Innovative Application of Metabolomics on Bioactive Ingredients of Foods
Abstract
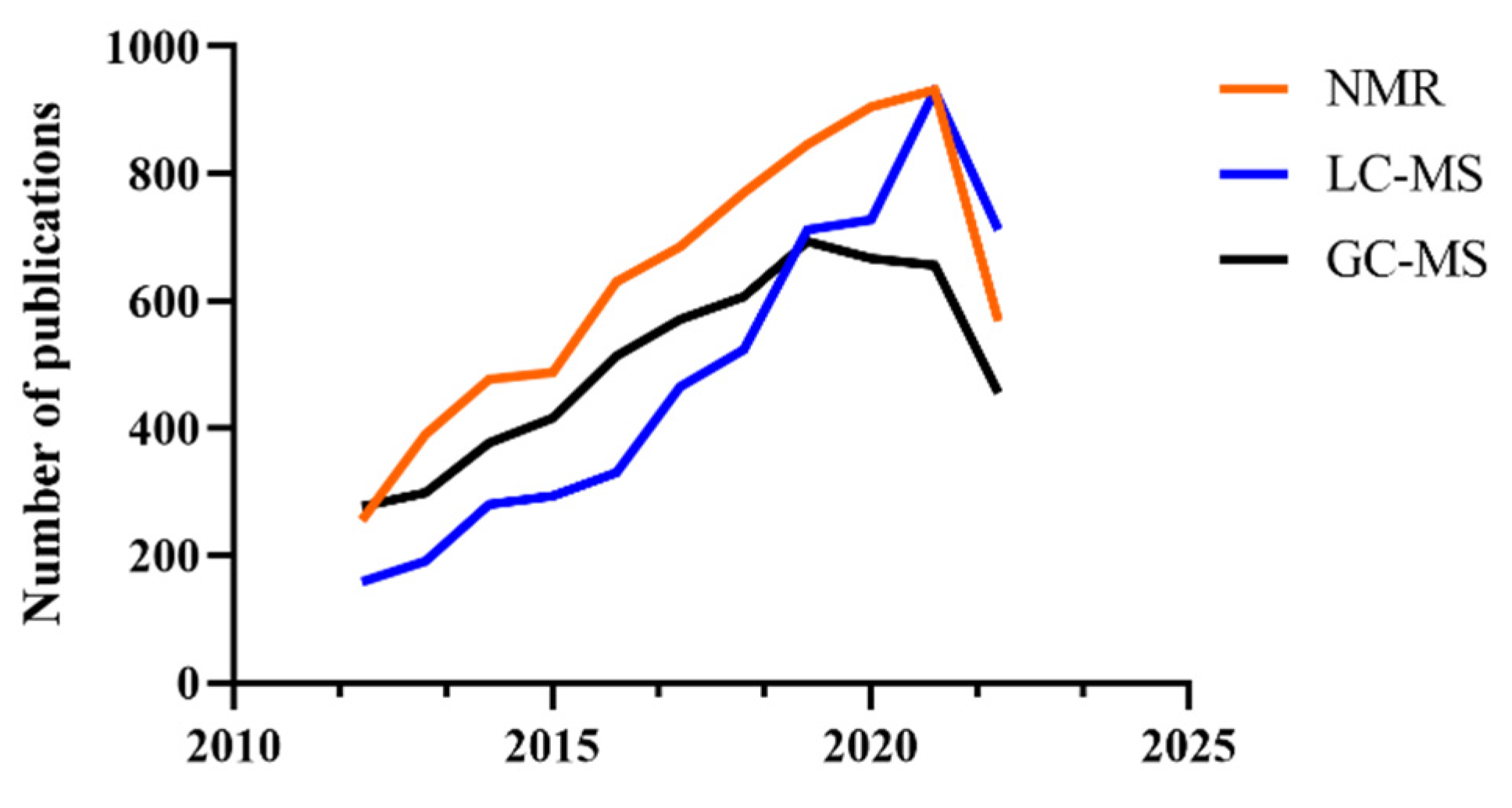
1. Introduction
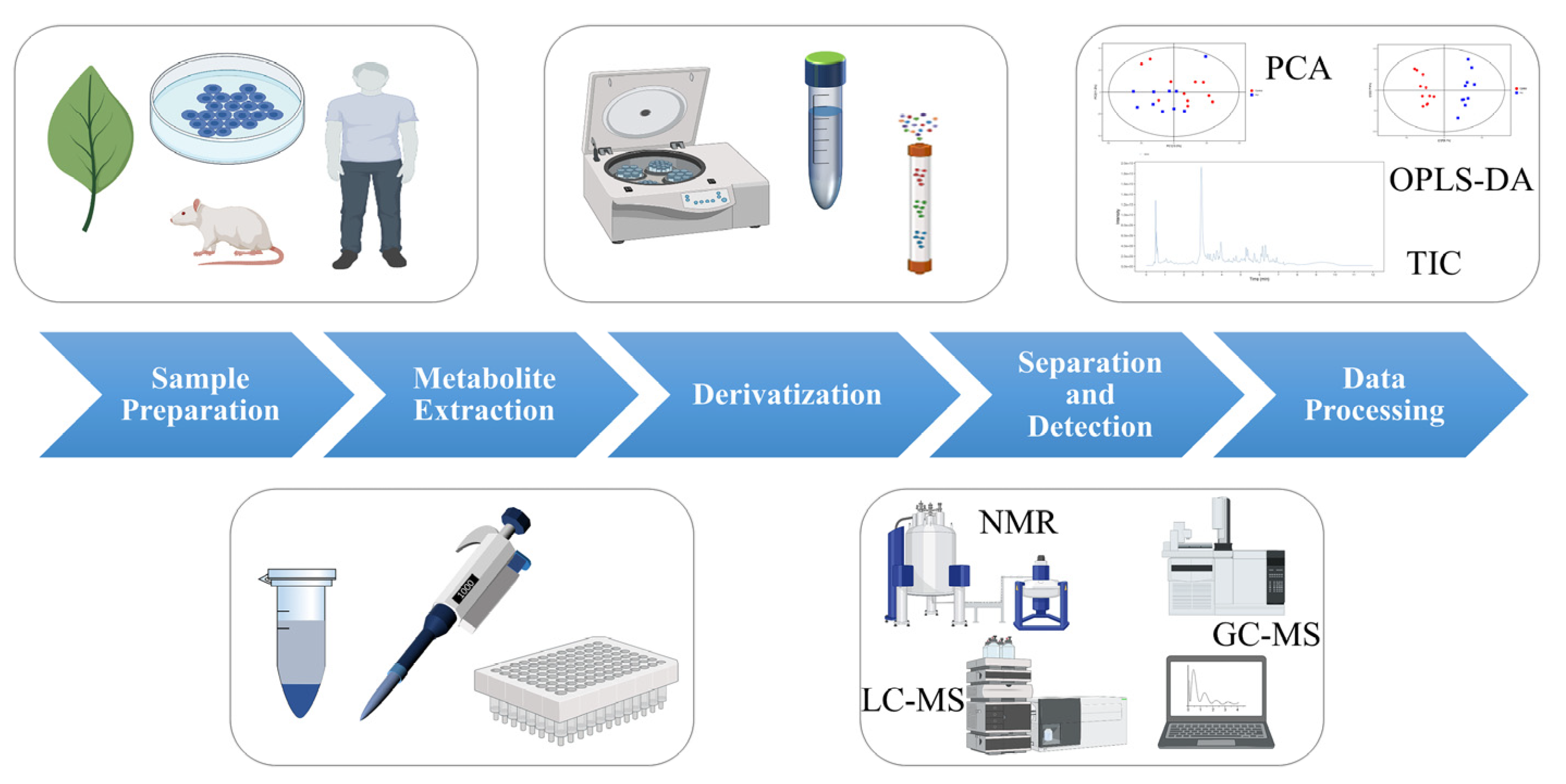
2. Process of Metabolomics Analysis
2.1. Sample Preparation
2.2. Metabolite Extraction
2.3. Derivatization
2.4. Separation and Detection
2.5. Data Processing
3. Application of Metabolomics in Nutrition and Health
3.1. Application of Metabolomics in the Discovery of Bioactive Substances in Plants
3.2. Application of Metabolomics in the Effect of Bioactives In Vitro
3.3. Application of Metabolomics in the Effect of Bioactives in Animals
3.4. Application of Metabolomics in the Screening of Bioactives for Human Trials
4. Conclusions
5. Challenges and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rizo, J.; Guillen, D.; Farres, A.; Diaz-Ruiz, G.; Sanchez, S.; Wacher, C.; Rodriguez-Sanoja, R. Omics in traditional vegetable fermented foods and beverages. Crit. Rev. Food Sci. Nutr. 2020, 60, 791–809. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ’Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhang, A.H.; Miao, J.H.; Sun, H.; Han, Y.; Yan, G.L.; Wu, F.F.; Wang, X.J. Metabolomics biotechnology, applications, and future trends: A systematic review. RSC Adv. 2019, 9, 37245–37257. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.M.; Lyu, Y.; Pletcher, S.D.; Promislow, D.E.L. Proteomics and metabolomics in ageing research: From biomarkers to systems biology. Essays Biochem. 2017, 61, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- Ribbenstedt, A.; Ziarrusta, H.; Benskin, J.P. Development, characterization and comparisons of targeted and non-targeted metabolomics methods. PLoS ONE 2018, 13, e0207082. [Google Scholar] [CrossRef]
- Feng, J.; Xu, B.; Ma, D.; Hao, Z.; Jia, Y.; Wang, C.; Wang, L. Metabolite identification in fresh wheat grains of different colors and the influence of heat processing on metabolites via targeted and non-targeted metabolomics. Food Res. Int. 2022, 160, 111728. [Google Scholar] [CrossRef]
- Pezzatti, J.; Boccard, J.; Codesido, S.; Gagnebin, Y.; Joshi, A.; Picard, D.; Gonzalez-Ruiz, V.; Rudaz, S. Implementation of liquid chromatography-high resolution mass spectrometry methods for untargeted metabolomic analyses of biological samples: A tutorial. Anal. Chim. Acta 2020, 1105, 28–44. [Google Scholar] [CrossRef]
- Larive, C.K.; Barding, G.A.; Dinges, M.M. NMR spectroscopy for metabolomics and metabolic profiling. Anal. Chem. 2015, 87, 133–146. [Google Scholar] [CrossRef]
- Collins, C.; McNamara, A.E.; Brennan, L. Role of metabolomics in identification of biomarkers related to food intake. Proc. Nutr. Soc. 2019, 78, 189–196. [Google Scholar] [CrossRef]
- Li, S.; Tian, Y.; Jiang, P.; Lin, Y.; Liu, X.; Yang, H. Recent advances in the application of metabolomics for food safety control and food quality analyses. Crit. Rev. Food Sci. Nutr. 2021, 61, 1448–1469. [Google Scholar] [CrossRef] [PubMed]
- Utpott, M.; Rodrigues, E.; Rios, A.D.; Mercali, G.D.; Flores, S.H. Metabolomics: An analytical technique for food processing evaluation. Food Chem. 2022, 366, 130685. [Google Scholar] [CrossRef] [PubMed]
- Segers, K.; Declerck, S.; Mangelings, D.; Vander Heyden, Y.; Van Eeckhaut, A. Analytical techniques for metabolomic studies: A review. Bioanalysis 2019, 11, 2297–2318. [Google Scholar] [CrossRef] [PubMed]
- Kaluarachchi, M.; Boulange, C.L.; Karaman, I.; Lindon, J.C.; Ebbels, T.M.D.; Elliott, P.; Tracy, R.P.; Olson, N.C. A comparison of human serum and plasma metabolites using untargeted (1)H NMR spectroscopy and UPLC-MS. Metabolomics 2018, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Misra, B.B. New software tools, databases, and resources in metabolomics: Updates from 2020. Metabolomics 2021, 17, 49. [Google Scholar] [CrossRef]
- Wishart, D.S.; Cheng, L.L.; Copie, V.; Edison, A.S.; Eghbalnia, H.R.; Hoch, J.C.; Gouveia, G.J.; Pathmasiri, W.; Powers, R.; Schock, T.B.; et al. NMR and Metabolomics-A Roadmap for the Future. Metabolites 2022, 12, 678. [Google Scholar] [CrossRef]
