Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Starting Coordinates of Rosemary Compounds
2.2. In Silico Determination of ADME Properties
2.3. Inverse Molecular Docking
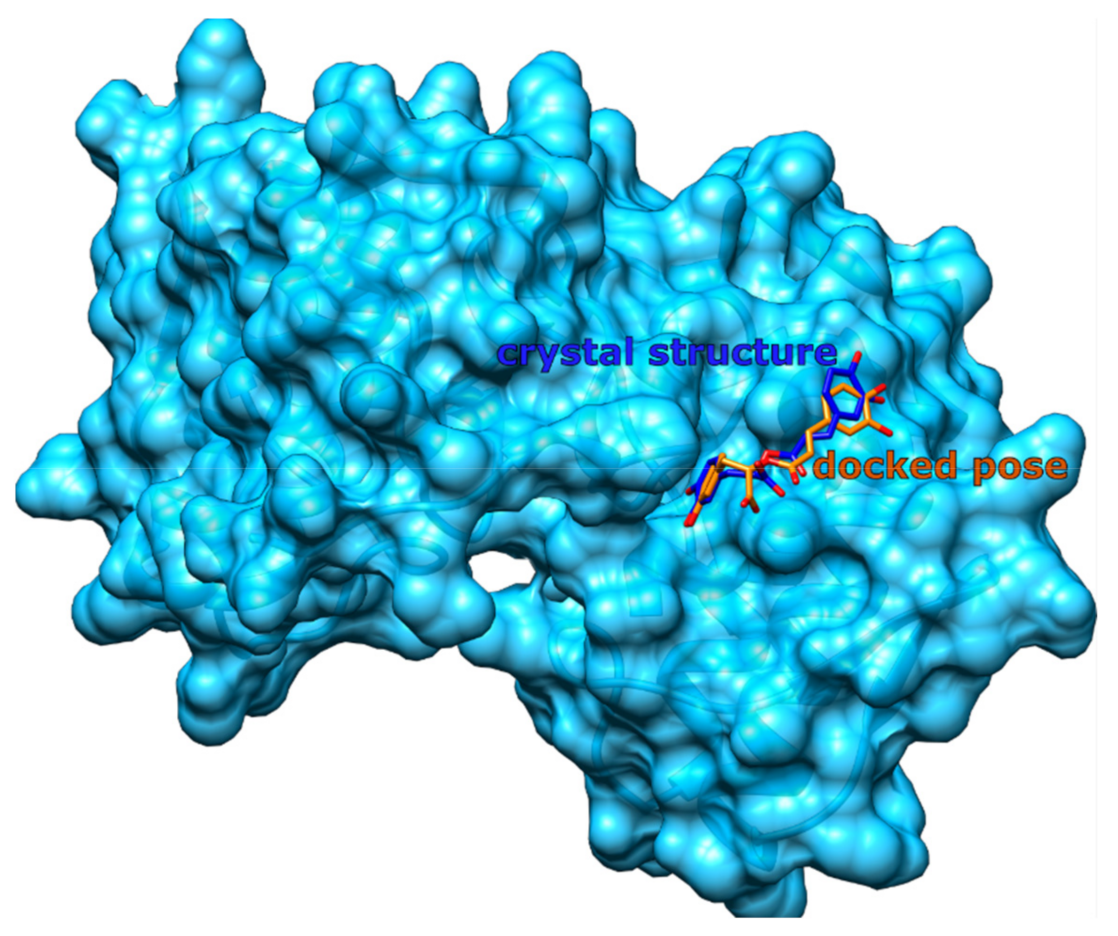
2.4. Method Validation
3. Results
3.1. Inverse Molecular Docking of Diterpenes
3.1.1. K-Ras
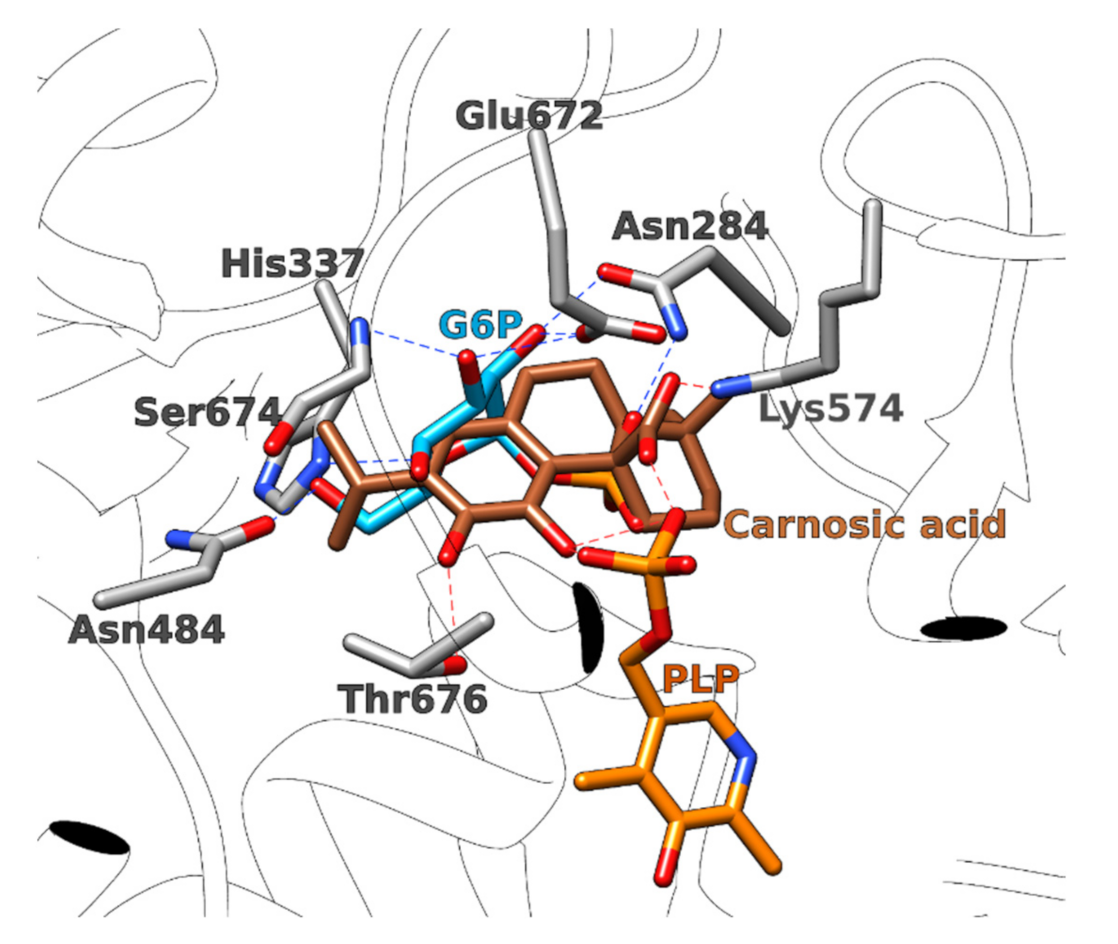
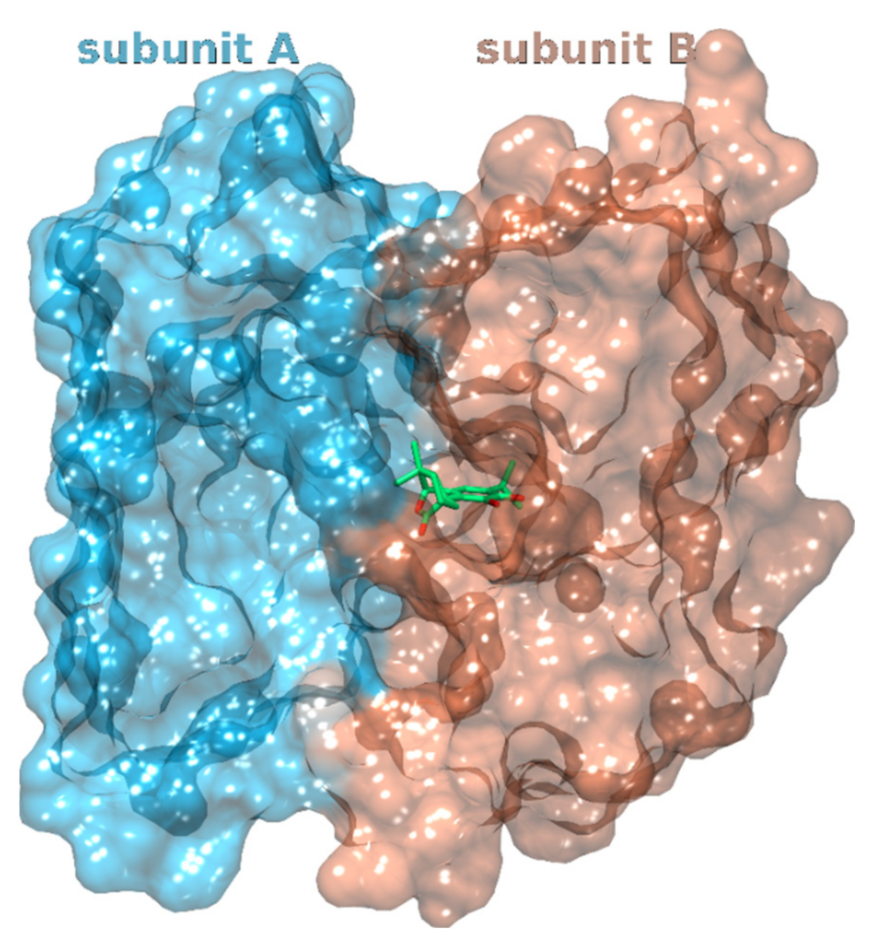
3.1.2. Glucosamine/Fructose-6-Phosphate Aminotransferase
3.1.3. Pyruvate Kinase 2–Muscle Isoform
3.1.4. Hemagglutinin HA1
3.1.5. HIV-1 and HIV-2 Protease
3.1.6. Enhanced Intra-Cellular Survival Protein
3.1.7. Peroxisome Proliferator-Activated Receptor δ
3.1.8. Glycogen Phosphorylase
3.1.9. Tubulin
3.1.10. Phosholipase A2
3.1.11. Vascular Endothelial Growth Factor Receptor 2
3.1.12. Aspartate Carbamoyltransferase
3.2. Inverse Docking of Rosmarinic Acid
3.2.1. Coagulation Factor X
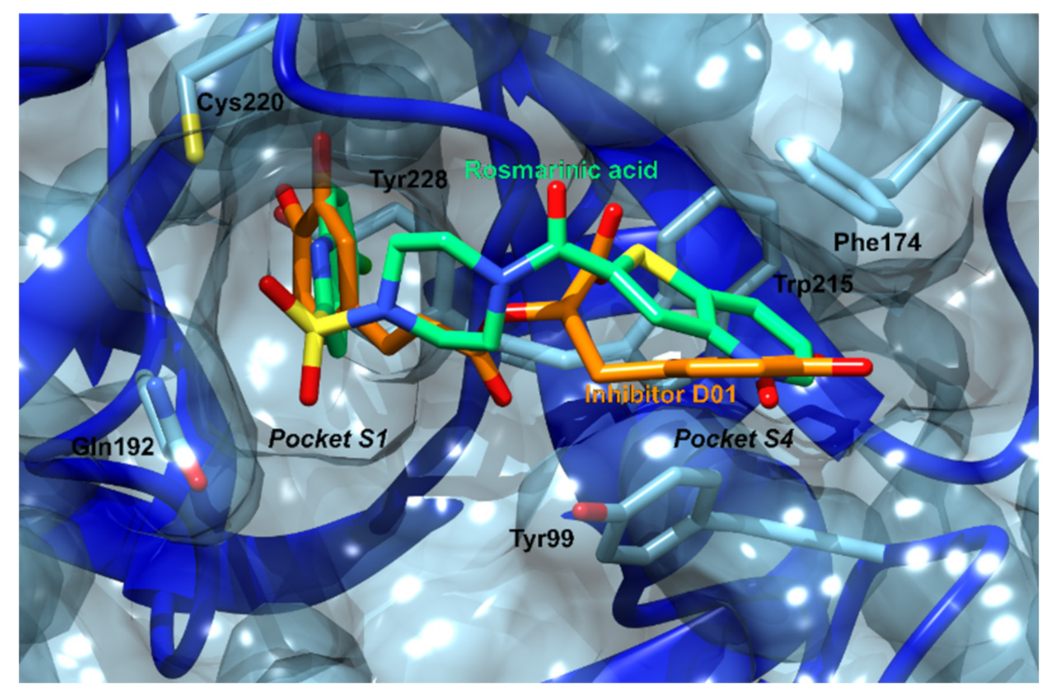
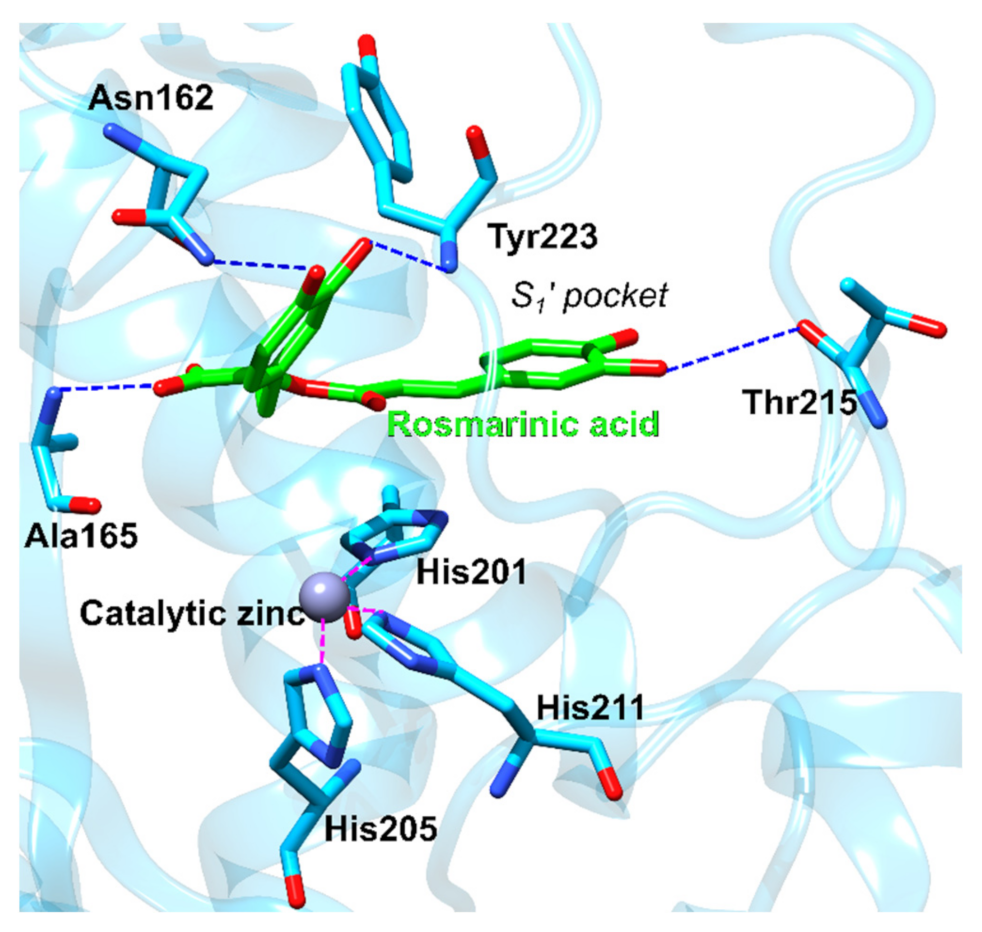
3.2.2. Phospholipase A2
3.2.3. Matrix Metalloproteinase-3
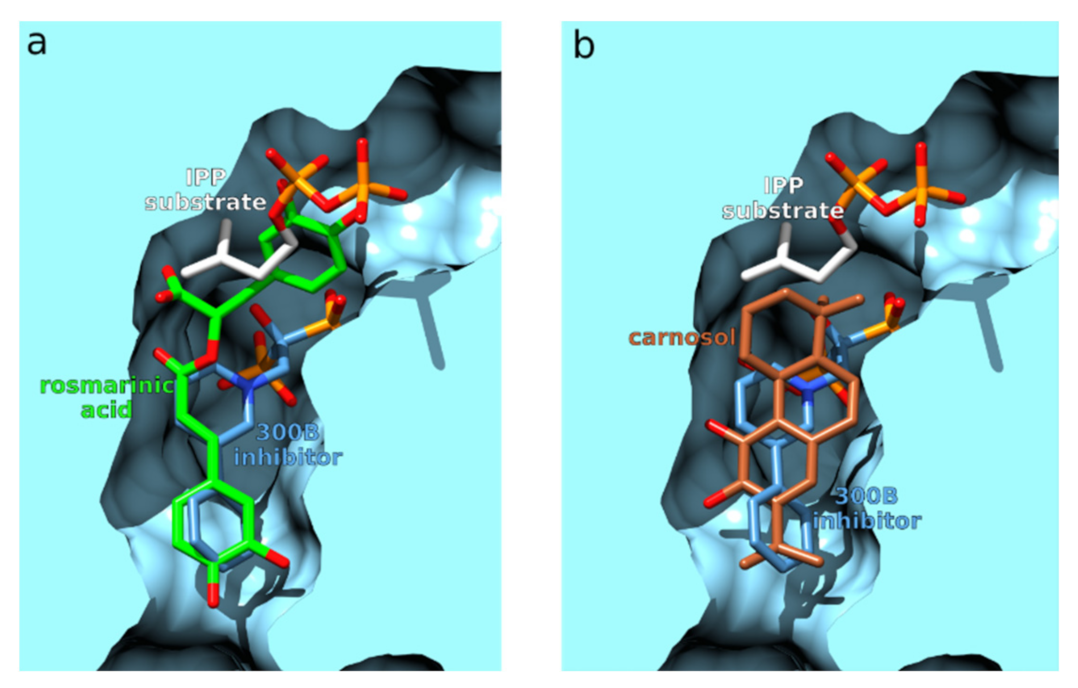
3.2.4. Farnesyl Pyrophosphate Synthase
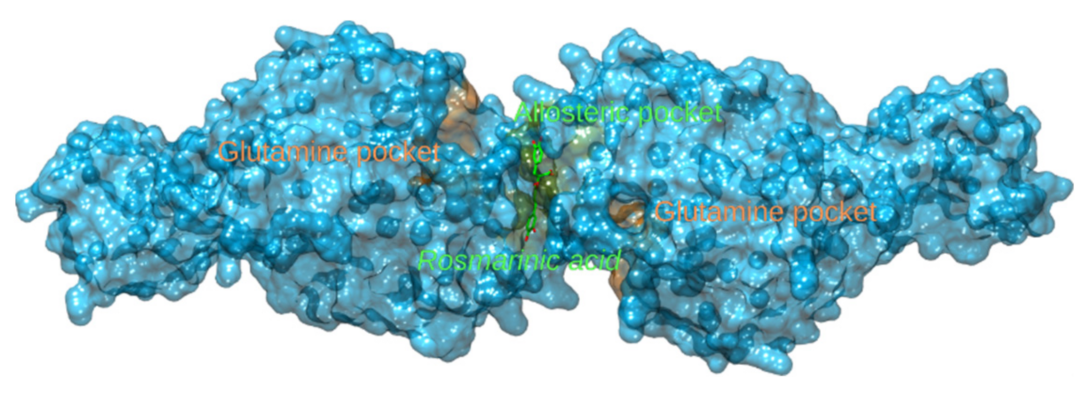
3.2.5. Glutamate Dehydrogenase 1 and Glutaminase
3.3. Method Validation
3.3.1. Redocking Procedure
3.3.2. Validation Using ROC, EF, and PC
3.4. In Silico Prediction of Pharmacokinetic Properties
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SMILES | Simplified Molecular-Input Line-Entry System |
| CANDOCK | Chemical Atomic Network based Docking |
| CHARMM | Chemistry at Harvard Macromolecular Mechanics |
| ROC | Receiver operating characteristics curve |
| PC | Predictiveness curve |
| TPF | True positive fraction |
| FPF | False positive fraction |
| PDB | Protein Data Bank |
| ROC AUC | Area under the receiver operating characteristics curve |
| EF | Enrichment factor |
| BEDROC | Boltzmann-enhanced discrimination of ROC |
| RIE | Robust initial enhancement |
| TG | Total gain |
| Eis | Enhanced intracellular survival |
| HIV | Human immunodeficiency virus |
| FIV | Feline immunodeficiency virus |
| RAS/MAPK | Rat sarcoma/mitogen-activated protein kinase |
| GTP | Guanosine triphosphate |
| GlmS | Glucosamine/fructose-6-phosphate aminotransferase |
| PKM | Pyruvate kinase M |
| PEP | Phosphoenolpyruvate |
| ADP | Adenosine diphosphate |
| ATP | Adenosine triphosphate |
| HA | Hemagglutinin |
| AIDS | Acquired immunodeficiency syndrome |
| HAART | Highly active antiretroviral therapy |
| PPAR | Peroxisome proliferator-activated receptor |
| COX-2 | Cyclooxygenase-2 |
| EC50 | Half maximal effective concentration |
| GP | Glycogen phosphorylase |
| PLA2 | Phospholipase A2 |
| VEGFR | Vascular endothelial growth factor receptor |
| VEGF | Vascular endothelial growth factor |
| MMP | Matrix metalloproteinase |
| FPPS | Farnesyl pyrophosphate synthase |
| IPP | Isopentenyl pyrophosphate |
| GDH1 | Glutamate dehydrogenase 1 |
| RMSD | Root-mean-square deviation |
References
- Begum, A.; Sandhya, S.; Ali, S.S.; Vinod, K.R.; Reddy, S.; Banji, D. An in-depth review on the medicinal flora Rosmarinus officinalis (Lamiaceae). Acta Sci. Pol. Technol. Aliment. 2013, 12, 61–74. [Google Scholar]
- Al-Sereiti, M.R.; Abu-Amer, K.M.; Sena, P. Pharmacology of rosemary (Rosmarinus officinalis Linn.) and its therapeutic potentials. IJEB 1999, 37, 124–130. [Google Scholar]
- Perez-Fons, L.; Garzon, M.T.; Micol, V. Relationship between the antioxidant capacity and effect of rosemary (Rosmarinus officinalis L.) polyphenols on membrane phospholipid order. J. Agric. Food Chem. 2009, 58, 161–171. [Google Scholar] [CrossRef]
- Yu, M.-H.; Choi, J.-H.; Chae, I.-G.; Im, H.-G.; Yang, S.-A.; More, K.; Lee, I.-S.; Lee, J. Suppression of LPS-induced inflammatory activities by Rosmarinus officinalis L. Food Chem. 2013, 136, 1047–1054. [Google Scholar] [CrossRef]
- Bakırel, T.; Bakırel, U.; Keleş, O.Ü.; Ülgen, S.G.; Yardibi, H. In vivo assessment of antidiabetic and antioxidant activities of rosemary (Rosmarinus officinalis) in alloxan-diabetic rabbits. J. Ethnopharmacol. 2008, 116, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Bozin, B.; Mimica-Dukic, N.; Samojlik, I.; Jovin, E. Antimicrobial and antioxidant properties of rosemary and sage (Rosmarinus officinalis L. and Salvia officinalis L., Lamiaceae) essential oils. J. Agric. Food Chem. 2007, 55, 7879–7885. [Google Scholar] [CrossRef]
- Tai, J.; Cheung, S.; Wu, M.; Hasman, D. Antiproliferation effect of Rosemary (Rosmarinus officinalis) on human ovarian cancer cells in vitro. Phytomedicine 2012, 19, 436–443. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.; García-Cañas, V.; Rocamora-Reverte, L.; Gómez-Martínez, Á.; Ferragut, J.A.; Cifuentes, A. Effect of rosemary polyphenols on human colon cancer cells: Transcriptomic profiling and functional enrichment analysis. Genes Nutr. 2013, 8, 43–60. [Google Scholar] [CrossRef] [Green Version]
