Prevalence of Multidrug-Resistant Salmonella enterica Serovars in Buffalo Meat in Egypt
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation and Identification of Salmonella spp.
2.3. Molecular Analysis
2.4. Antimicrobial Susceptibility Tests
3. Results and Discussion
3.1. Prevalence of the Isolated Salmonella enterica Serovars in Buffalo Meat
3.2. Distribution of Salmonella serovars Isolated from Buffalo Meat Samples
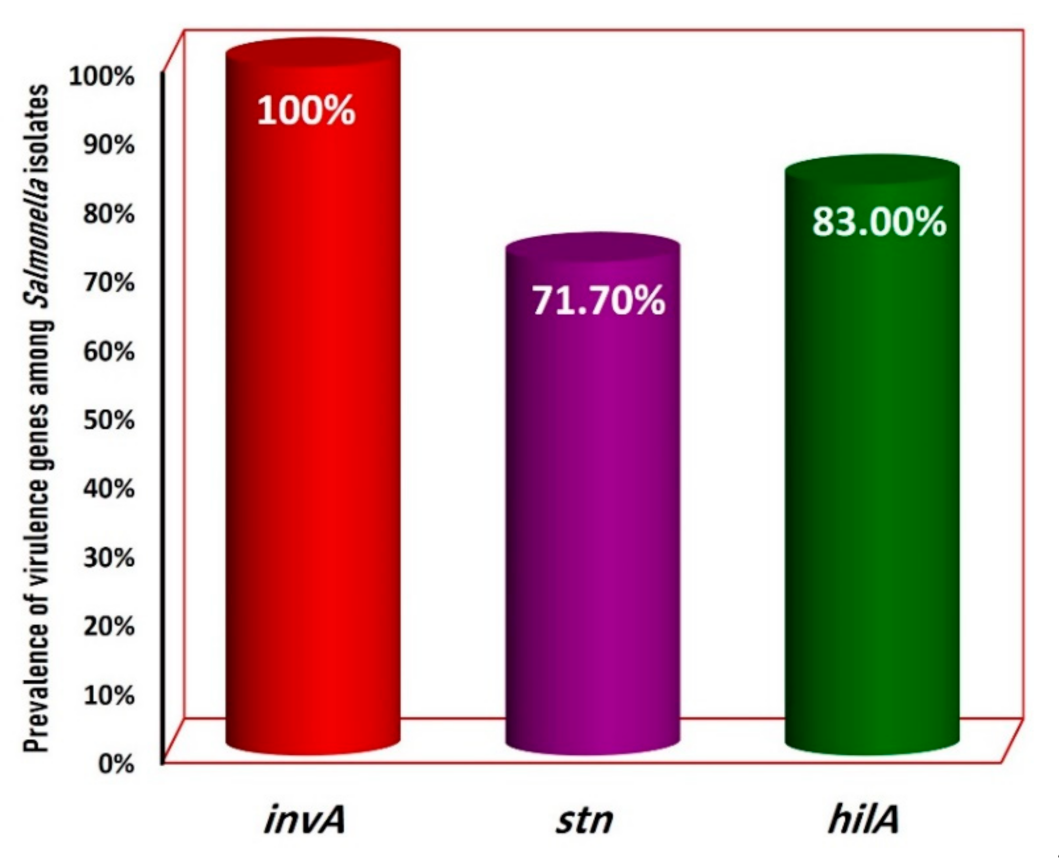
3.3. PCR Confirmation of the Salmonella Isolates
3.4. Antimicrobial Resistance of Salmonella Isolates
3.5. Antimicrobial Resistance Profiles of Salmonella Isolates
3.6. Categorization of Salmonella Isolates Based on Their Antimicrobial Resistance Profiles
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sethi, R.K. Breeding Strategies for Genetic Improvement in Buffaloes. Buffalo Bull. 2013, 32, 219–226. [Google Scholar]
- Kandeepan, G.; Mendiratta, S.K.; Shukla, V.; Vishnuraj, M.R. Processing characteristics of buffalo meat—A review. J. Meat sci. Technol. 2013, 1, 1–11. Available online: http://www.jakraya.com/journal/pdf/jmstArticle.pdf (accessed on 19 July 2022).
- Food and Drug Administration (FDA). Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins, Second edition. 2012. Available online: https://www.fda.gov/food/foodborne-pathogens/bad-bug-book-second-edition (accessed on 14 May 2022).
