Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus acidophilus NCFM to Freeze-Drying
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
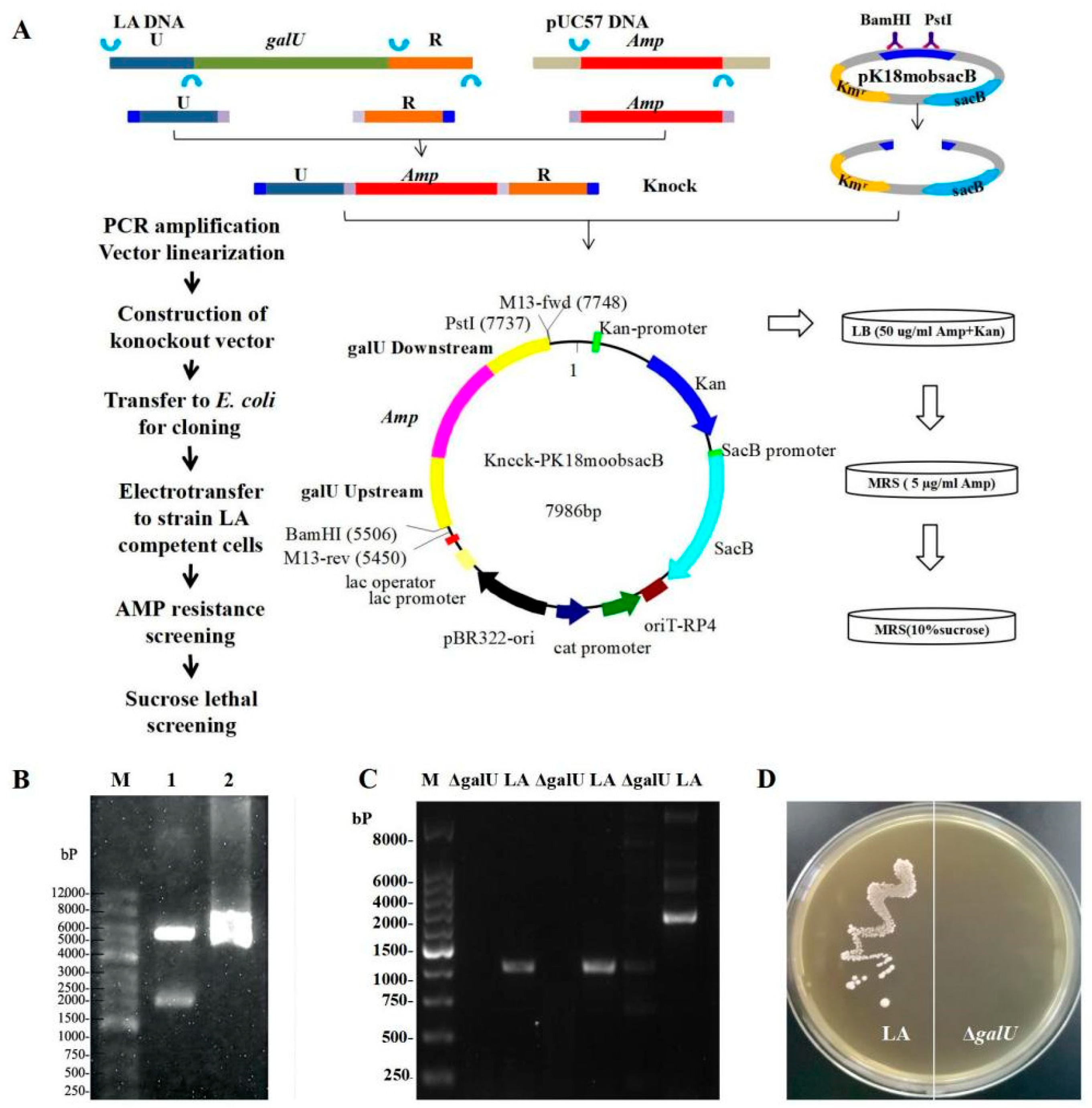
2.2. Knockout of galU
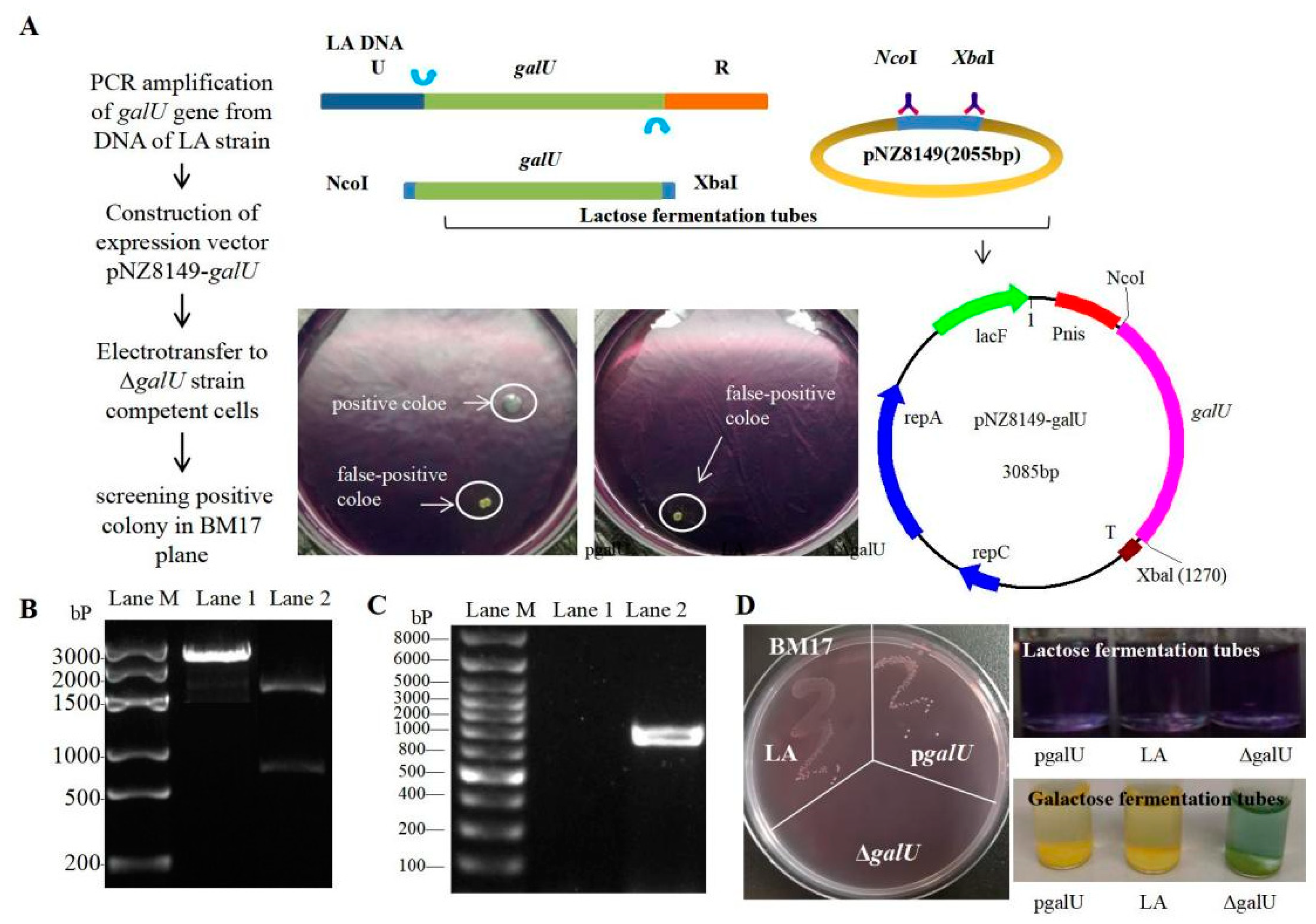
2.3. Expression of galU in ΔgalU
2.4. Determination of GalU Activity
2.5. Effect of Freeze-Drying on Bacterial Survival Rate
2.6. Transmission Electron Microscopy (TEM) to Assess the Cell Structure
2.7. Transcriptome Sequencing
3. Results
3.1. Acquisition of the galU Knockout Strain ΔgalU
3.2. Acquisition of the galU Re-Expression Strain pgalU
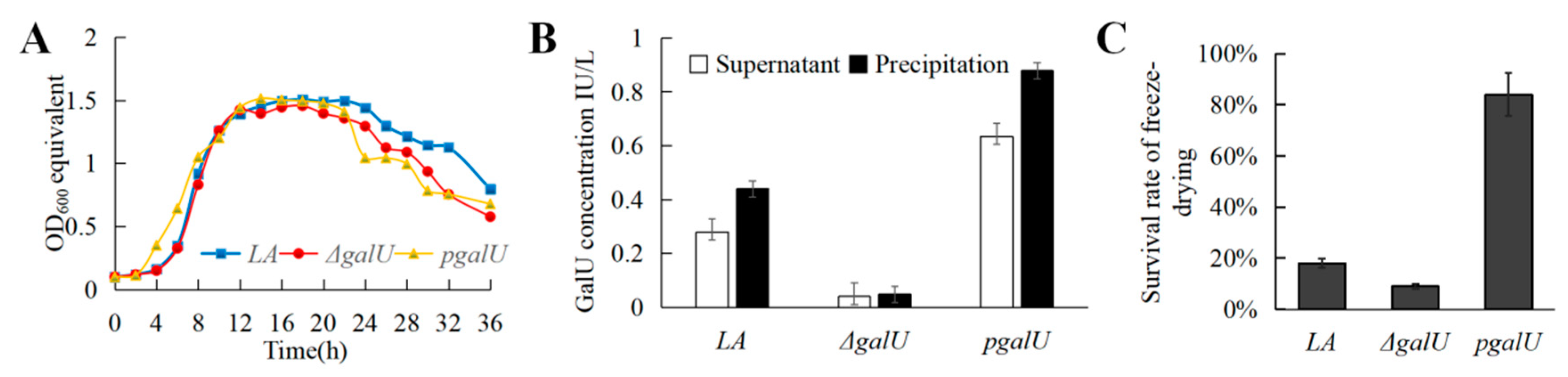
3.3. GalU Activity of the LA, ΔgalU, and pgalU Strains
3.4. Effect of galU on Freeze-Drying Survival Rate
3.5. TEM of LA, ΔgalU, and pgalU Strains
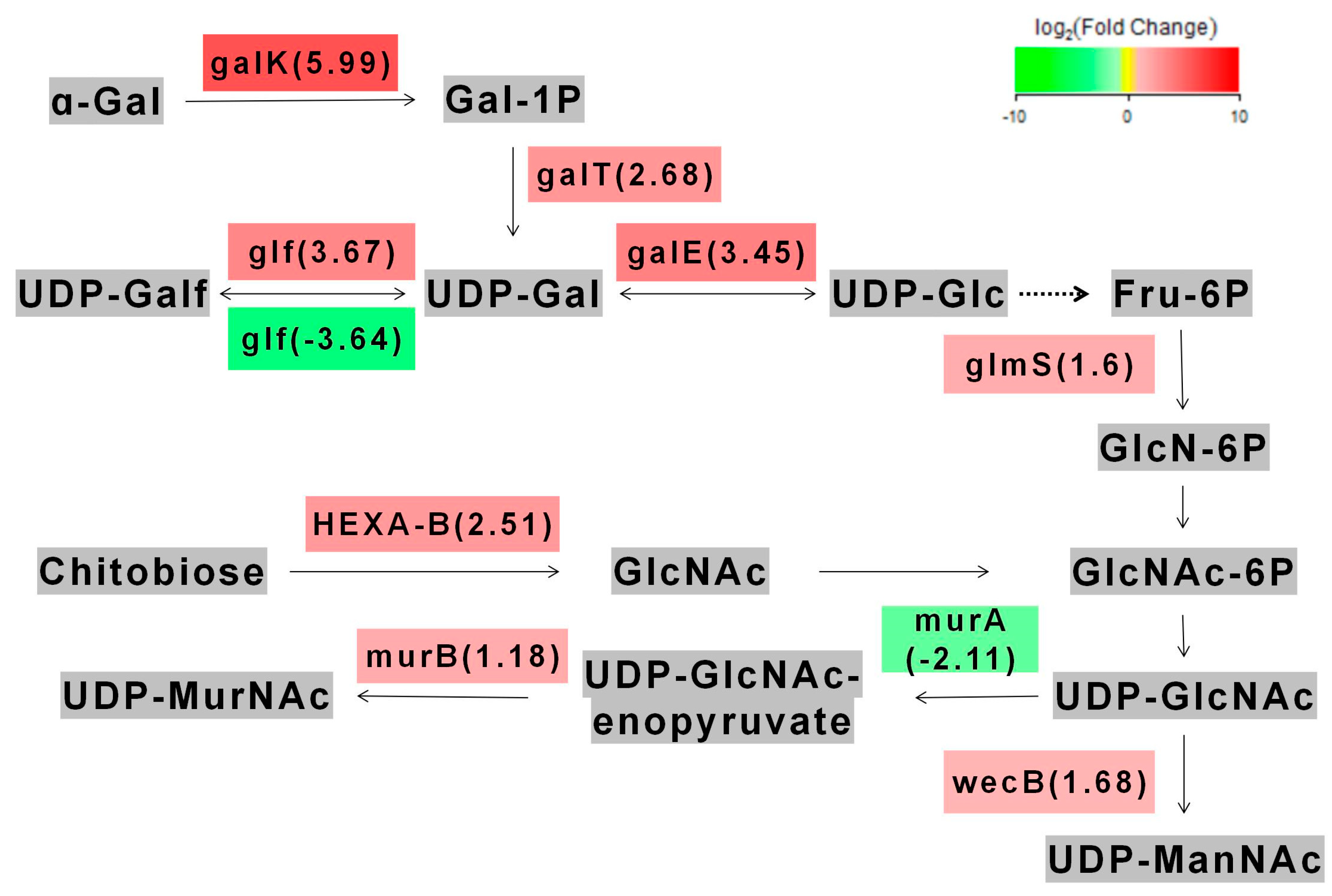
3.6. Regulation of Metabolic Pathways by galU
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Altermann, E.; Russell, W.; Azcarate, M.; Barrangou, R.; Buck, B.; Mcauliffe, O.; Souther, N.; Dobson, A.; Duong, T.; Callanan, M.; et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 2005, 102, 3906–3912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouwehand, C.; Tiihonen, K.; Saarinen, M.; Putaala, H.; Rautonen, N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: Intestinal and immune parameters. Br. J. Nutr. 2008, 101, 367–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christel, R.; Xavier, T.; Agathe, G.; Nicolas, B.; Christel, N.; Laurent, D.; Caroline, D.; Emilie, M.; Karen, G.; Mathias, C. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat. Med. 2007, 13, 35–37. [Google Scholar] [CrossRef]