- Yesil-Celiktas, O.; Sevimli, C.; Bedir, E.; Vardar-Sukan, F. Inhibitory effects of rosemary extracts, carnosic acid and rosmarinic acid on the growth of various human cancer cell lines. Plant Foods Hum. Nutr. 2010, 65, 158–163. [Google Scholar] [CrossRef]
- Singletary, K.; MacDonald, C.; Wallig, M. Inhibition by rosemary and carnosol of 7,12-dimethylbenz[a]anthracene (DMBA)-induced rat mammary tumorigenesis and in vivo DMBA-DNA adduct formation. Cancer Lett. 1996, 104, 43–48. [Google Scholar] [CrossRef]
- Huang, M.-T.; Ho, C.-T.; Wang, Z.Y.; Ferraro, T.; Lou, Y.-R.; Stauber, K.; Ma, W.; Georgiadis, C.; Laskin, J.D.; Conney, A.H. Inhibition of Skin Tumorigenesis by Rosemary and Its Constituents Carnosol and Ursolic Acid. Cancer Res. 1994, 54, 701–708. [Google Scholar]
- Lešnik, S.; Furlan, V.; Bren, U. Rosemary (Rosmarinus officinalis L.): Extraction techniques, analytical methods and health-promoting biological effects. Phytochem. Rev. 2021, 20, 1273–1328. [Google Scholar] [CrossRef]
- Okamura, N.; Fujimoto, Y.; Kuwabara, S.; Yagi, A. High-performance liquid chromatographic determination of carnosic acid and carnosol in Rosmarinus officinalis and Salvia officinalis. J. Chromatogr. A 1994, 679, 381–386. [Google Scholar] [CrossRef]
- Johnson, J.J. Carnosol: A promising anti-cancer and anti-inflammatory agent. Cancer Lett. 2011, 305, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Inaba, Y.; Maekawa, T.; Takeda, Y.; Tamura, H.; Yamaguchi, H. Recovery Mechanism of the Antioxidant Activity from Carnosic Acid Quinone, an Oxidized Sage and Rosemary Antioxidant. J. Agric. Food Chem. 2002, 50, 5863–5869. [Google Scholar] [CrossRef]
- Collins, M.A.; Charles, H.P. Antimicrobial activity of Carnosol and Ursolic acid: Two anti-oxidant constituents of Rosmarinus officinalis L. Food Microbiol. 1987, 4, 311–315. [Google Scholar] [CrossRef]
- Shin, H.-B.; Choi, M.-S.; Ryu, B.; Lee, N.-R.; Kim, H.-I.; Choi, H.-E.; Chang, J.; Lee, K.-T.; Jang, D.S.; Inn, K.-S. Antiviral activity of carnosic acid against respiratory syncytial virus. Virol. J. 2013, 10, 303. [Google Scholar] [CrossRef] [Green Version]
- Pukl, M.; Umek, A.; Pariš, A.; Štrukelf, B.; Renko, M.; Korant, B.D.; Turk, V. Inhibitory effect of carnosolic acid on HIV-1 protease. Planta Med. 1992, 58, 632. [Google Scholar] [CrossRef]
- Bai, N.; He, K.; Roller, M.; Lai, C.-S.; Shao, X.; Pan, M.-H.; Ho, C.-T. Flavonoids and Phenolic Compounds from Rosmarinus officinalis. J. Agric. Food Chem. 2010, 58, 5363–5367. [Google Scholar] [CrossRef]
- Kuhlmann, A.; Röhl, C. Phenolic Antioxidant Compounds Produced by in Vitro. Cultures of Rosemary (Rosmarinus officinalis) and Their Anti-inflammatory Effect on Lipopolysaccharide-Activated Microglia. Pharm. Biol. 2006, 44, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Visanji, J.M.; Thompson, D.G.; Padfield, P.J. Induction of G2/M phase cell cycle arrest by carnosol and carnosic acid is associated with alteration of cyclin A and cyclin B1 levels. Cancer Lett. 2006, 237, 130–136. [Google Scholar] [CrossRef] [PubMed]
- González-Vallinas, M.; Molina, S.; Vicente, G.; Zarza, V.; Martín-Hernández, R.; García-Risco, M.R.; Fornari, T.; Reglero, G.; De Molina, A.R. Expression of MicroRNA-15b and the Glycosyltransferase GCNT3 Correlates with Antitumor Efficacy of Rosemary Diterpenes in Colon and Pancreatic Cancer. PLoS ONE 2014, 9, e98556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, Q.; Ma, Y.; Dong, J.; Shen, R. Carnosic acid induces apoptosis associated with mitochondrial dysfunction and Akt inactivation in HepG2 cells. Int. J. Food Sci. Nutr. 2015, 66, 76–84. [Google Scholar] [CrossRef]
- Kar, S.; Palit, S.; Ball, W.B.; Das, P.K. Carnosic acid modulates Akt/IKK/NF-κB signaling by PP2A and induces intrinsic and extrinsic pathway mediated apoptosis in human prostate carcinoma PC-3 cells. Apoptosis 2012, 17, 735–747. [Google Scholar] [CrossRef]
- Barni, M.V.; Carlini, M.J.; Cafferata, E.G.; Puricelli, L.; Moreno, S. Carnosic acid inhibits the proliferation and migration capacity of human colorectal cancer cells. Oncol. Rep. 2012, 27, 1041–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aliebrahimi, S.; Kouhsari, S.M.; Arab, S.S.; Shadboorestan, A.; Ostad, S.N. Phytochemicals, withaferin A and carnosol, overcome pancreatic cancer stem cells as c-Met inhibitors. Biomed. Pharmacother. 2018, 106, 1527–1536. [Google Scholar] [CrossRef]
- Lo, A.-H.; Liang, Y.-C.; Lin-Shiau, S.-Y.; Ho, C.-T.; Lin, J.-K. Carnosol, an antioxidant in rosemary, suppresses inducible nitric oxide synthase through down-regulating nuclear factor-κB in mouse macrophages. Carcinogenesis 2002, 23, 983–991. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.-C.; Lee, M.-F.; Tsai, M.-L.; Lai, C.-S.; Lee, J.H.; Ho, C.-T.; Pan, M.-H. Rosmanol potently induces apoptosis through both the mitochondrial apoptotic pathway and death receptor pathway in human colon adenocarcinoma COLO 205 cells. Food Chem. Toxicol. 2011, 49, 485–493. [Google Scholar] [CrossRef]