- Sallam, K.I.; Abd-Elghany, S.M.; Hussein, M.A.; Imre, K.; Morar, A.; Morshdy, A.E.; Sayed-Ahmed, M.Z. Microbial decontamination of beef carcass surfaces by lactic acid, acetic acid, and trisodium phosphate sprays. BioMed Res. Int. 2020, 2020, 2324358. [Google Scholar] [CrossRef]
- Elmali, M.; Yaman, H. Microbiological quality of raw meat balls produced and sold in the eastern of Turkey. Pakistan J. Nutr. 2005, 4, 197–201. [Google Scholar] [CrossRef]
- Sallam, K.I.; Abd-Elghany, S.M.; Imre, K.; Morar, A.; Herman, V.; Hussein, M.A.; Mahros, M.A. Ensuring safety and improving keeping quality of meatballs by addition of sesame oil and sesamol as natural antimicrobial and antioxidant agents. Food Microbiol. 2021, 99, 103834. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.A.; Henard, C.A.; Liu, L.; Dieckman, L.; Vázquez-Torres, A.; Bourret, T.J. Salmonella enterica serovars Typhimurium has three transketolase enzymes contributing to the pentose phosphate pathway. J. Biol. Chem. 2018, 293, 11271–11282. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.; O’Brien, S.J.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef]
- Elshebrawy, H.A.; Mahros, M.A.; Abd-Elghany, S.M.; Elgazzar, M.M.; Hayashidani, H.; Sallam, K.I. Prevalence and molecular characterization of multidrug-resistant and β-lactamase producing Salmonella enterica serovars isolated from duck, pigeon, and quail carcasses in Mansoura, Egypt. LWT-Food Sci. Technol. 2021, 149, 111834. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention (CDC). Salmonella. Atlanta, GA. 2014. Available online: http://www.cdc.gov/Salmonella/ (accessed on 21 June 2016).
- Center for Disease Control and Prevention (CDC). Salmonellosis–General Information. 2012. Available online: http://www.cdc.gov/salmonella/general/ (accessed on 9 September 2022).
- Hammack, T. Salmonella species. In Bad Bug Book, Foodborne Pathogenic Microorganisms and Natural Toxins, 2nd ed.; Lampel, K.A., Al-Khaldi, S., Cahill, S.M., Eds.; U. S. Food and Drug Administration: Silver Spring, MD, USA, 2012; pp. 12–16. Available online: https://www.fda.gov/media/83271/download (accessed on 4 August 2022).
- Threlfall, E.J. Antimicrobial drug resistance in Salmonella: Problems and perspectives in food and water-borne infection. FEMS Microbiol. Rev. 2002, 26, 141–148. [Google Scholar] [CrossRef]
- Pezzotti, G.; Serafin, A.; Luzzi, I.; Mioni, R.; Milan, M.; Perin, R. Occurrence and resistance to antibiotics of Campylobacter jejuni and Campylobacter coli in animals and meat in northeastern Italy. Int. J. Food Microbiol. 2003, 82, 281–287. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nut. 2013, 53, 11–48. [Google Scholar] [CrossRef] [PubMed]
- Eltanani, G.; Gad, T.; Darwish, W.; El-Ghareeb, W.; Ismail, H. Prevalence of multidrug resistant Salmonella Typhimurium in retailed buffalo meat and offal with a reduction trial using rosemary and olive oils. Damanhour J. Vet. Sci. 2021, 6, 7–10. [Google Scholar] [CrossRef]
- Mohamed, W.S.; Ibrahim, M.A. Isolation on Salmonella species from buffalo calves meat and preliminary evaluation of its irradiated vaccine. Assiut Vet. Med. J. 2014, 61, 18–23. [Google Scholar] [CrossRef]
- International Organization for Standardization (ISO). Microbiology of Food Chain—Horizontal Method for Detection, Enumeration and Serotyping of SALMONELLA—Part 1: Detection of Salmonella spp., 1st ed.; ISO 6579-1:2017; The International Organization for Standardization: Geneva, Switzerland, 2017. [Google Scholar]
- Abd-Elghany, S.M.; Sallam, K.I. Occurrence and molecular identification of Vibrio parahaemolyticus in retail shellfish in Mansoura. Egypt. Food Cont. 2013, 33, 399–405. [Google Scholar] [CrossRef]
- Nayak, R.; Stewart, T.; Wang, R.F.; Lin, J.; Cerniglia, C.E.; Kenney, P.B. Genetic diversity and virulence gene determinants of antibiotic-resistant Salmonella isolated from preharvest Turkey production sources. Int. J. Food Microbiol. 2004, 91, 51–62. [Google Scholar] [CrossRef]
- Murugkar, H.V.; Rahman, H.; Dutta, P.K. Distribution of virulence genes in Salmonella serovars isolated from man & animals. Indian J. Med. Res. 2003, 117, 66–70. [Google Scholar] [PubMed]
- Cardona-Castro, N.; Restrepo-Pineda, E.; Correa-Ochoa, M. Detection of hilA gene sequences in serovars of Salmonella enterica subspecies enterica. Memórias Inst. Oswaldo Cruz. 2002, 97, 1153–1156. [Google Scholar] [CrossRef]
- CLSI (Clinical and Laboratory Standards Institute). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; M07USA; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; Available online: https://clsi.org/media/1928/m07ed11_sample.pdf (accessed on 1 July 2022).