- Konstantinov, S.; Hauke, S.; Vos, W.; Bruijns, S.; Satwinder, S.; Florence, V.; Daniel, M.; Sylvie, L.; Eric, A.; Klaenhammer, T.; et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. USA 2008, 105, 19474–19479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreasen, S.; Larsen, N.; Pedersen-Skovsgaard, T.; Berg, R.; Meller, K.; Svendsen, K.; Jakobsen, M.; Pedersen, B. Effects of Lactobacillus acidophilus NCFM on insulin sensitivity and the systemic inflammatory response in human subjects. Br. J. Nutr. 2010, 104, 1831–1838. [Google Scholar] [CrossRef] [Green Version]
- D’Alessandro, M.; Pisanu, F.; Baldo, D.; Parolin, C.; Filippini, G.; Vitali, B.; Lanciotti, R.; Patrignani, F. Unravelling the functional and technological potential of soy milk based microencapsulated Lactobacillus crispatus and Lactobacillus gasseri. J. Funct. Foods 2021, 87, 1756–4646. [Google Scholar] [CrossRef]
- Delphine, M.A.; Jennifer, K.; Glenn, G.; James, V. Mechanisms of probiosis and prebiosis: Considerations for enhanced functional foods. Curr. Opin. Biotechnol. 2009, 20, 135–141. [Google Scholar] [CrossRef]
- Canducci, F.; Armuzzi, A.; Cremonini, F.; Cammarota, G.; Gasbarrini, A. A lyophilized and inactivated culture of Lactobacillus acidophilus increases Helicobacter pylori eradication rates. Aliment. Pharmacol. Ther. 2001, 14, 1625–1629. [Google Scholar] [CrossRef]
- Giulio, B.D.; Orlando, P.; Barba, G.; Coppola, R.; Rosa, M.D.; Sada, A.; Prisco, P.P.D.; Nazzaro, F. Use of alginate and cryo-protective sugars to improve the viability of lactic acid bacteria after freezing and freeze-drying. World J. Microbiol. Biotechnol. 2005, 21, 736–749. [Google Scholar] [CrossRef]
- Bosca, S.; Barresi, A.; Fissore, D. On the use of model-based tools to optimize in-line a pharma ceuticals freeze-drying process. Dry. Technol. 2016, 37, 937–955. [Google Scholar] [CrossRef]
- Mingxue, L.; Xiaoqun, Z.; Yating, H.; Chaoran, X.; Lu, C.; Zhen, W.; Hangzhen, L.; Daodong, P. iTRAQ-based quantitative proteomic analysis of the effect of heat shock on freeze-drying of Lactobacillus acidophilus ATCC4356. Int. J. Food Sci. Technol. 2021, 56, 5569–5580. [Google Scholar] [CrossRef]