- Anand, A.; Sharma, A.; Kaur Saini, H.; Sharma, S.; Sharma, R.; Thakur, C.; Priyanka; Atanassova, M.; Caruso, G.; Pasdaran, A.; et al. Profiling of Plant Derived Natural Constituents by Using Magnetic Resonance Techniques. Concepts Magn. Reson. Part A 2022, 2022, 5705637. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Overview of NMR Spectroscopy-Based Metabolomics: Opportunities and Challenges. Methods Mol. Biol. 2019, 2037, 3–14. [Google Scholar] [CrossRef]
- Geier, F.M.; Leroi, A.M.; Bundy, J.G. (13)C Labeling of Nematode Worms to Improve Metabolome Coverage by Heteronuclear Nuclear Magnetic Resonance Experiments. Front. Mol. Biosci. 2019, 6, 27. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef]
- Markley, J.L.; Bruschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Eskandari, R.; Park, S.M.; Alvarez, J.; Tee, S.S.; Weissleder, R.; Kharas, M.G.; Lee, H.; Keshari, K.R. Real-time quantitative analysis of metabolic flux in live cells using a hyperpolarized micromagnetic resonance spectrometer. Sci. Adv. 2017, 3, e1700341. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Dominguez, R.; Sayago, A.; Fernandez-Recamales, A. Metabolomics in Alzheimer’s disease: The need of complementary analytical platforms for the identification of biomarkers to unravel the underlying pathology. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1071, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Ulaszewska, M.M.; Weinert, C.H.; Trimigno, A.; Portmann, R.; Andres Lacueva, C.; Badertscher, R.; Brennan, L.; Brunius, C.; Bub, A.; Capozzi, F.; et al. Nutrimetabolomics: An Integrative Action for Metabolomic Analyses in Human Nutritional Studies. Mol. Nutr. Food Res. 2019, 63, e1800384. [Google Scholar] [CrossRef]
- Yan, S.C.; Chen, Z.F.; Zhang, H.; Chen, Y.; Qi, Z.; Liu, G.; Cai, Z. Evaluation and optimization of sample pretreatment for GC/MS-based metabolomics in embryonic zebrafish. Talanta 2020, 207, 120260. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, Y.; Ji, H.; Wang, Y.; Zhang, Z.; Lu, H. Enhancing coverage in LC-MS-based untargeted metabolomics by a new sample preparation procedure using mixed-mode solid-phase extraction and two derivatizations. Anal. Bioanal. Chem. 2019, 411, 6189–6202. [Google Scholar] [CrossRef]
- Yanibada, B.; Boudra, H.; Debrauwer, L.; Martin, C.; Morgavi, D.P.; Canlet, C. Evaluation of sample preparation methods for NMR-based metabolomics of cow milk. Heliyon 2018, 4, e00856. [Google Scholar] [CrossRef]
- Reyes-Garcés, N.; Gionfriddo, E. Recent developments and applications of solid phase microextraction as a sample preparation approach for mass-spectrometry-based metabolomics and lipidomics. Trends Analyt. Chem. 2019, 113, 172–181. [Google Scholar] [CrossRef]
- Vasiljevic, T.; Singh, V.; Pawliszyn, J. Miniaturized SPME tips directly coupled to mass spectrometry for targeted determination and untargeted profiling of small samples. Talanta 2019, 199, 689–697. [Google Scholar] [CrossRef]
- Chan, W.; Zhao, Y.; Zhang, J. Evaluating the performance of sample preparation methods for ultra-performance liquid chromatography/mass spectrometry based serum metabonomics. Rapid Commun. Mass Spectrom. 2019, 33, 561–568. [Google Scholar] [CrossRef]
- Yen, S.; Bolte, E.; Aucoin, M.; Allen-Vercoe, E. Metabonomic Evaluation of Fecal Water Preparation Methods: The Effects of Ultracentrifugation. Curr. Metab. 2018, 6, 57–63. [Google Scholar] [CrossRef]
- Vuckovic, D. Sample preparation in global metabolomics of biological fluids and tissues. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Academic Press: London, UK, 2020; pp. 53–83. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Perez de Souza, L.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37. [Google Scholar] [CrossRef]