- Machado, D.G.; Cunha, M.P.; Neis, V.B.; Balen, G.O.; Colla, A.; Bettio, L.E.B.; Oliveira, Á.; Pazini, F.L.; Dalmarco, J.B.; Simionatto, E.L.; et al. Antidepressant-like effects of fractions, essential oil, carnosol and betulinic acid isolated from Rosmarinus officinalis L. Food Chem. 2013, 136, 999–1005. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, K.; El Omri, A.; Kondo, S.; Han, J.; Isoda, H. Rosmarinus officinalis polyphenols produce anti-depressant like effect through monoaminergic and cholinergic functions modulation. Behav. Brain Res. 2013, 238, 86–94. [Google Scholar] [CrossRef]
- Romo Vaquero, M.; Garcia Villalba, R.; Larrosa, M.; Yáñez-Gascón, M.J.; Fromentin, E.; Flanagan, J.; Roller, M.; Tomás-Barberán, F.A.; Espín, J.C.; García-Conesa, M.-T. Bioavailability of the major bioactive diterpenoids in a rosemary extract: Metabolic profile in the intestine, liver, plasma, and brain of Zucker rats. Mol. Nutr. Food Res. 2013, 57, 1834–1846. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.R. The dietary components carnosic acid and carnosol as neuroprotective agents: A mechanistic view. Mol. Neurobiol. 2016, 53, 6155–6168. [Google Scholar] [CrossRef]
- Rasoolijazi, H.; Azad, N.; Joghataei, M.; Kerdari, M.; Nikbakht, F.; Soleimani, M. The protective role of carnosic acid against beta-amyloid toxicity in rats. Sci. World J. 2013, 2013, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M. Rosmarinic acid: New aspects. Phytochem. Rev. 2013, 12, 207–227. [Google Scholar] [CrossRef]
- De Souza Gil, E.; Adrian Enache, T.; Maria Oliveira-Brett, A. Redox behaviour of verbascoside and rosmarinic acid. Comb. Chem. High Throughput Screen. 2013, 16, 92–97. [Google Scholar] [CrossRef]
- Fadel, O.; El Kirat, K.; Morandat, S. The natural antioxidant rosmarinic acid spontaneously penetrates membranes to inhibit lipid peroxidation in situ. Biochim. Biophys. Acta 2011, 1808, 2973–2980. [Google Scholar] [CrossRef] [Green Version]
- Kimura, Y.; Okuda, H.; Okuda, T.; Hatano, T.; Arichi, S. Studies on the activities of tannins and related compounds, X. Effects of caffeetannins and related compounds on arachidonate metabolism in human polymorphonuclear leukocytes. J. Nat. Prod. 1987, 50, 392–399. [Google Scholar] [CrossRef]
- Lucarini, R.; Bernardes, W.A.; Ferreira, D.S.; Tozatti, M.G.; Furtado, R.; Bastos, J.K.; Pauletti, P.M.; Januário, A.H.; Silva, M.L.A.; Cunha, W.R. In vivo analgesic and anti-inflammatory activities of Rosmarinus officinalis aqueous extracts, rosmarinic acid and its acetyl ester derivative. Pharm. Biol. 2013, 51, 1087–1090. [Google Scholar] [CrossRef] [Green Version]
- Amaral, G.P.; Mizdal, C.R.; Stefanello, S.T.; Mendez, A.S.L.; Puntel, R.L.; de Campos, M.M.A.; Soares, F.A.A.; Fachinetto, R. Antibacterial and antioxidant effects of Rosmarinus officinalis L. extract and its fractions. J. Tradit. Complement. Med. 2019, 9, 383–392. [Google Scholar] [CrossRef]
- Radziejewska, I.; Supruniuk, K.; Nazaruk, J.; Karna, E.; Poplawska, B.; Bielawska, A.; Galicka, A. Rosmarinic acid influences collagen, MMPs, TIMPs, glycosylation and MUC1 in CRL-1739 gastric cancer cell line. Biomed. Pharmacother. 2018, 107, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.-J.; Yan, H.; Wang, Y.-J.; Yang, Y.; Li, X.-B.; Shi, A.-C.; Jing-Wen, X.; Yu-Bao, L.; Li, L.; Wang, X.-X. Proteomics analysis demonstrating rosmarinic acid suppresses cell growth by blocking the glycolytic pathway in human HepG2 cells. Biomed. Pharmacother. 2018, 105, 334–349. [Google Scholar] [CrossRef]
- Cui, H.-Y.; Zhang, X.-J.; Yang, Y.; Zhang, C.; Zhu, C.-H.; Miao, J.-Y.; Chen, R. Rosmarinic acid elicits neuroprotection in ischemic stroke via Nrf2 and heme oxygenase 1 signaling. Neural Regen. Res. 2018, 13, 2119–2128. [Google Scholar]
- Wang, J.; Li, G.; Rui, T.; Kang, A.; Li, G.; Fu, T.; Li, J.; Di, L.; Cai, B. Pharmacokinetics of rosmarinic acid in rats by LC-MS/MS: Absolute bioavailability and dose proportionality. RSC Adv. 2017, 7, 9057–9063. [Google Scholar] [CrossRef] [Green Version]
- da Silva, S.B.; Amorim, M.; Fonte, P.; Madureira, R.; Ferreira, D.; Pintado, M.; Sarmento, B. Natural extracts into chitosan nanocarriers for rosmarinic acid drug delivery. Pharm. Biol. 2015, 53, 642–652. [Google Scholar] [CrossRef] [Green Version]
- Madureira, A.R.; Campos, D.A.; Fonte, P.; Nunes, S.; Reis, F.; Gomes, A.M.; Sarmento, B.; Pintado, M.M. Characterization of solid lipid nanoparticles produced with carnauba wax for rosmarinic acid oral delivery. RSC Adv. 2015, 5, 22665–22673. [Google Scholar] [CrossRef]
- Xu, X.; Huang, M.; Zou, X. Docking-based inverse virtual screening: Methods, applications, and challenges. Biochem. Biophys. Rep. 2018, 4, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Warrier, S.B.; Kharkar, P.S. Inverse Virtual Screening in Drug Repositioning: Detailed Investigation and Case Studies. In Crystallizing Ideas–The Role of Chemistry; Springer: Berlin/Heidelberg, Germany, 2016; pp. 71–83. [Google Scholar]
- Konc, J. Identification of neurological disease targets of natural products by computational screening. Neural Regen. Res. 2019, 14, 2075. [Google Scholar] [CrossRef]