- Wiley, H.; Leveille, A. Influence of Periodicity of Eating on the Activity of Adipose Tissue and Muscle Glycogen Synthesizing Enzymes in the Rat. J. Nutr. 1970, 100, 85–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alberto, A.; Jack, P. Bacterial glycogen and plant starch biosynthesis. Biochem. Educ. 1992, 20, 196–203. [Google Scholar] [CrossRef]
- Cools, F.; Torfs, E.; Vanhoutte, B.; Bidart, D.; Bonofiglio, L.; Mollerach, M.; Maes, L.; Caljon, G. Streptococcus pneumoniae galU gene mutation has a direct effect on biofilm growth, adherence and phagocytosis in vitro and pathogenicity in vivo. Pathog. Dis. 2018, 76, 1120–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munch, C.; Petersen, A.; Nygaard, P.; Jespersen, K. Mutants constitutive for nucleoside-catabolizing enzymes in Escherichia coli K12 isolation, charactrization and mapping. Eur. J. Biochem. 2010, 27, 208–215. [Google Scholar] [CrossRef]
- Sulek, K.; Smedsgaard, J.; Skov, T.; Wilcks, A.; Licht, T. Metabolic footprint of Lactobacillus acidophilus NCFM at different pH. Metabolomics 2012, 8, 244–252. [Google Scholar] [CrossRef]
- Kim, H.; Shim, J.E.; Shin, J.; Lee, I. Ecolinet: A database of cofunctional gene network for Escherichia coli. Database 2015, 2015, bav001. [Google Scholar] [CrossRef]
- Choonia, H.S.; Saptarshi, S.D.; Lele, S.S. Release of intracellular β-galactosidase from Lactobacillus acidophilus and L-asparaginase from Pectobacterium carotovorum by high-pressure homogenization. Chem. Eng. Commun. 2013, 200, 1415–1424. [Google Scholar] [CrossRef]
- Guimont, C.; Gaillard, J. Comparative study of the protein compositionof three strains of Streptococcus thermophilus grown either in M17 medium or in milk. Dairy Sci. Technol. 