- Singh, S.; Yadav, A.S.; Singh, S.M.; Bharti, P. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food Res. Int. 2010, 43, 2027–2030. [Google Scholar] [CrossRef]
- Mezali, L.; Hamdi, T.M. Prevalence and antimicrobial resistance of Salmonella isolated from meat and meat products in Algiers (Algeria). Foodborne Pathog. Dis. 2012, 9, 522–529. [Google Scholar] [CrossRef]
- Boonmar, S.; Morita, Y.; Pulsrikarn, C.; Chaichana, P.; Pornruagwong, S.; Chaunchom, S.; Sychanh, T.; Khounsy, T.; Sisavath, D.; Yamamoto, S.; et al. Salmonella prevalence in meat at retail markets in Pakse, Champasak Province, Laos, and antimicrobial susceptibility of isolates. J. Glob. Antimicrob. Resist. 2013, 1, 157–161. [Google Scholar] [CrossRef]
- Hassan, M.; Lutful Kabir, S.M.; Tanvir Rahman, M.D.; Sarker, Y.A. Bacteriological quality assessment of buffalo meat collected from different districts of Bangladesh with particular emphasis on the molecular detection and antimicrobial resistance of the isolated Salmonella species. Asian Australas. J. Food Saf. Secur. 2018, 2, 12–20. [Google Scholar] [CrossRef]
- Bahiraie, A.; Moghaddam, A. Isolation and identification of salmonellae serotypes from buffalo and bovine meat which were slaughtered in the Ahvaz slaughterhouse. In Proceedings of the Seventh World Buffalo Congress, Manila, Philippines, 20–23 October 2004; pp. 10–11. [Google Scholar]
- Sychanh, T.; Chaunchom, S.; Pulsrikarn, C.; Pornreongwong, S.; Chaichana, P.; Boonmar, S. Salmonella prevalence in slaughtered buffaloes and cattle in Champasak province, Lao People’s Democratic Republic. Kasetsart J. (Nat. Sci.) 2013, 47, 561–570. [Google Scholar]
- Elsayed, M.S.; Abdeen, E.; Akiela, M.A.; Farouk, T.; Zahran, R. Real time and conventional PCR for characterization of Salmonella spp. from imported meat to Egypt. Adv. Anim. Vet. Sci. 2014, 2, 199–203. [Google Scholar] [CrossRef]
- Singh, S.; Kshirsagar, D.P.; Brahmbhatt, M.N.; Nayak, J.B.; Chatur, Y.A. Isolation and characterization of Salmonella spp. from buffalo meat samples. Buffalo Bull. 2015, 34, 301–312. Available online: https://www.researchgate.net/publication/282641324 (accessed on 4 July 2022).
- Saud, B.; Paudel, G.; Khichaju, S.; Bajracharya, D.; Dhungana, G.; Awasthi, M.; Shrestha, V. Multidrug-Resistant Bacteria from Raw Meat of Buffalo and Chicken, Nepal. Vet. Med. Int. 2019, 2019, 7960268. [Google Scholar] [CrossRef] [PubMed]
- Mannion, C.; Fanning, J.; McLernon, J.; Lendrum, L.; Gutierrez, M.; Duggan, S.; Egan, J. The role of transport, lairage and slaughter processes in the dissemination of Salmonella spp. in pigs in Ireland. Food Res. Int. 2012, 45, 871–879. [Google Scholar] [CrossRef]
- Oliveira, F.A.; Brandelli, A.; Tondo, E.C. Antimicrobial resistance in Salmonella enteritidis from foods involved in human salmonellosis outbreaks in southern Brazil. New Microbiol. 2006, 29, 49–54. Available online: https://www.researchgate.net/publication/7172228 (accessed on 16 June 2022).