- Martineau, E.; Tea, I.; Loaec, G.; Giraudeau, P.; Akoka, S. Strategy for choosing extraction procedures for NMR-based metabolomic analysis of mammalian cells. Anal. Bioanal. Chem. 2011, 401, 2133–2142. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Kolho, K.L.; Pessia, A.; Jaakkola, T.; de Vos, W.M.; Velagapudi, V. Faecal and Serum Metabolomics in Paediatric Inflammatory Bowel Disease. J. Crohns Colitis 2017, 11, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Seeger, K. Simple and Rapid (Extraction) Protocol for NMR Metabolomics-Direct Measurement of Hydrophilic and Hydrophobic Metabolites Using Slice Selection. Anal. Chem. 2021, 93, 1451–1457. [Google Scholar] [CrossRef]
- Karu, N.; Deng, L.; Slae, M.; Guo, A.C.; Sajed, T.; Huynh, H.; Wine, E.; Wishart, D.S. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Anal. Chim. Acta 2018, 1030, 1–24. [Google Scholar] [CrossRef]
- Erarpat, S.; Bodur, S.; Ozturk Er, E.; Bakirdere, S. Combination of ultrasound-assisted ethyl chloroformate derivatization and switchable solvent liquid-phase microextraction for the sensitive determination of l-methionine in human plasma by GC-MS. J. Sep. Sci. 2020, 43, 1100–1106. [Google Scholar] [CrossRef]
- Zhao, S.; Li, L. Dansylhydrazine Isotope Labeling LC-MS for Comprehensive Carboxylic Acid Submetabolome Profiling. Anal. Chem. 2018, 90, 13514–13522. [Google Scholar] [CrossRef]
- Zhu, S.; Zheng, Z.; Peng, H.; Sun, J.; Zhao, X.E.; Liu, H. Quadruplex stable isotope derivatization strategy for the determination of panaxadiol and panaxatriol in foodstuffs and medicinal materials using ultra high performance liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2020, 1616, 460794. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Su, S.; Yan, P.; Shang, J.; Wang, J.; Yan, C.; Li, J.; Wang, Q.; Xiong, X.; Xu, H. Integrative analyses of widely targeted metabolomic profiling and derivatization-based LC-MS/MS reveals metabolic changes of Zingiberis Rhizoma and its processed products. Food Chem. 2022, 389, 133068. [Google Scholar] [CrossRef]
- Bian, X.; Li, N.; Tan, B.; Sun, B.; Guo, M.Q.; Huang, G.; Fu, L.; Hsiao, W.L.W.; Liu, L.; Wu, J.L. Polarity-Tuning Derivatization-LC-MS Approach for Probing Global Carboxyl-Containing Metabolites in Colorectal Cancer. Anal. Chem. 2018, 90, 11210–11215. [Google Scholar] [CrossRef] [PubMed]
- Harrieder, E.M.; Kretschmer, F.; Bocker, S.; Witting, M. Current state-of-the-art of separation methods used in LC-MS based metabolomics and lipidomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2022, 1188, 123069. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. NMR metabolomics: A look ahead. J. Magn. Reson. 2019, 306, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Agin, A.; Heintz, D.; Ruhland, E.; de la Barca, J.M.C.; Zumsteg, J.; Moal, V.; Gauehez, A.S.; Namer, I.J. Metabolomics—An overview. From basic principles to potential biomarkers (part 1). Med. Nucl.-Imag. Fonct. Metab. 2016, 40, 4–10. [Google Scholar] [CrossRef]
- Zhao, D.S.; Wu, Z.T.; Li, Z.Q.; Wang, L.L.; Jiang, L.L.; Shi, W.; Li, P.; Li, H.J. Liver-specific metabolomics characterizes the hepatotoxicity of Dioscorea bulbifera rhizome in rats by integration of GC-MS and (1)H-NMR. J. Ethnopharmacol. 2018, 226, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Arumugam, S.; Lorkiewicz, P.K.; Higashi, R.M.; Laulhe, S.; Nantz, M.H.; Moseley, H.N.; Fan, T.W. Chemoselective detection and discrimination of carbonyl-containing compounds in metabolite mixtures by 1H-detected 15N nuclear magnetic resonance. Magn. Reson. Chem. 2015, 53, 337–343. [Google Scholar] [CrossRef]
- Rubert, J.; Righetti, L.; Stranska-Zachariasova, M.; Dzuman, Z.; Chrpova, J.; Dall’Asta, C.; Hajslova, J. Untargeted metabolomics based on ultra-high-performance liquid chromatography-high-resolution mass spectrometry merged with chemometrics: A new predictable tool for an early detection of mycotoxins. Food Chem. 2017, 224, 423–431. [Google Scholar] [CrossRef]