2002, 82, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.; Chen, J.; Du, G.; Li, H.; Zhang, J.; Dong, Z. Codon and propeptide optimizations to improve the food-grade expression of bile salt hydrolase in Lactococcus lactis. Protein Pept. Lett. 2015, 22, 727–735. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Gou, K.; Luo, Y. Heterologous expression of stearoyl-CoA desaturase-1 in Lactococcus lactis NZ3900. Chin. J. Biotechnol. 2012, 28, 1106–1117. [Google Scholar] [CrossRef]
- Guo, M.; Yi, S.; Guo, Y.; Zhang, S.; Niu, J.; Wang, K.; Hu, G. Construction of a recombinant Lactococcus lactis strain expressing a variant porcine epidemic diarrhea virus S1 gene and its immunogenicity analysis in mice. Viral Immunol. 2019, 141, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Zhu, J.; Liu, L.; Cong, Y.; Hu, F.; Li, J.; Yu, X. Functional identification of a putative β-galactosidase gene in the special lac gene cluster of Lactobacillus acidophilus. Curr. Microbiol. 2010, 60, 172–180. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, R.; Duan, G.; Shi, J. Food-grade expression of helicobacter pylori UreB subunit in Lactococcus lactis and its Immunoreactivity. Curr. Microbiol. 2011, 62, 1726–1731. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.; Mcconville, K.; Mcreynolds, C.; Collins, M. Electrotransformation of Lactobacillus plantarum using linearized plasmid DNA. Lett. Appl. Microbiol. 2007, 25, 419–425. [Google Scholar] [CrossRef] [Green Version]
- Thu-Ha, N.; Barbara, S.; Stanimira, K.; Wolfgang, K.; Kulbe, D.; Christina, D.; Dietmar, H. Characterization and molecular cloning of a heterodimeric β-galactosidase from the probiotic strain Lactobacillus acidophilus R22. Fems Microbiol. Lett. 2007, 269, 136–144. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.J.; Zhang, R.G.; Duan, G.C.; Fan, Q.T. Expression of Helicobacter pylori ureB gene in Lactococcus lactis NZ3900 and its immunoreactivity. Chin. J. Zoonoses 2011, 79, 699–703. [Google Scholar] [CrossRef]