- Alemu, S.; Zawde, B.M. Prevalence and antimicrobial resistance profiles of Salmonella enterica serovars isolated from slaughtered cattle in Bahir Dar, Ethiopia. Trop. Anim. Health Prod. 2002, 44, 595–600. [Google Scholar] [CrossRef]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- Zou, Q.-H.; Li, R.-Q.; Liu, G.-R.; Liu, S.-L. Genotyping of Salmonella with lineage-specific genes: Correlation with serotyping. Int. J. Infect. Dis. 2016, 49, 134–140. [Google Scholar] [CrossRef][Green Version]
- Imre, A.; Olasz, F.; Nagy, B. Development of a PCR system for the characterisation of Salmonella flagellin genes. Acta Vet. Hung. 2005, 53, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, S.; Butaye, P.; Maliehe, T.S.; Magwedere, K.; Basson, A.K.; Madoroba, E. Virulence Factors and Antimicrobial Resistance in Salmonella Species Isolated from Retail Beef in Selected KwaZulu-Natal Municipality Areas, South Africa. Appl. Sci. 2022, 12, 2843. [Google Scholar] [CrossRef]
- Sallam, K.I.; Mohammed, M.A.; Hassan, M.A.; Tamura, T. Prevalence, molecular identification and antimicrobial resistance profile of Salmonella serovars isolated from retail beef products in Mansoura, Egypt. Food Cont. 2014, 38, 209–214. [Google Scholar] [CrossRef]
- Thung, T.Y.; Radu, S.; Mahyudin, N.A.; Rukayadi, Y.; Zakaria, Z.; Mazlan, N.; Tan, B.H.; Lee, E.; Yeoh, S.L.; Chin, Y.Z.; et al. Prevalence, Virulence Genes and Antimicrobial Resistance Profiles of Salmonella Serovars from Retail Beef in Selangor, Malaysia. Front. Microbiol. 2018, 11, 2697. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Zhu, L.; Mao, Y.; Liang, R.; Niu, L.; Zhang, Y.; Li, K.; Luo, X. Prevalence and profile of Salmonella from samples along the production line in Chinese beef processing plants. Food Control 2014, 38, 54–60. [Google Scholar] [CrossRef]
- Yanestria, S.M.; Rahmaniar, R.P.; Wibisono, F.J.; Effendi, M.H. Detection of invA gene of Salmonella from milkfish (Chanos chanos) at Sidoarjo wet fish market, Indonesia, using polymerase chain reaction technique. Vet World 2019, 12, 170–175. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 8, 268–281. [Google Scholar] [CrossRef]
- Miranda, J.M.; Mondrago, N.A.; Martinez, B.; Guarddon, M.; Rodriguez, J.A. Prevalence and Antimicrobial Resistance Patterns of Salmonella from Different Raw Foods in Mexico. J. Food Prot. 2009, 72, 966–971. [Google Scholar] [CrossRef]
- Fluit, A.C. Towards more virulent and antibiotic-resistant Salmonella? FEMS Immunol. Med. Microbiol. 2005, 43, 1–11. [Google Scholar] [CrossRef]
