC-FISH (cells/mL) | |||
|---|---|---|---|---|---|---|
| Oysters | Mussels | Oysters | Mussels | Oysters | Mussels | |
| 0 | 9.50 × 106 | 1.65 × 107 | 1.80 × 107 | 1.97 × 108 | 7.04 × 106 | 2.23 × 107 |
| 1 | 6.24 × 106 | 1.02 × 107 | 6.09 × 106 | 1.57 × 108 | 3.93 × 106 | 1.74 × 107 |
| 5 | 2.00 × 105 | 8.10 × 105 | 1.78 × 106 | 8.54 × 107 | 1.36 × 106 | 8.54 × 106 |
| 24 | 1.57 × 104 | 2.06 × 105 | 8.80 × 105 | 9.76 × 106 | 1.40 × 105 | 1.44 × 106 |
| 48 | <10 | 4.50 × 103 | 1.68 × 104 | 3.50 × 105 | 6.40 × 103 | 7.80 × 104 |
| 96 | <10 | <10 | 1.30 × 103 | 6.50 × 104 | 6.20 × 102 | 7.30 × 103 |
| Time (Hours) | mTSA Plate Count (cfu/mL) | FISH (cells/mL) | DVC-FISH (cells/mL) | |||
|---|---|---|---|---|---|---|
| Oysters | Mussels | Oysters | Mussels | Oysters | Mussels | |
| 0 | 1.65 × 108 | 1.78 × 107 | 1.90 × 108 | 7.80 × 108 | 2.00 × 108 | 1.83 × 107 |
| 1 | 1.03 × 108 | 1.50 × 106 | 1.41 × 108 | 7.19 × 107 | 1.61 × 108 | 7.32 × 106 |
| 5 | 8.50 × 106 | 1.70 × 105 | 4.80 × 107 | 6.34 × 107 | 1.31 × 107 | 1.44 × 106 |
| 24 | 2.26 × 106 | 7.70 × 104 | 6.30 × 106 | 3.53 × 107 | 2.40 × 106 | 1.22 × 106 |
| 48 | 3.50 × 105 | 4.70 × 104 | 1.30 × 106 | 7.32 × 106 | 8.30 × 105 | 1.30 × 105 |
| 96 | 1.98 × 104 | 1.10 × 104 | 2.11 × 105 | 1.22 × 106 | 7.60 × 104 | 8.20 × 104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Hernández, J.; Hernández, M.; Moreno, Y. Combination of Direct Viable Count and Fluorescent In Situ Hybridization (DVC-FISH) as a Potential Method for Identifying Viable Vibrio parahaemolyticus in Oysters and Mussels. Foods 2021, 10, 1502. https://doi.org/10.3390/foods10071502
García-Hernández J, Hernández M, Moreno Y. Combination of Direct Viable Count and Fluorescent In Situ Hybridization (DVC-FISH) as a Potential Method for Identifying Viable Vibrio parahaemolyticus in Oysters and Mussels. Foods. 2021; 10(7):1502. https://doi.org/10.3390/foods10071502
Chicago/Turabian StyleGarcía-Hernández, Jorge, Manuel Hernández, and Yolanda Moreno. 2021. "Combination of Direct Viable Count and Fluorescent In Situ Hybridization (DVC-FISH) as a Potential Method for Identifying Viable Vibrio parahaemolyticus in Oysters and Mussels" Foods 10, no. 7: 1502. https://doi.org/10.3390/foods10071502
APA StyleGarcía-Hernández, J., Hernández, M., & Moreno, Y. (2021). Combination of Direct Viable Count and Fluorescent In Situ Hybridization (DVC-FISH) as a Potential Method for Identifying Viable Vibrio parahaemolyticus in Oysters and Mussels. Foods, 10(7), 1502. https://doi.org/10.3390/foods10071502