- Song, J.; Jiaping, L.; Wang, Z.; Zhang, L.; Zhang, S.; Xianbao, H. Effect of centrifugation conditions on the survival rate of freeze-drying culture. Sci. Technol. Food Ind. 2009, 30, 82–84. [Google Scholar] [CrossRef]
- Hood, S.; Zottola, A. Electron microscopic study of the adherence properties of Lactobacillus acidophilus. J. Food Sci. 2006, 52, 791–792. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Zhen, N.; Zeng, X.; Wang, H.; Yu, J.; Guo, Y. Effects of heat shock treatment on the survival rate of Lactobacillus acidophilus after freeze-drying. Food Res. Int. 2020, 136, 109507. [Google Scholar] [CrossRef] [PubMed]
- Carević, M.; Vukašinović-Sekulić, M.; Ćorović, M.; Rogniaux, H.; Ropartz, D.; Veličković, D.; Bezbradica, D. Evaluation of β-galactosidase from Lactobacillus acidophilus as biocatalyst for galacto-oligosaccharides synthesis: Product structural characterization and enzyme immobilization. J. Biosci. Bioeng. 2018, 86, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Beerthuyzen, M.; Vaughan, E.; Vos, W.; Kuipers, P. Controlled gene expression systems for lactic acid bacteria: Transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 1997, 63, 4581–4584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broach, B.; Gu, X.; Bar-Peled, M. Biosynthesis of UDP-glucuronic acid and UDP-galacturonic acid in Bacillus cereus subsp. cytotoxis NVH 391-98. Febs J. 2012, 279, 1831–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geiger, B.; Nguyen, H.-M.; Wenig, S.; Lorenz, C.; Kittl, R.; Mathiesen, G.; Eijsink, V.G.; Haltrich, D.; Nguyen, T.-H. From by-product to valuable components: Efficient enzymatic conversion of lactose in whey using β-galactosidase from Streptococcus thermophilus. Biochem. Eng. J. 2016, 96, 1024–1037. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Sun, Z.; Zhong, J.; Zhang, Q.; Huan, L. Adverse effect of nisin resistance protein on nisin-induced expression system in Lactococcus lactis. Microbiol. Res. 2010, 165, 458–465. [Google Scholar] [CrossRef]