- Cha, S.Y.; Kang, M.; Yoon, R.H.; Park, C.K.; Moon, O.K.; Jang, H.K. Prevalence and antimicrobial susceptibility of Salmonella isolates in Pekin ducks from South Korea. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 473–479. [Google Scholar] [CrossRef]
- Kijima, M.; Shirakawa, T.; Uchiyama, M.; Kawanishi, M.; Ozawa, M.; Koike, R. Trends in the serovar and antimicrobial resistance in clinical isolates of Salmonella enterica from cattle and pigs between 2002 and 2016 in Japan. J. Appl. Microbiol. 2019, 127, 1869–1875. [Google Scholar] [CrossRef]
- Rao, S.; Linke, L.; Doster, E.; Hyatt, D.; Burgess, B.A.; Magnuson, R.; Morley, P.S. Genomic diversity of class I integrons from antimicrobial resistant strains of Salmonella Typhimurium isolated from livestock, poultry and humans. PLoS ONE 2020, 15, e0243477. [Google Scholar] [CrossRef]
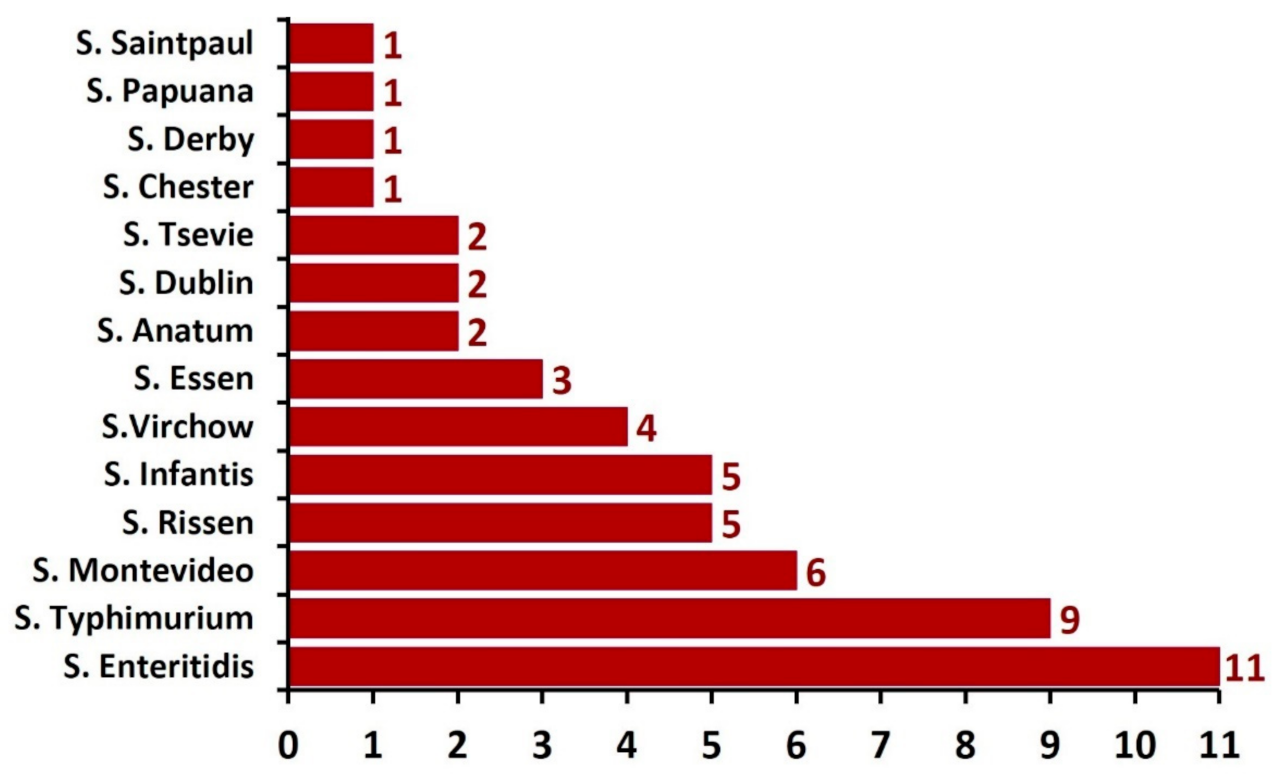

| Serovars (Number) | Positivity of the Salmonella Serovars for the Tested Virulence Genes | ||
|---|---|---|---|
| invA | stn | hilA | |
| S. Enteritidis (9) | + | + | + |
| S. Enteritidis (1) | + | − | + |
| S. Enteritidis (1) | + | + | − |
| S. Typhimurium (7) | + | + | + |
| S. Typhimurium (1) | + | − | + |
| S. Typhimurium (1) | + | + | − |
| S. Montevideo (3) | + | + | − |
| S. Montevideo (3) | + | + | + |
| S. Rissen (5) | + | − | + |
| S. Infantis (4) | + | + | + |
| S. Infantis (1) | + | − | + |
| S. Virchow (3) | + | − | + |
| S. Virchow (1) | + | + | − |
| S. Essen (2) | + | + | − |
| S. Essen (1) | + | + | + |
| S. Anatum (2) | + | − | + |
| S. Dublin (2) | + | + | + |
| S. Tsevie (1) | + | + | + |
| S. Tsevie (1) | + | − | + |
| S. Chester (1) | + | + | + |
| S. Derby (1) | + | + | + |
| S. Papuana (1) | + | − | + |
| S. Saintpaul (1) | + | + | − |
| Total (53) | 53 (100%) | 38 (71.7%) | 44 (83.0%) |
| Antimicrobial Agents | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| E | 0 (0%) | 0 (0%) | 53 (100%) |
| S | 0 (0%) | 1 (1.9%) | 52 (98.1%) |
| CM | 1 (1.9%) | 2 (3.8%) | 50 (94.3%) |
| CPM | 7 (13.2%) | 5 (9.4%) | 41 (77.4%) |
| NA | 15 (28.3%) | 3 (5.7%) | 35 (66.0%) |
| STX | 21 (39.6%) | 3 (5.7%) | 29 (54.7%) |
| CET | 24 (45.3%) | 2 (3.8%) | 27 (50.9%) |
| AMP | 30 (56.6%) | 1 (1.9%) | 22 (41.5%) |
| TE | 31 (58.5%) | 4 (7.5%) | 18 (34.0%) |
| ENR | 37 (69.8%) | 0 (0%) | 16 (30.2%) |
| CIP | 39 (73.6%) | 3 (5.6%) | 11 (20.8%) |
| MEM | 44 (83.0%) | 2 (3.8%) | 7 (13.2%) |
| CTX | 46 (86.8%) | 2 (3.8%) | 5 (9.4%) |
| GEN | 49 (92.5%) | 0 (0%) | 4 (7.5%) |
| IPM | 50 (94.3%) | 1 (1.9%) | 2 (3.8%) |
| AMK | 52 (98.1%) | 0 (0%) | 1 (1.9%) |
| Salmonella Strains (n = Number) | Antimicrobial Resistance Profile | MAR Index |
|---|---|---|
| S. Enteritidis | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN, IPM, AMK | 1 |