- Kores, K.; Lešnik, S.; Bren, U.; Janežič, D.; Konc, J. Discovery of novel potential human targets of resveratrol by inverse molecular docking. J. Chem. Inf. Model. 2019, 59, 2467–2478. [Google Scholar] [CrossRef] [PubMed]
- Furlan, V.; Konc, J.; Bren, U. Inverse molecular docking as a novel approach to study anticarcinogenic and anti-neuroinflammatory effects of curcumin. Molecules 2018, 23, 3351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterling, T.; Irwin, J.J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Petersson, G.; Nakatsuji, H. Gaussian 16; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Duraán-Iturbide, N.A.; Díaz-Eufracio, B.r.I.; Medina-Franco, J.L. In silico ADME/Tox profiling of natural products: A focus on BIOFACQUIM. ACS Omega 2020, 5, 16076–16084. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Fine, J.; Konc, J.; Samudrala, R.; Chopra, G. Candock: Chemical atomic network-based hierarchical flexible docking algorithm using generalized statistical potentials. J. Chem. Inf. Model. 2020, 60, 1509–1527. [Google Scholar] [CrossRef]
- Konc, J.; Lešnik, S.; Škrlj, B.; Janežič, D. ProBiS-Dock Database: A Web Server and Interactive Web Repository of Small Ligand–Protein Binding Sites for Drug Design. J. Chem. Inf. Model. 2021, 61, 4097–4107. [Google Scholar] [CrossRef]
- Konc, J.; Janežič, D. An improved branch and bound algorithm for the maximum clique problem. MATCH Commun. Math. Comput. Chem. 2007, 4, 569–590. [Google Scholar]
- Konc, J.; Miller, B.T.; Štular, T.; Lešnik, S.; Woodcock, H.L.; Brooks, B.R.; Janežič, D. ProBiS-CHARMMing: Web interface for prediction and optimization of ligands in protein binding sites. J. Chem. Inf. Model. 2015, 55, 2308–2314. [Google Scholar] [CrossRef] [PubMed]
- Triballeau, N.; Acher, F.; Brabet, I.; Pin, J.-P.; Bertrand, H.-O. Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J. Med. Chem. 2005, 48, 2534–2547. [Google Scholar] [CrossRef] [PubMed]
- Truchon, J.-F.; Bayly, C.I. Evaluating virtual screening methods: Good and bad metrics for the “early recognition” problem. J. Chem. Inf. Model. 2007, 47, 488–508. [Google Scholar] [CrossRef] [PubMed]
- Empereur-Mot, C.; Guillemain, H.; Latouche, A.; Zagury, J.-F.; Viallon, V.; Montes, M. Predictiveness curves in virtual screening. J. Cheminform. 2015, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Gaulton, A.; Bellis, L.J.; Bento, A.P.; Chambers, J.; Davies, M.; Hersey, A.; Light, Y.; McGlinchey, S.; Michalovich, D.; Al-Lazikani, B. ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. [Google Scholar] [CrossRef] [Green Version]
- Sheridan, R.P.; Singh, S.B.; Fluder, E.M.; Kearsley, S.K. Protocols for bridging the peptide to nonpeptide gap in topological similarity searches. J. Chem. Inf. Comput. Sci. 2001, 41, 1395–1406. [Google Scholar] [CrossRef]
- Empereur-Mot, C.; Zagury, J.-F.; Montes, M. Screening explorer–An interactive tool for the analysis of screening results. J. Chem. Inf. Model. 2016, 56, 2281–2286. [Google Scholar] [CrossRef]
- Ahmad, H.H.; Hamza, A.H.; Hassan, A.Z.; Sayed, A.H. Promising therapeutic role of Rosmarinus officinalis successive methanolic fraction against colorectal cancer. Int. J. Pharm. Pharm. Sci 2013, 5, 164–170. [Google Scholar]
- Sacco, C.; Bellumori, M.; Santomauro, F.; Donato, R.; Capei, R.; Innocenti, M.; Mulinacci, N. An in vitro evaluation of the antibacterial activity of the non-volatile phenolic fraction from rosemary leaves. Nat. Prod. Res. 2015, 29, 1537–1544. [Google Scholar] [CrossRef]
- Moreno, S.; Scheyer, T.; Romano, C.S.; Vojnov, A.A. Antioxidant and antimicrobial activities of rosemary extracts linked to their polyphenol composition. Free Radic. Res. 2006, 40, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Pavić, V.; Jakovljević, M.; Molnar, M.; Jokić, S. Extraction of carnosic acid and carnosol from sage (Salvia officinalis L.) leaves by supercritical fluid extraction and their antioxidant and antibacterial activity. Plants 2019, 8, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pariš, A.; Štrukelj, B.; Renko, M.; Turk, V.; Pukl, M.; Umek, A.; Korant, B.D. Inhibitory Effect of Carnosolic Acid on HIV-1 Protease in Cell-Free Assays. J. Nat. Prod. 1993, 56, 1426–1430. [Google Scholar] [CrossRef]
- Falo, M.; Fillmore, H.; Reeves, T.; Phillips, L. Matrix metalloproteinase-3 expression profile differentiates adaptive and maladaptive synaptic plasticity induced by traumatic brain injury. J. Neurosci. Res. 2006, 84, 768–781. [Google Scholar] [CrossRef] [PubMed]
- López-Jiménez, A.; García-Caballero, M.; Medina, M.Á.; Quesada, A.R. Anti-angiogenic properties of carnosol and carnosic acid, two major dietary compounds from rosemary. Eur. J. Nutr. 2013, 52, 85–95. [Google Scholar] [CrossRef]
- Kranenburg, O. The KRAS oncogene: Past, present, and future. Biochim. Biophys. Acta 2005, 1756, 81–82. [Google Scholar] [CrossRef]