- Platteeuw, C.; van Alen-Boerrigter, I.N.; van Schalkwijk, S.A.; De Vos, W.M. Food-grade cloning and expression system for Lactococcus lactis. Appl. Environ. Microbiol. 1996, 62, 1008–1013. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Wang, H.; Yang, X.; Zhang, A.; Yang, F. Lactococcus lactis Anchoring avian infectious bronchitis virus multi-epitope peptide epiC induced specific immune responses in chickens. J. Agric. Chem. Soc. Jpn. 2013, 77, 1831–1839. [Google Scholar] [CrossRef]
- Wang, P.; Baiyuan, L.; Li, B.; Cai, X.; Zeng, Z.; Chen, X.; Wang, X. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas. Microb. Cell Factories 2015, 32, 156–170. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Sagaram, U.; Kim, J.; Wang, N. Requirement of the galU gene for polysaccharide production by and pathogenicity and growth in planta of Xanthomonas citri, subspecies citri. Appl. Environ. Microbiol. 2010, 76, 2234–2242. [Google Scholar] [CrossRef] [Green Version]
- Zechel, L.; Withers, S. Glycosidase mechanisms: Anatomy of a finely tuned catalyst. Acc. Chem. Res. 2000, 33, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Belogurov, A.; Artsimovitch, I. Regulation of transcript elongation. Annu. Rev. Microbiol. 2015, 69, 49–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Gao, Y.; Gao, G.; Lou, Y. Oral administration of recombinant Lactococcus lactis expressing the cellulase gene increases digestibility of fiber in geese. Curr. Microbiol. 2015, 71, 693–698. [Google Scholar] [CrossRef]
- Yi, Z.; Saixiang, F.; Chenggang, X.; Bin, Z.; Suming, Z.; Lingyun, Z.; Xianhui, H.; Jingyi, L.; Zhen, Y.; Ming, L. The role of galU and galE of Haemophilus parasuis SC096 in serum resistance and biofilm formation. Vet. Microbiol. 2013, 162, 278–284. [Google Scholar] [CrossRef]