| S. Enteritidis | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN | 0.875 |
| S. Enteritidis | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR | 0.625 |
| S. Enteritidis | E, S, CM, CPM, NA, SXT, CET, AMP, TE | 0.563 |
| S. Enteritidis (n = 2) | E, S, CM, CPM, NA, SXT, CET | 0.438 |
| S. Enteritidis | E, S, CM, CPM, NA, SXT | 0.375 |
| S. Enteritidis | E, S, CM, CPM, NA | 0.312 |
| S. Enteritidis | E, S, CM, CPM | 0.250 |
| S. Enteritidis (n = 2) | E, S, CM | 0.187 |
| S. Typhimurium | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN, IPM | 0.938 |
| S. Typhimurium | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM | 0.750 |
| S. Typhimurium | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR | 0.625 |
| S. Typhimurium (n = 2) | E, S, CM, CPM, NA, SXT, CET, AMP | 0.500 |
| S. Typhimurium | E, S, CM, CPM, NA, SXT | 0.375 |
| S. Typhimurium | E, S, CM | 0.187 |
| S. Typhimurium | E, S | 0.125 |
| S. Montevideo | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN | 0.875 |
| S. Montevideo | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR | 0.625 |
| S. Montevideo | E, S, CM, CPM, NA, SXT, CET | 0.438 |
| S. Montevideo (n = 2) | E, S, CM, CPM, NA | 0.312 |
| S. Montevideo | E, S | 0.125 |
| S. Rissen | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX | 0.812 |
| S. Rissen | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP | 0.687 |
| S. Rissen | E, S, CM, CPM, NA, SXT, CET, AMP | 0.500 |
| S. Rissen | E, S, CM, CPM | 0.250 |
| S. Rissen | E, S, CM | 0.187 |
| S. Infantis | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM | 0.750 |
| S. Infantis | E, S, CM, CPM, NA, SXT, CET, AMP, TE | 0.563 |
| S. Infantis | E, S, CM, CPM, NA, SXT, CET | 0.438 |
| S. Infantis | E, S, CM, CPM, NA | 0.312 |
| S. Infantis | E, S, CM, CPM | 0.250 |
| S. Virchow | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP | 0.687 |
| S. Virchow | E, S, CM, CPM, NA, SXT, CET, AMP | 0.500 |
| S. Virchow | E, S, CM, CPM, NA | 0.312 |
| S. Virchow | E, S | 0.125 |
| S. Essen | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP | 0.687 |
| S. Essen | E, S, CM, CPM, NA, SXT, CET | 0.438 |
| S. Essen | E, S, CM | 0.187 |
| S. Tsevie | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP | 0.687 |
| S. Tsevie | E, S, CM, CPM, NA | 0.312 |
| S. Dublin | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR | 0.625 |
| S. Dublin | E, S, CM, CPM | 0.250 |
| S. Anatum | E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR | 0.625 |
| S. Anatum | E, S | 0.125 |
| S. Derby | E, S, CM, CPM | 0.250 |
| S. Papuana | E, S, CM, CPM | 0.250 |
| S. Saintpaul | E, S, CM | 0.187 |
| S. Chester | E, S | 0.125 |