- Alboniga, O.E.; Gonzalez, O.; Alonso, R.M.; Xu, Y.; Goodacre, R. Optimization of XCMS parameters for LC-MS metabolomics: An assessment of automated versus manual tuning and its effect on the final results. Metabolomics 2020, 16, 14. [Google Scholar] [CrossRef]
- McLean, C.; Kujawinski, E.B. AutoTuner: High Fidelity and Robust Parameter Selection for Metabolomics Data Processing. Anal. Chem. 2020, 92, 5724–5732. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.Q.; Chong, J.; Li, S.Z.; Xia, J.G. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhong, F.Y.; Zhu, J.J. Bridging Targeted and Untargeted Mass Spectrometry-Based Metabolomics via Hybrid Approaches. Metabolites 2020, 10, 348. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Erban, A.; Weber, R.J.M.; Creek, D.J.; Brown, M.; Breitling, R.; Hankemeier, T.; Goodacre, R.; Neumann, S.; Kopka, J.; et al. Mass appeal: Metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics 2013, 9, S44–S66. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53, e1. [Google Scholar] [CrossRef] [PubMed]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Fang, H.; Zhang, A.H.; Sun, H.; Yu, J.B.; Wang, L.; Wang, X.J. High-throughput metabolomics screen coupled with multivariate statistical analysis identifies therapeutic targets in alcoholic liver disease rats using liquid chromatography-mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1109, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Uarrota, V.G.; Moresco, R.; Coelho, B.; Nunes Eda, C.; Peruch, L.A.; Neubert Ede, O.; Rocha, M.; Maraschin, M. Metabolomics combined with chemometric tools (PCA, HCA, PLS-DA and SVM) for screening cassava (Manihot esculenta Crantz) roots during postharvest physiological deterioration. Food Chem. 2014, 161, 67–78. [Google Scholar] [CrossRef]
- Eicher, T.; Kinnebrew, G.; Patt, A.; Spencer, K.; Ying, K.; Ma, Q.; Machiraju, R.; Mathe, A.E.A. Metabolomics and Multi-Omics Integration: A Survey of Computational Methods and Resources. Metabolites 2020, 10, 202. [Google Scholar] [CrossRef]
- Morais, C.; Lima, K. Principal Component Analysis with Linear and Quadratic Discriminant Analysis for Identification of Cancer Samples Based on Mass Spectrometry. J. Braz. Chem. Soc. 2017, 29, 472–481. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis--a marriage of convenience or a shotgun wedding. Anal. Chim. Acta. 2015, 879, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Daghir-Wojtkowiak, E.; Kaliszan, R.; Markuszewski, M.J. PLS-Based and Regularization-Based Methods for the Selection of Relevant Variables in Non-targeted Metabolomics Data. Front. Mol. Biosci. 2016, 3, 35. [Google Scholar] [CrossRef]
- Li, M.X.; Liu, M.Y.; Wang, B.Y.; Shi, L. Metabonomics Analysis of Stem Extracts from Dalbergia sissoo. Molecules 2022, 27, 1982. [Google Scholar] [CrossRef]
- Homayoun, S.B.; Shrikant, I.B.; Kazem, M.; Hemmat, M.; Reza, M. Compared application of the new OPLS-DA statistical model versus partial least squares regression to manage large numbers of variables in an injury case-control study. Sci. Res. Essays 2011, 6, 4369–4377. [Google Scholar] [CrossRef]
- Xue, W.; Zhang, H.; Liu, M.; Chen, X.; He, S.; Chu, Y. Metabolomics-based screening analysis of PPCPs in water pretreated with five different SPE columns. Anal. Methods 2021, 13, 4594–4603. [Google Scholar] [CrossRef] [PubMed]
- Arendse, E.; Fawole, O.A.; Magwaza, L.S.; Nieuwoudt, H.; Opara, U.L. Evaluation of biochemical markers associated with the development of husk scald and the use of diffuse reflectance NIR spectroscopy to predict husk scald in pomegranate fruit. Sci. Hortic. 2018, 232, 240–249. [Google Scholar] [CrossRef]
- Rohart, F.; Villa-Vialaneix, N.; Paris, A.; Canlet, C.; Sancristobal, M. Phenotypic Prediction Based on Metabolomic Data: LASSO vs. BOLASSO, Primary Data vs Wavelet Transformation. HAL Open Sci. 2010, 3–55. Available online: https://hal.archives-ouvertes.fr/hal-00658819 (accessed on 12 August 2022).