- Kulkarni, R.; Thomas, R.A.; Tucker, J.D. Expression of DNA repair and apoptosis genes in mitochondrial mutant and normal cells following exposure to ionizing radiation. Environ. Mol. Mutagenes. 2011, 52, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Post, A.E.M.; Bussink, J.; Sweep, F.C.G.J.; Span, P.N. A Changes in DNA damage repair gene expression and cell cycle gene expression do not explain radioresistance in tamoxifen-resistant breast cancer. Oncol. Res. 2019, 43, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yang, H.; Miller, J.; Shih, D.; Hicks, G.; Xie, J. Cells deficient in oxidative DNA damage repair genes Myhand Ogg1are sensitive to oxidants with increased G2/M arrest and multinucleation. Carcinogenesis 2008, 55, 456–458. [Google Scholar] [CrossRef] [Green Version]

| Strain or Plasmid | Relevant Characteristic (s) | Source or Reference |
|---|---|---|
| Strains | ||
| Escherichia coli Trans1 T1 | F−φ80 (lacZ) ΔM15ΔlacX74hsdR (rk−, mk+) ΔrecA1398endA1tonA | TransGen Biotech |
| Escherichia coli DH10BT1 | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15ΔlacX74 recA1 endA1araD139Δ (ara, leu) 7697 galU galKλ- rpsL nupG tonA | Biovector NTCC |
| Lactobacillus acidophilus NCFM (LA) | Wild-type strain | ATCC |
| ΔgalU | LA strain with galU deleted | This work |
| NZ3900 | lacF−, pepN: nisR nisK | Biovector NTCC |
| pgalU | ΔgalU strain with plasmid pNZ8149-galU | This work |
| Plasmid | ||
| pUC57 | Apr; lacZ/MCS; pMB11 ori | Biovector NTCC |
| pK18mobsacB | Kmr; lacZ/MCS; pBR322 ori; sacB | Laboratory collection |
| Knock-PK18mobsaB | Kmr; MCS (with BamHI and PstI); sacB | This work |
| pNZ8149 | lacF; nisA; nisC; MCS | Biovector NTCC |
| pNZ8149-galU | lacF; nisA; nisC; MCS (with NcoI and XhoI) | This work |
| Primer | Sequence | Position in Chromosome |
|---|---|---|
| galU-1-F (LA) | 5′-ggatccGCGAACAACTCTTTCACAA | 610275 |