| S. Chester | E | 0.0625 |
| n = 53 | Average MAR: 0.436 |
| Antimicrobial Resistance Phenotype | Number and (%) of Isolates | MAR Index 1 | Classification of Strains | |
|---|---|---|---|---|
| Type of Resistance | Number and (%) of Isolates | |||
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN, IPM, AMK | 1 (1.9%) | 1 | Pandrug-resistance | 1 (1.9%) |
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN, IPM | 1 (1.9%) | 0.938 | Extensively drug-resistant | 4 (7.5%) |
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX, GEN | 2 (3.8%) | 0.875 | ||
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM, CTX | 1 (1.9%) | 0.8125 | ||
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP, MEM | 2 (3.8%) | 0.750 | Multi-drug resistant | 42 (79.2%) |
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR, CIP | 4 (7.54%) | 0.687 | ||
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, ENR | 5 (9.4%) | 0.625 | ||
| E, S, CM, CPM, NA, SXT, CET, AMP, TE, | 2 (3.8%) | 0.562 | ||
| E, S, CM, CPM, NA, SXT, CET, AMP | 4 (7.5%) | 0.500 | ||
| E, S, CM, CPM, NA, SXT, CET | 5 (9.4%) | 0.437 | ||
| E, S, CM, CPM, NA, SXT | 2 (3.8%) | 0.375 | ||
| E, S, CM, CPM, NA | 6 (11.3%) | 0.312 | ||
| E, S, CM, CPM | 6 (11.3%) | 0.250 | ||
| E, S, CM | 6 (11.3%) | 0.187 | ||
| E, S | 5 (9.4%) | 0.125 | Low-drug resistant | 6 (11.3%) |
| E | 1 (1.9%) | 0.062 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd-Elghany, S.M.; Fathy, T.M.; Zakaria, A.I.; Imre, K.; Morar, A.; Herman, V.; Pașcalău, R.; Șmuleac, L.; Morar, D.; Imre, M.; et al. Prevalence of Multidrug-Resistant Salmonella enterica Serovars in Buffalo Meat in Egypt. Foods 2022, 11, 2924. https://doi.org/10.3390/foods11182924
Abd-Elghany SM, Fathy TM, Zakaria AI, Imre K, Morar A, Herman V, Pașcalău R, Șmuleac L, Morar D, Imre M, et al. Prevalence of Multidrug-Resistant Salmonella enterica Serovars in Buffalo Meat in Egypt. Foods. 2022; 11(18):2924. https://doi.org/10.3390/foods11182924
Chicago/Turabian StyleAbd-Elghany, Samir Mohammed, Takwa Mohammed Fathy, Amira Ibrahim Zakaria, Kálmán Imre, Adriana Morar, Viorel Herman, Raul Pașcalău, Laura Șmuleac, Doru Morar, Mirela Imre, and et al. 2022. "Prevalence of Multidrug-Resistant Salmonella enterica Serovars in Buffalo Meat in Egypt" Foods 11, no. 18: 2924. https://doi.org/10.3390/foods11182924
APA StyleAbd-Elghany, S. M., Fathy, T. M., Zakaria, A. I., Imre, K., Morar, A., Herman, V., Pașcalău, R., Șmuleac, L., Morar, D., Imre, M., & Sallam, K. I. (2022). Prevalence of Multidrug-Resistant Salmonella enterica Serovars in Buffalo Meat in Egypt. Foods, 11(18), 2924. https://doi.org/10.3390/foods11182924