- Anowar, F.; Sadaoui, S.; Selim, B. Conceptual and empirical comparison of dimensionality reduction algorithms (PCA, KPCA, LDA, MDS, SVD, LLE, ISOMAP, LE, ICA, t-SNE). Comput. Sci. Rev. 2021, 40, 100378. [Google Scholar] [CrossRef]
- Mazzilli, K.M.; McClain, K.M.; Lipworth, L.; Playdon, M.C.; Sampson, J.N.; Clish, C.B.; Gerszten, R.E.; Freedman, N.D.; Moore, S.C. Identification of 102 Correlations between Serum Metabolites and Habitual Diet in a Metabolomics Study of the Prostate, Lung, Colorectal, and Ovarian Cancer Trial. J. Nutr. 2020, 150, 694–703. [Google Scholar] [CrossRef]
- Ruoppolo, M.; Caterino, M.; Albano, L.; Pecce, R.; Di Girolamo, M.G.; Crisci, D.; Costanzo, M.; Milella, L.; Franconi, F.; Campesi, I. Targeted metabolomic profiling in rat tissues reveals sex differences. Sci. Rep. 2018, 8, 4663. [Google Scholar] [CrossRef]
- Galanakis, C.M. Functionality of Food Components and Emerging Technologies. Foods 2021, 10, 128. [Google Scholar] [CrossRef] [PubMed]
- Fraga, C.G.; Croft, K.D.; Kennedy, D.O.; Tomas-Barberan, F.A. The effects of polyphenols and other bioactives on human health. Food Funct. 2019, 10, 514–528. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, S.; Guha, S.; Majumder, K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients 2018, 10, 1738. [Google Scholar] [CrossRef]
- Fasakhodi, M.T.; Abed-Elmdoust, A.; Mirvaghefi, A.; Hosseini, S.V.; Tavabe, K.R. Changes of extracted bioactive compounds from brown algae (Cystoseira indica) after conversion to mill and tablet using a quantitative metabolomics approach. Aquacu. Int. 2021, 29, 2793–2804. [Google Scholar] [CrossRef]
- Bi, W.; Zhao, G.; Zhou, Y.; Xia, X.; Wang, J.; Wang, G.; Lu, S.; He, W.; Bi, T.; Li, J. Metabonomics analysis of flavonoids in seeds and sprouts of two Chinese soybean cultivars. Sci. Rep. 2022, 12, 5541. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Tang, H. Quantitative Metabonomic Analysis Reveals the Germination-Associated Dynamic and Systemic Biochemical Changes for Mung-Bean (Vigna radiata) Seeds. J. Proteome Res. 2020, 19, 2457–2470. [Google Scholar] [CrossRef]
- Xu, C.; Liang, L.; Yang, T.; Feng, L.; Mao, X.; Wang, Y. In-vitro bioactivity evaluation and non-targeted metabolomic analysis of green tea processed from different tea shoot maturity. LWT 2021, 152, 112234. [Google Scholar] [CrossRef]
- Hellal, K.; Mediani, A.; Ismail, I.S.; Tan, C.P.; Abas, F. H-1 NMR-based metabolomics and UHPLC-ESI-MS/MS for the investigation of bioactive compounds from Lupinus albus fractions. Food Res. Int. 2021, 140, 110046. [Google Scholar] [CrossRef]
- Cui, Y.; Du, K.; Hou, S.; Yang, R.; Qi, L.; Li, J.; Chang, Y. A comprehensive strategy integrating metabolomics with multiple chemometric for discovery of function related active markers for assessment of foodstuffs: A case of hawthorn (Crataegus cuneata) fruits. Food Chem. 2022, 383, 132464. [Google Scholar] [CrossRef]
- Tomas, M.; Zhang, L.; Zengin, G.; Rocchetti, G.; Capanoglu, E.; Lucini, L. Metabolomic insight into the profile, in vitro bioaccessibility and bioactive properties of polyphenols and glucosinolates from four Brassicaceae microgreens. Food Res. Int. 2021, 140, 110039. [Google Scholar] [CrossRef]
- Rocchetti, G.; Tomas, M.; Zhang, L.; Zengin, G.; Lucini, L.; Capanoglu, E. Red beet (Beta vulgaris) and amaranth (Amaranthus sp.) microgreens: Effect of storage and in vitro gastrointestinal digestion on the untargeted metabolomic profile. Food Chem. 2020, 332, 127415. [Google Scholar] [CrossRef]
- Gupta, A.J.; Hageman, J.A.; Wierenga, P.A.; Boots, J.W.; Gruppen, H. Chemometric analysis of soy protein hydrolysates used in animal cell culture for IgG production—An untargeted metabolomics approach. Process Biochem. 2014, 49, 309–317. [Google Scholar] [CrossRef]
- Richardson, J.; Shah, B.; Bondarenko, P.V.; Bhebe, P.; Zhang, Z.; Nicklaus, M.; Kombe, M.C. Metabolomics analysis of soy hydrolysates for the identification of productivity markers of mammalian cells for manufacturing therapeutic proteins. Biotechnol. Prog. 2015, 31, 522–531. [Google Scholar] [CrossRef]
- Nie, J.H.; Huang, J.X.; Wu, Q.R.; Qin, X.M.; Li, Z.Y. Uncovering the anti-proliferation mechanism and bioactive compounds in red kidney bean coat against B16-F10 melanoma cells by metabolomics and network pharmacology analysis. Food Funct. 2019, 10, 912–924. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gong, X.; Quan, Y.; Zhou, Y.; Li, Y.; Peng, C. A Cell-Based Metabonomics Approach to Investigate the Varied Influences of Chrysophanol-8-O-beta-D-Glucoside With Different Concentrations on L-02 Cells. Front. Pharmacol. 2018, 9, 1530. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; Chen, Z.; Huang, C.; Liu, X.; Chen, C.; Liu, X.; Huang, J.; Liu, L.; Lin, D. Metabonomic analysis of the therapeutic effect of exendin-4 for the treatment of tBHP-induced injury in mouse glomerulus mesangial cells. Free Radic. Res. 2018, 52, 544–555. [Google Scholar] [CrossRef]
- Li, H.; Lu, Y.; Geng, Y. Analysis of the Effect of Vitamin C at IC50 on RAW264.7 and K562 Cells Based on 1H NMR Metabonomics. ACS Food Sci. Technol. 2021, 1, 1120–1129. [Google Scholar] [CrossRef]
- Chen, C.; Gao, J.; Wang, T.S.; Guo, C.; Yan, Y.J.; Mao, C.Y.; Gu, L.W.; Yang, Y.; Li, Z.F.; Liu, A. NMR-based Metabolomic Techniques Identify the Toxicity of Emodin in HepG2 Cells. Sci. Rep. 2018, 8, 9379. [Google Scholar] [CrossRef]