- Matikas, A.; Mistriotis, D.; Georgoulias, V.; Kotsakis, A. Targeting KRAS mutated non-small cell lung cancer: A history of failures and a future of hope for a diverse entity. Crit. Rev. Oncol. Hematol. 2017, 110, 1–12. [Google Scholar] [CrossRef]
- Fell, J.B.; Fischer, J.P.; Baer, B.R.; Blake, J.F.; Bouhana, K.; Briere, D.M.; Brown, K.D.; Burgess, L.E.; Burns, A.C.; Burkard, M.R.; et al. Identification of the clinical development candidate MRTX849, a covalent KRASG12C inhibitor for the treatment of cancer. J. Med. Chem. 2020, 63, 6679–6693. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, Z.; Abdeslam-Hassan, M.; Ouassila, L.; Danielle, B. Extraction and modeling of Algerian rosemary essential oil using supercritical CO2: Effect of pressure and temperature. Energy Procedia 2012, 18, 1038–1046. [Google Scholar] [CrossRef] [Green Version]
- Balskus, E.P. Colibactin: Understanding an elusive gut bacterial genotoxin. Nat. Prod. Rep. 2015, 32, 1534–1540. [Google Scholar] [CrossRef]
- Teplyakov, A.; Obmolova, G.; Badet-Denisot, M.-A.; Badet, B.; Polikarpov, I. Involvement of the C terminus in intramolecular nitrogen channeling in glucosamine 6-phosphate synthase: Evidence from a 1.6\AA crystal structure of the isomerase domain. Structure 1998, 6, 1047–1055. [Google Scholar] [CrossRef] [Green Version]
- Bearne, S.L.; Blouin, C. Inhibition of Escherichia coli Glucosamine-6-phosphate Synthase by Reactive Intermediate Analogues: The Role of the 2-amino function in Catalysis. J. Biol. Chem. 2000, 275, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Fadaka, A.; Ajiboye, B.; Ojo, O.; Adewale, O.; Olayide, I.; Emuowhochere, R. Biology of glucose metabolization in cancer cells. J. Oncol. Sci. 2017, 3, 45–51. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Christofk, H.R.; Schuman, E.; Subtelny, A.O.; Sharfi, H.; Harlow, E.E.; Xian, J.; Cantley, L.C. Identification of small molecule inhibitors of pyruvate kinase M2. Biochem. Pharmacol. 2010, 79, 1118–1124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.-S.; Webby, R.J. Traditional and new influenza vaccines. Clin. Microbiol. Rev. 2013, 26, 476–492. [Google Scholar] [CrossRef] [Green Version]
- Kadam, R.U.; Wilson, I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proc. Natl. Acad. Sci. USA 2017, 114, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Chu, Y.; Wang, Y. HIV protease inhibitors: A review of molecular selectivity and toxicity. HIV AIDS 2015, 7, 95. [Google Scholar]
- Malhotra, S.; Dhundial, R.; Bhatia, N.; Duggal, N. HIV-2 Infections from a Tertiary Care Hospital in India-A Case Report. J. Hum. Virol. Retrovirol. 2018, 5, 2–6. [Google Scholar]
- De Silva, T.; Weiss, R.A. HIV-2 goes global: An unaddressed issue in Indian anti-retroviral programmes. Indian J. Med. Res. 2010, 132, 660. [Google Scholar]
- Visseaux, B.; Damond, F.; Matheron, S.; Descamps, D.; Charpentier, C. HIV-2 molecular epidemiology. Infect. Genet. Evol. 2016, 46, 233–240. [Google Scholar] [CrossRef]
- De Silva, T.I.; van Tienen, C.; Rowland-Jones, S.L.; Cotten, M. Dual infection with HIV-1 and HIV-2: Double trouble or destructive interference? HIV Ther. 2010, 4, 305–323. [Google Scholar]
- Annabel, B.; Anna, D.; Hannah, M. Global Tuberculosis Report 2019; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Garzan, A.; Willby, M.J.; Green, K.D.; Tsodikov, O.V.; Posey, J.E.; Garneau-Tsodikova, S. Discovery and optimization of two Eis inhibitor families as kanamycin adjuvants against drug-resistant M. tuberculosis. ACS Med. Chem. Lett. 2016, 7, 1219–1221. [Google Scholar] [CrossRef] [Green Version]
- Houghton, J.L.; Biswas, T.; Chen, W.; Tsodikov, O.V.; Garneau-Tsodikova, S. Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase Eis. Chembiochem 2013, 14, 2127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.-C.; Baiga, T.J.; Downes, M.; La Clair, J.J.; Atkins, A.R.; Richard, S.B.; Fan, W.; Stockley-Noel, T.A.; Bowman, M.E.; Noel, J.P.; et al. Structural basis for specific ligation of the peroxisome proliferator-activated receptor δ. Proc. Natl. Acad. Sci. USA 2017, 114, E2563–E2570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dressel, U.; Allen, T.L.; Pippal, J.B.; Rohde, P.R.; Lau, P.; Muscat, G.E. The peroxisome proliferator-activated receptor β/δ agonist, GW501516, regulates the expression of genes involved in lipid catabolism and energy uncoupling in skeletal muscle cells. Mol. Endocrinol. 2003, 17, 2477–2493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Colby, J.K.; Zuo, X.; Jaoude, J.; Wei, D.; Shureiqi, I. The role of PPAR-δ in metabolism, inflammation, and cancer: Many characters of a critical transcription factor. Int. J. Mol. Sci. 2018, 19, 3339. [Google Scholar] [CrossRef] [Green Version]
- Waku, T.; Shiraki, T.; Oyama, T.; Maebara, K.; Nakamori, R.; Morikawa, K. The nuclear receptor PPARγ individually responds to serotonin-and fatty acid-metabolites. EMBO J. 2010, 29, 3395–3407. [Google Scholar] [CrossRef] [Green Version]