| galU-1-R (LA) | 5′-GAAATGTTGAATACTCATGATAACGCCAGCCAACCAA | 610898 |
| galU-2-F (LA) | 5′-CTGATTAAGCATTGGTAATGGCTCGTCAAGTTGCTCT | 612410 |
| galU-2-R (LA) | 5′-ggaattccCTGGCACCGTCAGTAAGAG | 612957 |
| amp-F (pUC57) | 5′-TTGGTTGGCTGGCGTTATCATGAGTATTCAACATTTC | 1650 |
| amp-R (pUC57) | 5′-AGAGCAACTTGACGAGCCATTACCAATGCTTAATCAG | 2492 |
| galU-4-F (LA) | 5′-TCCATAACCGAGTAGGAGA | 611061 |
| galU-4-R (LA) | 5′-TAAAGACATGGGCAAATAC | 611953 |
| galU-5-F (LA) | 5′-GCTGGTCGAATTGCTAACT | 611093 |
| galU-5-R (LA) | 5′-GTATCAATGGCATCAGTTAA | 611915 |
| galU-6-F (LA) | 5′-TTGGCTGGCGTTATCATTT | 612396 |
| galU-6-R (LA) | 5′-GACCGTCATTAAGCATTGTAC | 614143 |
| galU-7-F (LA) | 5′-ATTATAAGGAGGCACTCACCATGGGCAGAAAGTGTATATATA | 611190 |
| galU-7-R (LA) | 5′-CAAAGAAAGCTTGAGCTCTCTAGATTTATTTTTTCGCTTATC | 612125 |
| galU-8-F (pNZ) | 5′-ATTATAAGGAGGCACTCAccatgg | 184 |
| galU-8-R (pNZ) | 5′-tctagaGAGCTCAAGCTTTCTTTG | 236 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, Z.; Zeng, X.; Guo, Y.; Wu, Z.; Cai, Z.; Pan, D. Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus acidophilus NCFM to Freeze-Drying. Foods 2022, 11, 1719. https://doi.org/10.3390/foods11121719
Zeng Z, Zeng X, Guo Y, Wu Z, Cai Z, Pan D. Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus acidophilus NCFM to Freeze-Drying. Foods. 2022; 11(12):1719. https://doi.org/10.3390/foods11121719
Chicago/Turabian StyleZeng, Zhidan, Xiaoqun Zeng, Yuxing Guo, Zhen Wu, Zhendong Cai, and Daodong Pan. 2022. "Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus acidophilus NCFM to Freeze-Drying" Foods 11, no. 12: 1719. https://doi.org/10.3390/foods11121719
APA StyleZeng, Z., Zeng, X., Guo, Y., Wu, Z., Cai, Z., & Pan, D. (2022). Determining the Role of UTP-Glucose-1-Phosphate Uridylyltransferase (GalU) in Improving the Resistance of Lactobacillus acidophilus NCFM to Freeze-Drying. Foods, 11(12), 1719. https://doi.org/10.3390/foods11121719