- Dallons, M.; Schepkens, C.; Dupuis, A.; Tagliatti, V.; Colet, J.M. New Insights About Doxorubicin-Induced Toxicity to Cardiomyoblast-Derived H9C2 Cells and Dexrazoxane Cytoprotective Effect: Contribution of In Vitro (1)H-NMR Metabonomics. Front. Pharmacol. 2020, 11, 79. [Google Scholar] [CrossRef]
- Zhang, R.; Zhai, Q.; Yu, Y.; Li, X.; Zhang, F.; Hou, Z.; Cao, Y.; Feng, J.; Xue, P. Safety assessment of crude saponins from Chenopodium quinoa willd. husks: 90-day oral toxicity and gut microbiota & metabonomics study in rats. Food Chem. 2022, 375, 131655. [Google Scholar] [CrossRef]
- Dong, W.; Zhao, Y.; Hao, Y.; Sun, G.; Huo, J.; Wang, W. Integrated molecular biology and metabonomics approach to understand the mechanism underlying reduction of insulin resistance by corn silk decoction. J. Ethnopharmacol. 2022, 284, 114756. [Google Scholar] [CrossRef]
- Jiang, W.; Si, L.; Li, P.; Bai, B.; Qu, J.; Hou, B.; Zou, H.; Fan, X.; Liu, Z.; Liu, Z.; et al. Serum metabonomics study on antidiabetic effects of fenugreek flavonoids in streptozotocin-induced rats. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2018, 1092, 466–472. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, L.; Wu, J.; Qiao, S.; Xu, W.; Ma, S.; Zhao, B.; Wang, X. Intervention of resistant starch 3 on type 2 diabetes mellitus and its mechanism based on urine metabonomics by liquid chromatography-tandem mass spectrometry. Biomed. Pharmacother. 2020, 128, 110350. [Google Scholar] [CrossRef]
- Ren, F.; Chen, Q.; Meng, C.; Chen, H.; Zhou, Y.; Zhang, H.; Chen, W. Serum metabonomics revealed the mechanism of Ganoderma amboinense polysaccharides in preventing non-alcoholic fatty liver disease (NAFLD) induced by high-fat diet. J. Funct. Foods 2021, 82, 104496. [Google Scholar] [CrossRef]
- Xu, W.; Hu, B.; Cheng, Y.; Guo, Y.; Yao, W.; Qian, H. Material basis research for Echinacea purpurea (L.) Moench against hepatocellular carcinoma in a mouse model through integration of metabonomics and molecular docking. Phytomedicine 2022, 98, 153948. [Google Scholar] [CrossRef]
- Guo, Z.T.; Hu, B.; Zhu, L.Y.; Yang, Y.L.; Liu, C.Y.; Liu, F.; Shi, Y.; Li, M.Y.; Gu, Z.H.; Xin, Y.; et al. Microbiome-metabolomics insights into the feces of high-fat diet mice to reveal the anti-obesity effects of yak (Bos grunniens) bone collagen hydrolysates. Food Res. Int. 2022, 156, 111024. [Google Scholar] [CrossRef]
- Song, R.; Xu, Y.; Jia, Z.; Liu, X.Y.; Zhang, X.X. Integration of intestinal microbiota and metabonomics to elucidate different alleviation impacts of non-saponification and saponification astaxanthin pre-treatment on paracetamol-induced oxidative stress in rats. Food Funct. 2022, 13, 1860–1880. [Google Scholar] [CrossRef]
- Trimigno, A.; Munger, L.; Picone, G.; Freiburghaus, C.; Pimentel, G.; Vionnet, N.; Pralong, F.; Capozzi, F.; Badertscher, R.; Vergeres, G. GC-MS Based Metabolomics and NMR Spectroscopy Investigation of Food Intake Biomarkers for Milk and Cheese in Serum of Healthy Humans. Metabolites 2018, 8, 26. [Google Scholar] [CrossRef]
- Esteban-Fernandez, A.; Ibanez, C.; Simo, C.; Bartolome, B.; Moreno-Arribas, M.V. An Ultrahigh-Performance Liquid Chromatography-Time-of-Flight Mass Spectrometry Metabolomic Approach to Studying the Impact of Moderate Red-Wine Consumption on Urinary Metabolome. J. Proteome Res. 2018, 17, 1624–1635. [Google Scholar] [CrossRef]
- Fernández-Ochoa, Á.; Borrás-Linares, I.; Baños, A.; García-López, J.D.; Guillamón, E.; Nuñez-Lechado, C.; Quirantes-Piné, R.; Segura-Carretero, A. A fingerprinting metabolomic approach reveals deregulation of endogenous metabolites after the intake of a bioactive garlic supplement. J. Funct. Foods 2018, 49, 137–145. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, A.; Sandhu, A.K.; Edirisinghe, I.; Burton-Freeman, B.M. Functional Deficits in Gut Microbiome of Young and Middle-Aged Adults with Prediabetes Apparent in Metabolizing Bioactive (Poly)phenols. Nutrients 2020, 12, 3595. [Google Scholar] [CrossRef] [PubMed]
- Songvut, P.; Chariyavilaskul, P.; Khemawoot, P.; Tansawat, R. Pharmacokinetics and metabolomics investigation of an orally modified formula of standardized Centella asiatica extract in healthy volunteers. Sci. Rep. 2021, 11, 6850. [Google Scholar] [CrossRef]
- Kumar, A.; Mosa, K.A.; Ji, L.; Kage, U.; Dhokane, D.; Karre, S.; Madalageri, D.; Pathania, N. Metabolomics-assisted biotechnological interventions for developing plant-based functional foods and nutraceuticals. Crit. Rev. Food Sci. Nutr. 2018, 58, 1791–1807. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.L.; Chen, Z.C.; Hong, J.; Shi, J.X. Promoting Human Nutrition and Health through Plant Metabolomics: Current Status and Challenges. Biology 2020, 10, 20. [Google Scholar] [CrossRef]
- Chon, S.U. Total Polyphenols and Bioactivity of Seeds and Sprouts in Several Legumes. Curr. Pharm. Des. 2013, 19, 6112–6124. [Google Scholar] [CrossRef]
- Huang, H.Q.; Jiang, X.J.; Xiao, Z.L.; Yu, L.; Pham, Q.; Sun, J.H.; Chen, P.; Yokoyama, W.; Yu, L.L.L.; Luo, Y.S.; et al. Red Cabbage Microgreens Lower Circulating Low-Density Lipoprotein (LDL), Liver Cholesterol, and Inflammatory Cytokines in Mice Fed a High-Fat Diet. J. Agric. Food Chem. 2016, 64, 9161–9171. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, P.R.; Barnes, D.M. Phytochemical composition and biological activity of 8 varieties of radish (Raphanus sativus L.) sprouts and mature taproots. J. Food Sci. 2011, 76, C185–C192. [Google Scholar] [CrossRef]