- Alexacou, K.-M.; Tenchiu, A.-C.; Chrysina, E.D.; Charavgi, M.-D.; Kostas, I.D.; Zographos, S.E.; Oikonomakos, N.G.; Leonidas, D.D. The binding of β-d-glucopyranosyl-thiosemicarbazone derivatives to glycogen phosphorylase: A new class of inhibitors. Biorg. Med. Chem. 2010, 18, 7911–7922. [Google Scholar] [CrossRef]
- Treadway, J.L.; Mendys, P.; Hoover, D.J. Glycogen phosphorylase inhibitors for treatment of type 2 diabetes mellitus. Expert Opin. Investig. Drugs 2001, 10, 439–454. [Google Scholar] [CrossRef]
- Spasov, A.; Chepljaeva, N.; Vorob’ev, E. Glycogen phosphorylase inhibitors in the regulation of carbohydrate metabolism in type 2 diabetes. Russ. J. Bioorganic Chem. 2016, 42, 133–142. [Google Scholar] [CrossRef]
- Martin, J.L.; Johnson, L.N.; Withers, S.G. Comparison of the binding of glucose and glucose 1-phosphate derivatives to T-state glycogen phosphorylase b. Biochemistry 1990, 29, 10745–10757. [Google Scholar] [CrossRef]
- Barbier, P.; Tsvetkov, P.O.; Breuzard, G.; Devred, F. Deciphering the molecular mechanisms of anti-tubulin plant derived drugs. Phytochem. Rev. 2014, 13, 157–169. [Google Scholar] [CrossRef]
- Nawrotek, A.; Knossow, M.; Gigant, B. The determinants that govern microtubule assembly from the atomic structure of GTP-tubulin. J. Mol. Biol. 2011, 412, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Bomalaski, J.S.; Clark, M.A. Phospholipase A2 and arthritis. Arthritis Rheum. 1993, 36, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.S. Phospholipase A2 as targets for anti-cancer drugs. Biochem. Pharmacol. 2007, 74, 949–959. [Google Scholar] [CrossRef]
- Mallat, Z.; Lambeau, G.; Tedgui, A. Lipoprotein-associated and secreted phospholipases A2 in cardiovascular disease: Roles as biological effectors and biomarkers. Circulation 2010, 122, 2183–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ong, W.-Y.; Farooqui, T.; Kokotos, G.; Farooqui, A.A. Synthetic and natural inhibitors of phospholipases A2: Their importance for understanding and treatment of neurological disorders. ACS Chem. Neurosci. 2015, 6, 814–831. [Google Scholar] [CrossRef]
- Ivy, S.P.; Wick, J.Y.; Kaufman, B.M. An overview of small-molecule inhibitors of VEGFR signaling. Nat. Rev. Clin. Oncol. 2009, 6, 569–579. [Google Scholar] [CrossRef]
- Kufareva, I.; Abagyan, R. Type-II kinase inhibitor docking, screening, and profiling using modified structures of active kinase states. J. Med. Chem. 2008, 51, 7921–7932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vásquez, A.F.; Reyes Munoz, A.; Duitama, J.; González Barrios, A. Discovery of new potential CDK2/VEGFR2 type II inhibitors by fragmentation and virtual screening of natural products. J. Biomol. Struct. Dyn. 2021, 39, 3285–3299. [Google Scholar] [CrossRef]
- Rathi, E.; Kumar, A.; Kini, S.G. Molecular dynamics guided insight, binding free energy calculations and pharmacophore-based virtual screening for the identification of potential VEGFR2 inhibitors. J. Recept. Signal Transduct. 2019, 39, 415–433. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Arora, N.; Murty, U.S.N. Aspartate carbamoyltransferase of Plasmodium falciparum as a potential drug target for designing anti-malarial chemotherapeutic agents. Med. Chem. Res. 2012, 21, 2480–2493. [Google Scholar] [CrossRef]
- Bosch, S.S.; Lunev, S.; Batista, F.A.; Linzke, M.; Kronenberger, T.; Dömling, A.S.; Groves, M.R.; Wrenger, C. Molecular Target Validation of Aspartate Transcarbamoylase from Plasmodium falciparum by Torin 2. ACS Infect. Dis. 2020, 6, 986–999. [Google Scholar] [CrossRef]
- Wang, J.; Stieglitz, K.A.; Cardia, J.P.; Kantrowitz, E.R. Structural basis for ordered substrate binding and cooperativity in aspartate transcarbamoylase. Proc. Natl. Acad. Sci. USA 2005, 102, 8881–8886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, J.I.; Cardoso, F.F.; Soares, A.M.; dal Pai Silva, M.; Gallacci, M.; Fontes, M.R. Structural and functional studies of a bothropic myotoxin complexed to rosmarinic acid: New insights into Lys49-PLA2 inhibition. PLoS ONE 2011, 6, e28521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Li, X.; Xia, Y.; Yu, Z.; Cai, N.; Malwal, S.R.; Han, X.; Oldfield, E.; Zhang, Y. Farnesyl Pyrophosphate Synthase as a Target for Drug Development: Discovery of Natural-Product-Derived Inhibitors and Their Activity in Pancreatic Cancer Cells. J. Med. Chem. 2019, 62, 10867–10896. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, M.; Monroe, D.; Oliver, J.; Roberts, H. Factors IXa and Xa play distinct roles in tissue factor-dependent initiation of coagulation. Blood 1995, 86, 1794–1801. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Zhou, L.; Zhang, Y.; Yi, D.; Liu, L.; Rao, W.; Wu, Y.; Ma, D.; Liu, X.; Zhou, X.-H.A.; et al. Risk factors of stroke in Western and Asian countries: A systematic review and meta-analysis of prospective cohort studies. BMC Public Health 2014, 14, 776. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Peng, Y.; Wang, X.; Qian, Y.; Xiang, P.; Wade, S.W.; Guo, H.; Lopez, J.A.G.; Herzog, C.A.; Handelsman, Y. Cardiovascular events and death after myocardial infarction or ischemic stroke in an older Medicare population. Clin. Cardiol. 2019, 42, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Perzborn, E.; Roehrig, S.; Straub, A.; Kubitza, D.; Mueck, W.; Laux, V. Rivaroxaban: A new oral factor Xa inhibitor. Atertio. Thromb. Vasc. Biol. 2010, 30, 376–381. [Google Scholar] [CrossRef]