- Kim, M.A.; Kim, M.J. Isoflavone profiles and antioxidant properties in different parts of soybean sprout. J. Food Sci. 2020, 85, 689–695. [Google Scholar] [CrossRef]
- Cuykx, M.; Rodrigues, R.M.; Laukens, K.; Vanhaecke, T.; Covaci, A. In vitro assessment of hepatotoxicity by metabolomics: A review. Arch. Toxicol. 2018, 92, 3007–3029. [Google Scholar] [CrossRef]
- Aragonès, G.; Danesi, F.; Del Rio, D.; Mena, P. The importance of studying cell metabolism when testing the bioactivity of phenolic compounds. Trends Food Sci. Technol. 2017, 69, 230–242. [Google Scholar] [CrossRef]
- Sorice, A.; Siano, F.; Capone, F.; Guerriero, E.; Picariello, G.; Budillon, A.; Ciliberto, G.; Paolucci, M.; Costantini, S.; Volpe, M.G. Potential Anticancer Effects of Polyphenols from Chestnut Shell Extracts: Modulation of Cell Growth, and Cytokinomic and Metabolomic Profiles. Molecules 2016, 21, 1411. [Google Scholar] [CrossRef] [PubMed]
- Di Nunzio, M.; Picone, G.; Pasini, F.; Caboni, M.F.; Gianotti, A.; Bordoni, A.; Capozzi, F. Olive oil industry by-products. Effects of a polyphenol-rich extract on the metabolome and response to inflammation in cultured intestinal cell. Food Res. Int. 2018, 113, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Recent and potential developments of biofluid analyses in metabolomics. J. Proteomics 2012, 75, 1079–1088. [Google Scholar] [CrossRef]
- Ammar, N.M.; Hassan, H.A.; Mohammed, M.A.; Serag, A.; Abd El-Alim, S.H.; Elmotasem, H.; El Raey, M.; El Gendy, A.N.; Sobeh, M.; Abdel-Hamid, A.Z. Metabolomic profiling to reveal the therapeutic potency of Posidonia oceanica nanoparticles in diabetic rats. RSC Adv. 2021, 11, 8398–8410. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.Y.; Liu, Y.; He, F.; An, R.; Du, Z.M. LC-MS based urinary metabolomics study of the intervention effect of aloe-emodin on hyperlipidemia rats. J. Pharm. Biomed. Anal. 2018, 156, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Chen, Q.; Di, L.; Li, N. Evaluation of Anti-Diabetic Potential of Corn Silk in High-Fat Diet/Streptozotocin-Induced Type 2 Diabetes Mice Model. Endocr. Metab. Immune Disord. Drug Targets 2021, 21, 131–138. [Google Scholar] [CrossRef]
- Xu, W.Q.; Hu, B.; Cheng, Y.L.; Guo, Y.H.; Yao, W.R.; Qian, H. Echinacea purpurea suppresses the cell survival and metastasis of hepatocellular carcinoma through regulating the PI3K/Akt pathway. Int. J. Biochem. Cell Biol. 2022, 142, 106115. [Google Scholar] [CrossRef]
- Clark, L.T.; Watkins, L.; Pina, I.L.; Elmer, M.; Akinboboye, O.; Gorham, M.; Jamerson, B.; McCullough, C.; Pierre, C.; Polis, A.B.; et al. Increasing Diversity in Clinical Trials: Overcoming Critical Barriers. Curr. Probl. Cardiol. 2019, 44, 148–172. [Google Scholar] [CrossRef]
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.D.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef]
- Kristensen, M.; Engelsen, S.B.; Dragsted, L.O. LC–MS metabolomics top-down approach reveals new exposure and effect biomarkers of apple and apple-pectin intake. Metabolomics 2011, 8, 64–73. [Google Scholar] [CrossRef]
- Cueva, C.; Gil-Sanchez, I.; Ayuda-Duran, B.; Gonzalez-Manzano, S.; Gonzalez-Paramas, A.M.; Santos-Buelga, C.; Bartolome, B.; Moreno-Arribas, M.V. An Integrated View of the Effects of Wine Polyphenols and Their Relevant Metabolites on Gut and Host Health. Molecules 2017, 22, 99. [Google Scholar] [CrossRef]
- Caesar, L.K.; Kellogg, J.J.; Kvalheim, O.M.; Cech, N.B. Opportunities and Limitations for Untargeted Mass Spectrometry Metabolomics to Identify Biologically Active Constituents in Complex Natural Product Mixtures. J. Nat. Prod. 2019, 82, 469–484. [Google Scholar] [CrossRef]
- Qaraei, M.; Abbaasi, S.; Ghiasi-Shirazi, K. Randomized non-linear PCA networks. Inf. Sci. 2021, 545, 241–253. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Guijas, C.; Benton, H.P.; Warth, B.; Siuzdak, G. METLIN MS (2) molecular standards database: A broad chemical and biological resource. Nat. Methods 2020, 17, 953–954. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.A.; Altaf-Ul-Amin, M.; Kanaya, S. Insight into Knapsack Metabolite Ecology Database: A Comprehensive Source of Species: Voc-Biological Activity Relationships. In Volatile Organic Compound Analysis in Biomedical Diagnosis Applications; Apple Academic Press: London, UK, 2018; pp. 225–247. [Google Scholar] [CrossRef]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Colin, K.; Alex, S.; Jessica, E.; Ciara, C.; Harry, S.; Yang, Z.; Yang, J.; Du, X. The Metabolome of Food Knowledge Database: Development of a Nutrition Database to Support Precision Nutrition. Curr. Dev. Nutr. 2022, 6, 53–83. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
| Main Metabolites | Sample (Sources) | Analytical Technique | Application of Metabolomics | Reference |
|---|---|---|---|---|
| Flavonoids | Soybean seeds | LC-ESI-MS/MS | Evaluated the dynamic changes of metabolites in soybean seeds before and after germination. | [77] |
| Amino acids, sugars, choline | Mung bean | NMR | Evaluated the dynamic changes of metabolites in mung bean at different germination stages. | [78] |