- Komoriya, S.; Kobayashi, S.; Osanai, K.; Yoshino, T.; Nagata, T.; Haginoya, N.; Nakamoto, Y.; Mochizuki, A.; Nagahara, T.; Suzuki, M. Design, synthesis, and biological activity of novel factor Xa inhibitors: Improving metabolic stability by S1 and S4 ligand modification. Biorg. Med. Chem. 2006, 14, 1309–1330. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, L.A.; Banci, L.; Bertini, I.; Cantini, F.; Donaire, A.; Gonnelli, L. Matrix metalloproteinase–inhibitor interaction: The solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors. J. Biol. Inorg. Chem. 2007, 12, 1197–1206. [Google Scholar] [CrossRef]
- World Helath Organization. Control of the Leishmaniases WHO Technical Report Series 949; World Health Organization: Geneva, Switzerland, 2010. [Google Scholar]
- Martin, M.B.; Grimley, J.S.; Lewis, J.C.; Heath, H.T.; Bailey, B.N.; Kendrick, H.; Yardley, V.; Caldera, A.; Lira, R.; Urbina, J.A.; et al. Bisphosphonates Inhibit the Growth of Trypanosoma b rucei, Trypanosoma c ruzi, Leishmania d onovani, Toxoplasma g ondii, and Plasmodium f alciparum: A Potential Route to Chemotherapy. J. Med. Chem. 2001, 44, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Li, D.; Alesi, G.N.; Fan, J.; Kang, H.-B.; Lu, Z.; Boggon, T.J.; Jin, P.; Yi, H.; Wright, E.R.; et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell 2015, 27, 257–270. [Google Scholar] [CrossRef] [Green Version]
- McDermott, L.A.; Iyer, P.; Vernetti, L.; Rimer, S.; Sun, J.; Boby, M.; Yang, T.; Fioravanti, M.; O’Neill, J.; Wang, L.; et al. Design and evaluation of novel glutaminase inhibitors. Biorg. Med. Chem. 2016, 24, 1819–1839. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Allen, A.; Kwagh, J.; Doliba, N.M.; Qin, W.; Najafi, H.; Collins, H.W.; Matschinsky, F.M.; Stanley, C.A.; Smith, T.J. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J. Biol. Chem. 2006, 281, 10214–10221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Li, M.; Chen, P.; Narayan, S.; Matschinsky, F.M.; Bennett, M.J.; Stanley, C.A.; Smith, T.J. Green tea polyphenols control dysregulated glutamate dehydrogenase in transgenic mice by hijacking the ADP activation site. J. Biol. Chem. 2011, 286, 34164–34174. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Smith, C.J.; Walker, M.T.; Smith, T.J. Novel Inhibitors Complexed with Glutamate Dehydrogenase Allosteric Regulation by Control of Protein Dynamics. J. Biol. Chem. 2009, 284, 22988–23000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLaBarre, B.; Gross, S.; Fang, C.; Gao, Y.; Jha, A.; Jiang, F.; Song, J.J.; Wei, W.; Hurov, J.B. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry 2011, 50, 10764–10770. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, K.; Pan, C.Q.; Karlberg, T.; Balaji, G.; Uttamchandani, M.; Suresh, V.; Schüler, H.; Low, B.C.; Sivaraman, J. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl. Acad. Sci. USA 2012, 109, 7705–7710. [Google Scholar] [CrossRef] [Green Version]
- Furlan, V.; Bren, U. Insight into Inhibitory Mechanism of PDE4D by Dietary Polyphenols Using Molecular Dynamics Simulations and Free Energy Calculations. Biomolecules 2021, 11, 479. [Google Scholar] [CrossRef]
- Kores, K.; Konc, J.; Bren, U. Mechanistic insights into side effects of troglitazone and rosiglitazone using a novel inverse molecular docking protocol. Pharmaceutics 2021, 13, 315. [Google Scholar] [CrossRef]
- Jukič, M.; Kores, K.; Janežič, D.; Bren, U. Repurposing of Drugs for SARS-CoV-2 Using Inverse Docking Fingerprints. Front. Chem. 2021, 9, 757826. [Google Scholar] [CrossRef]
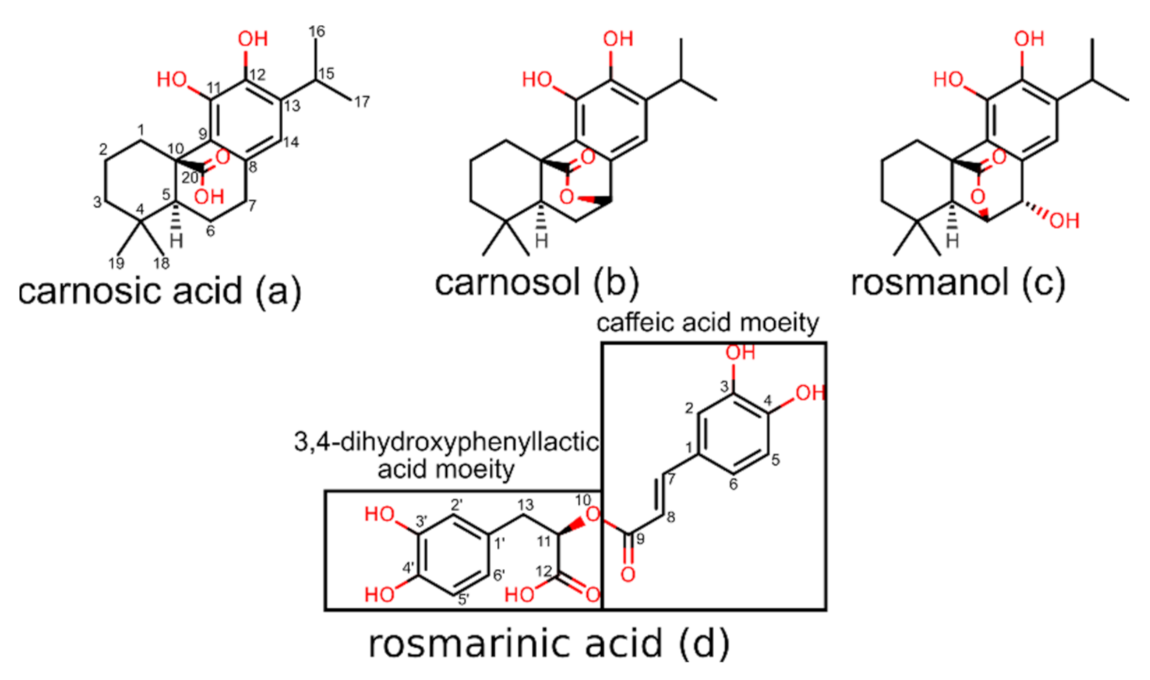

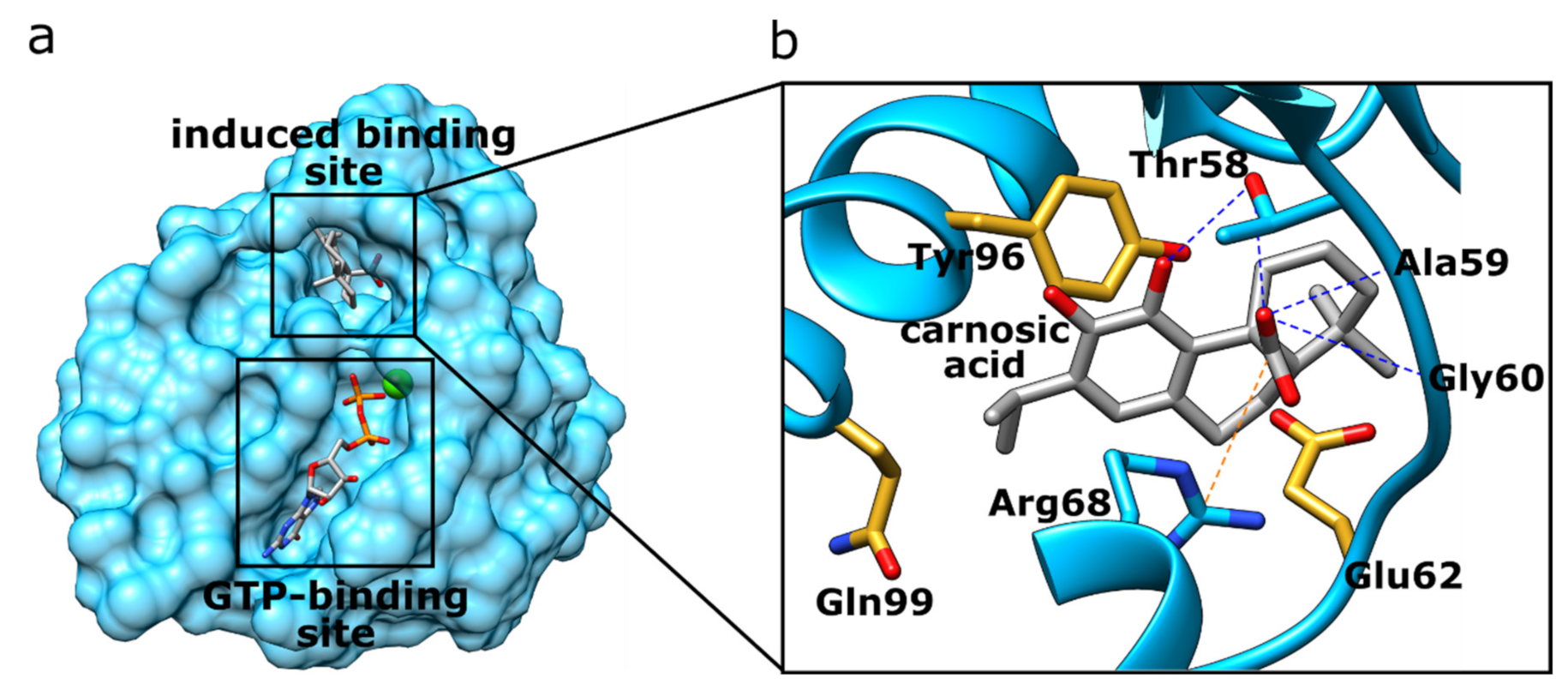

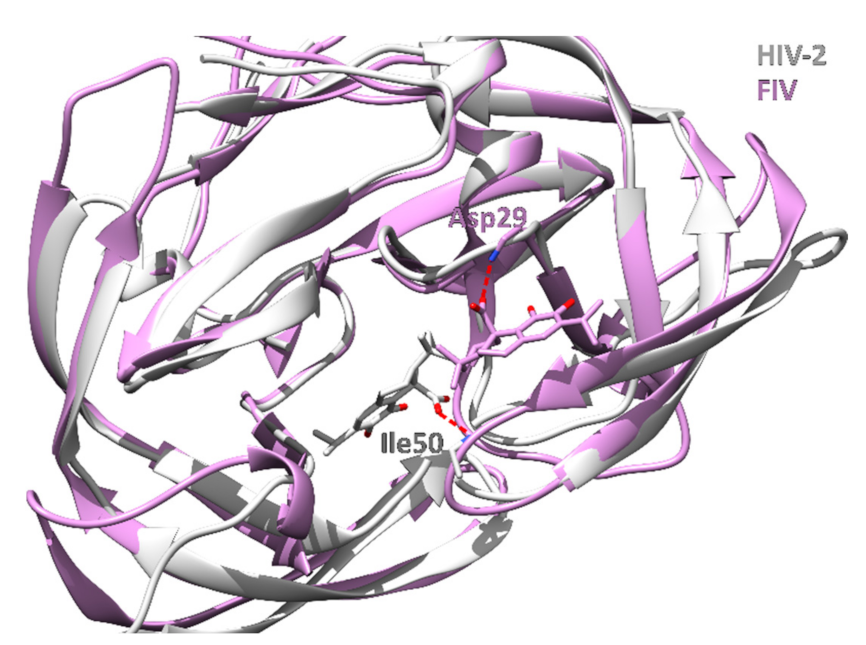
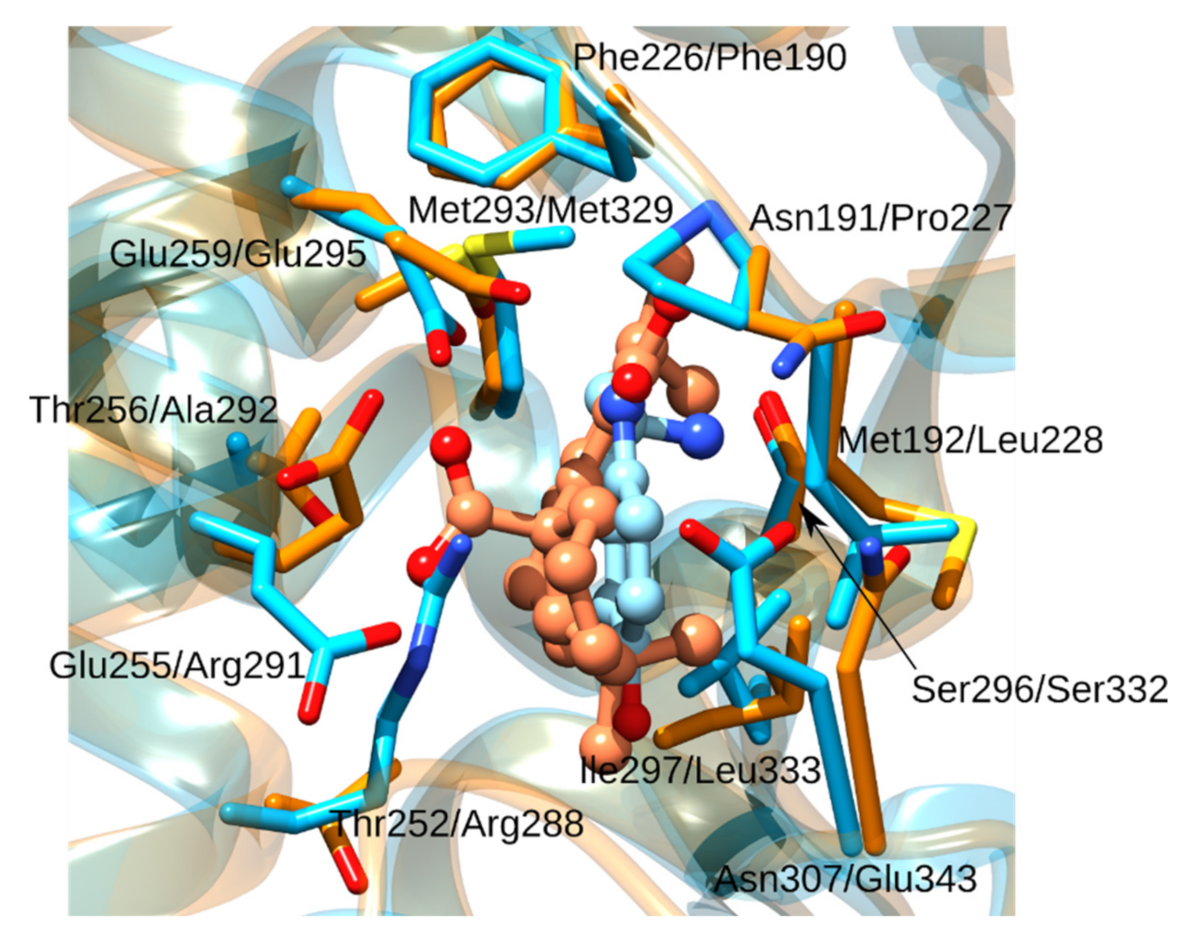


| Rank | PDB ID with Chain | Ligand | Predicted Ligand Docking Score (arb. Units) | Protein Name | Organism | Protein Function and Disease Correlation | Reported Experimental Correlation of Protein and Ligand |
|---|---|---|---|---|---|---|---|
| 1 | 4lucB | Carnosic acid | −69.9 | K-Ras G12C | Homo sapiens | Controls cell proliferation and differentiation. Its gene is a proto-oncogene. | [65] |
| 2 | 3oojA | Carnosic acid | −68.2 | Glucosamine- fructose-6-phosphate aminotransferase | Escherichia coli | Catalyzes the first step in hexosamine metabolism and is needed for E. coli growth and infection spread. | [66,67,68] |
| 3 | 3srdD | Carnosic acid | −68.1 | Pyruvate kinase M2 | Homo sapiens | Catalyzes the last step in the glycolysis. Important in providing ATP to cancer cells. | |