| Flavonoids and polyphenols | Green tea bud | UPLC-QTOF-MS | Combined the characteristic metabolites with in vitro biological activities to determine the health effects of natural metabolites. | [79] |
| Isoflavones and alkaloids | Lupinus albus fractions | 1H NMR UHPLC-ESI-MS/MS | Identified the effects of different extract components on the bioactivities of metabolites. | [80] |
| Procyanidin C1, orientin, quercetin, etc. | Hawthorn | UHPLC-Q-TOF/MS LC-MS/MS | Screened the metabolites with specific biological activities by combining various types of stoichiometry. | [81] |
| Polyphenols, glucosinolates and monomeric anthocyanins | Four Brassicaceae microgreens | UHPLC-QTOF | Compared the changes of metabolite concentrations before and after simulated gastrointestinal digestion in vitro. | [82] |
| Polyphenol | Red beet and amaranth | UHPLC-QTOF | Identified the effects of different storage periods on metabolite changes. | [83] |
| Phenyllactate and ferulate | Soybean protein hydrolysate | UHPLC/MS/MS2 GC/MS | Analyzed the compounds with significant effects on cell growth and IgG production. | [84] |
| Ornithine and citrulline | Soybean hydrolysates | LC-MS/MS | Screened productivity markers by comparing cell growth condition. | [85] |
| Phenolic substances | Red kidney bean extracts | NMR LC-MS | Analyzed the antiproliferative mechanism of different chemical components on B16-F10 melanoma cells. | [86] |
| Alanine, aspartate and glutamate | CG | UPLC-MS/MS | Studied the effects of different concentrations of CG on L-02 cells metabolism. | [87] |
| Glutamate and lactate | Exendin-4 | NMR | Investigated the mechanism of protective effect of exendin-4 on mouse glomerulus mesangial cells. | [88] |
| Glycerolipid, cyanomino acid, inositol phosphate, etc. | Vitamin C | 1H NMR | Determined the effect of half inhibitory concentration of Vitamin C on cell metabolism. | [89] |
| Alanine, Aspartate, glutamate, etc. | Emodin | 1H NMR | Evaluated the cytotoxic effects of high concentrations of emodin on cells. | [90] |
| Lactate and glucose | Doxorubicin and dexrazoxane | 1H NMR | Identified the important factor of dextroprazole induced cardiotoxicity. | [91] |
| Nicotinamide, nicotinic acid, Arginine, etc. | Quinoa saponins | UHPLC-MS | Combined the metabolomics with the changes of intestinal microbes in rats and identified the differential effects of quinoa saponins on different sexes. | [92] |
| Phosphatidylcholine and palmitic acid | Corn silk | UPLC-ESI-Q-TOF/MS | Identified the changes of diabetes markers through the differences of serum metabolites in rats. | [93] |
| Flavonoids | Fenugreek | UPLC-Q-TOF-MS | Investigated the function of fenugreek flavonoids in regulating blood glucose by serum metabolomics. | [94] |
| Valine, leucine, LPCs, etc. | RS3 | UHPLC-LTQ/Orbitrap MS | Identified the antidiabetic mechanism of RS3 by urine metabolomics. | [95] |
| Alanine, aspartate, glutamate, etc. | GAP | LC-MS | Studied the regulation of GAP on mice with nonalcoholic fatty liver by serum metabolomics. | [96] |
| Phenylalanine, tyrosine and tryptophan | EP | LC-MS | Combined the metabolomics with molecular docking technology to obtain effective bioactive components. | [97] |
| Arginine and proline | The hydrolysates of yak bone glue | UPLC-QTF/MS | Determined the anti-obesity mechanism of the hydrolysates of yak bone glue by fecal metabolomics. | [98] |
| Propionic acid, taurine, glutathione, etc. | Astaxanthin | LC-MS | Clarified the mechanism of astaxanthin alleviating oxidative stress in rats. | [99] |
| Galactose, galactonate and lactic | Cheese, milk and soy beverages | GC-MS 1H NMR | Explored possible food biomarkers of human intake by metabolomics. | [100] |
| 5-(dihydroxyphenyl)-γ-valerolactones and 4-hydroxyl-5-(phenyl)-valeric acids | Red wine | UHPLC−TOF-MS | Determined the health effects of moderate red wine consumption on human metabolism by urine metabolomics and fecal metabolomics. | [101] |
| Lysophosphatidylcholines, lysophosphatidylethanolamines and acylcarnitines | Garlic supplements | HPLC-ESI-QTOF-MS | Verified the function of garlic supplement in enhancing immunity by fingerprint metabolomics. | [102] |
| 3,8-dihydroxy-urolithin derivatives and phenyl-γ-valerolactones | A (poly) phenols-rich test drink | UHPLC-QQQ | Determined the regulating mechanism of polyphenol beverage on diabetes patients by blood and urine metabolomics. | [103] |
| Choline | ECa 233 | 1H NMR LC-MS/MS | Evaluated the drug bioavailability of ECa 233 by metabolomics. | [104] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, S.; Liu, C.; Liu, X. Innovative Application of Metabolomics on Bioactive Ingredients of Foods. Foods 2022, 11, 2974. https://doi.org/10.3390/foods11192974
Hu S, Liu C, Liu X. Innovative Application of Metabolomics on Bioactive Ingredients of Foods. Foods. 2022; 11(19):2974. https://doi.org/10.3390/foods11192974
Chicago/Turabian StyleHu, Sumei, Caiyu Liu, and Xinqi Liu. 2022. "Innovative Application of Metabolomics on Bioactive Ingredients of Foods" Foods 11, no. 19: 2974. https://doi.org/10.3390/foods11192974
APA StyleHu, S., Liu, C., & Liu, X. (2022). Innovative Application of Metabolomics on Bioactive Ingredients of Foods. Foods, 11(19), 2974. https://doi.org/10.3390/foods11192974