| 4 | 1kenA | Carnosic acid | −66.9 | Hemagglutinin HA1 | Influenza A virus | Enables viral entry into cells causing the flu. | |
| 5 | 2hpeA | Carnosic acid | −65.0 | HIV-2 protease | Human immunodeficiency virus 2 | Hydrolyzes peptide bonds leading to functional proteins essential for HIV infectivity. | [69] |
| 6 | 4jd6C | Carnosic acid | −64.8 | Enhanced intracellular survival protein | Mycobacterium tuberculosis | Acetylates amine groups in aminoglycoside drugs, thus preventing the binding to the ribosome, leading to M. tuberculosis resistance. | |
| 7 | 5u46A | Carnosic acid | −64.7 | Peroxisome proliferator activated receptor delta | Homo sapiens | Regulates lipid catabolism and its transport and storage and is also associated with insulin secretion and resistance. It is implicated in metabolic disorders and cancer. | γ isoform [70] |
| 8 | 3mt7A | Carnosic acid | −64.5 | Glycogen phosphorylase | Oryctolagus cuniculus | Breaks the non-reducing ends in the chain of glycogen that enables glucose production. Its inhibition can manage type II diabetes. | |
| 9 | 3rycC | Carnosic acid | −64.2 | Tubulin | Rattus norvegicus | Involved in cell division as it forms microtubules which in turn form mitotic spindles that pull chromosomes apart during cell division. Tubulin targeting is used in cancer treatment. | |
| 10 | 2j9kB | Carnosic acid | −63.5 | HIV-1 protease | Human immunodeficiency virus 1 | Hydrolyzes peptide bonds leading to functional proteins essential to HIV infectivity. | [69] |
| 11 | 1fxfB | Carnosol | −63.3 | Phospholipase A2 | Sus scrofa | Catalyzes the hydrolysis of glycerophospholipids thus releasing free fatty acids, including arachidonic acid. Its action is implicated in several inflammation-based diseases such as arthritis, coronary artery disease, Alzheimer’s and cancer. | |
| 12 | 3ogpA | Carnosic acid | −63.3 | FIV Protease | Feline immunodeficiency virus | Hydrolyzes peptide bonds leading to functional proteins essential to FIV infectivity in cats. | |
| 13 | 2p2hA | Carnosic acid | −63.1 | Vascular endothelial growth factor receptor 2 | Homo sapiens | Signal protein crucial in angiogenesis. Its inhibition is used in cancer treatment. | Negative: [71] |
| 14 | 5ilqC | Carnosic acid | −63.0 | Aspartate carbamoyltransferase | Plasmodium falciparum | Enzyme involved in pyrimidine biosynthesis, crucial for Plasmodium falciparum (causative agent of malaria) survival and replication. | |
| 15 | 4iv5A | Carnosic acid | −62.8 | Aspartate carbamoyltransferase | Trypanosoma cruzi | Enzyme involved in pyrimidine biosynthesis, crucial for Trypanosoma cruzi (causative agent of Chagas disease) survival and replication |
| Rank | PDB ID with Chain | Predicted Ligand Docking Score (arb. Units) | Protein Name | Organism | Protein Function and Disease Correlation | Reported Experimental Correlation of Protein and Ligand |
|---|---|---|---|---|---|---|
| 1 | 2d1jA | −86.1 | Coagulation factor X | Homo sapiens | Serine endopeptidase is involved in the coagulation cascade. Its deficiency leads to a bleeding disorder. Its inhibitors are popular anticoagulants. | |
| 2 | 1fxfB | −84.8 | Phospholipase A2 | Sus scrofa | Catalyzes the hydrolysis of glycerophospholipids thus releasing free fatty acids, including arachidonic acid. Its action is implicated in several inflammation-based disease such as arthritis, coronary artery disease, Alzheimer’s and cancer. | [113] |
| 3 | 2jt5A | −84.5 | Matrix metalloproteinase-3 | Homo sapiens | Zinc-dependent endopeptidase which is involved in the remodeling of the extracellular matrix. Involved in arthritis, multiple sclerosis, aneurysms, and the spread of metastatic cancer. After traumatic brain injury, matrix metalloproteinase-3 (MMP-3) concentrations increase and lead to additional damage to the blood–brain barrier. | |
| 4 | 4jzbA | −83.2 | Farnesyl pyrophosphate synthase | Leishmania major | Farnesyl pyrophosphate synthase (FPPS) is an essential enzyme involved in the biosynthesis of ergosterol in leishmania parasites, the causative agents of leishmaniasis. | [114] |
| 5 | 3qmuB | −80.2 | Glutamate dehydrogenase 1 | Bos Taurus | Part of the glutaminolysis pathway, playing a crucial role in nitrogen and carbon metabolism. Inhibition leads to in vivo and in vitro reduced viability of cancer cells. | |
| 6 | 5fi6A | −77.6 | Glutaminase | Homo sapiens |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lešnik, S.; Bren, U. Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking. Foods 2022, 11, 67. https://doi.org/10.3390/foods11010067
Lešnik S, Bren U. Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking. Foods. 2022; 11(1):67. https://doi.org/10.3390/foods11010067
Chicago/Turabian StyleLešnik, Samo, and Urban Bren. 2022. "Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking" Foods 11, no. 1: 67. https://doi.org/10.3390/foods11010067
APA StyleLešnik, S., & Bren, U. (2022). Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking. Foods, 11(1), 67. https://doi.org/10.3390/foods11010067







