Simultaneous Monitoring of the Evolution of Chemical Parameters in the Fermentation Process of Pineapple Fruit Wine Using the Liquid Probe for Near-Infrared Coupled with Chemometrics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pineapple Wine
2.1.1. Yeast Culture Preparation
2.1.2. Preparation of Pineapple Must
2.1.3. Pineapple Juice Fermentation
2.2. NIR Liquid Probe Spectral Acquisition
2.3. Pineapple Wine Chemical Analysis for Ethanol, Total Soluble Solids, Total Acidity, and Total Volatile Acids Using the Conventional Reference Methods
2.4. NIR Data Analysis
2.4.1. Preprocessing Method
2.4.2. Searching Combination Moving Window Partial Least Squares (SCMWPLS) Analysis
Step 1: MWPLSR Calculation
Step 2: SCMWPLS Calculation
2.4.3. Calibration Development
2.5. Evaluation of the Predictive Ability of PLS and SCMWPLS Models
3. Results and Discussion
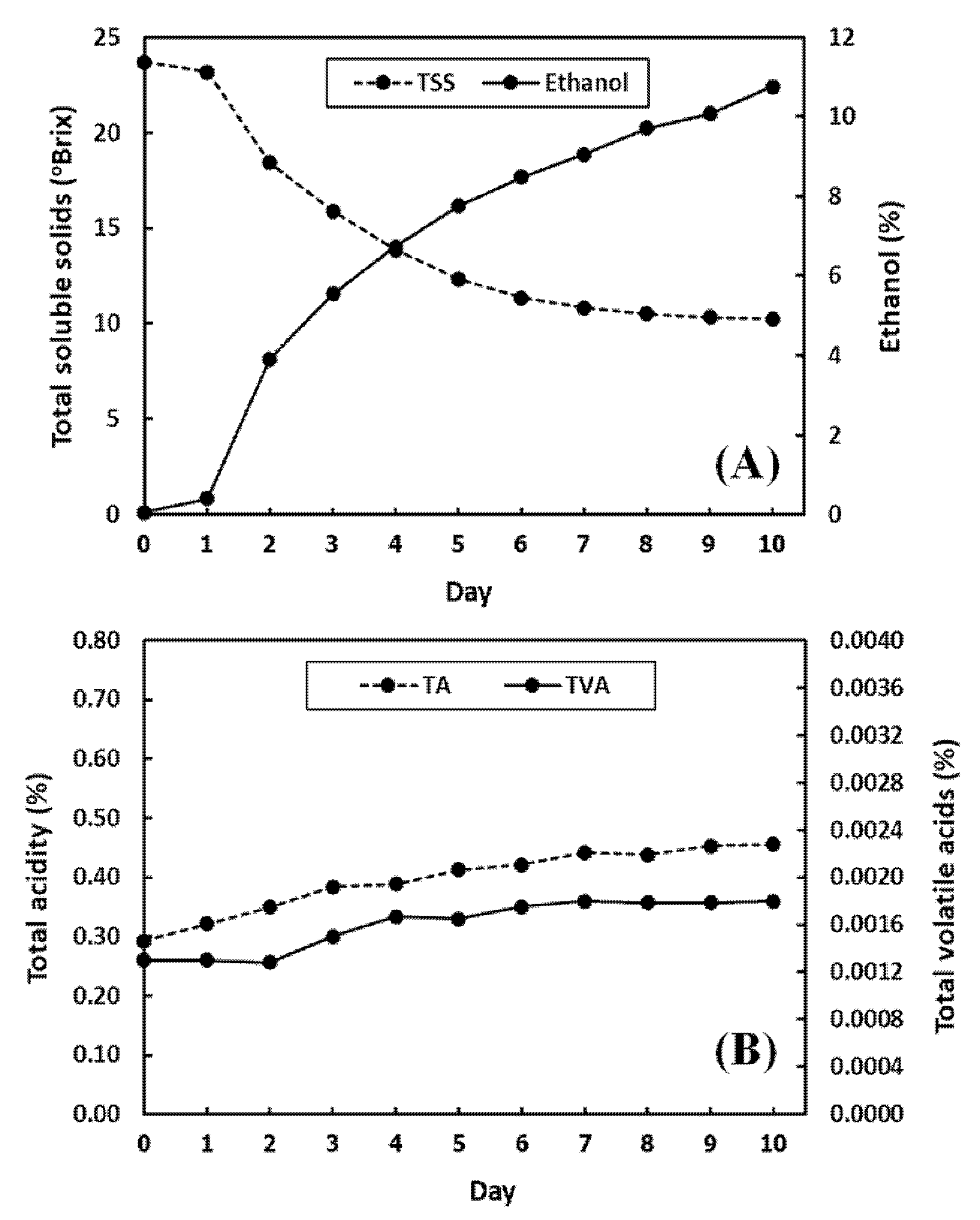
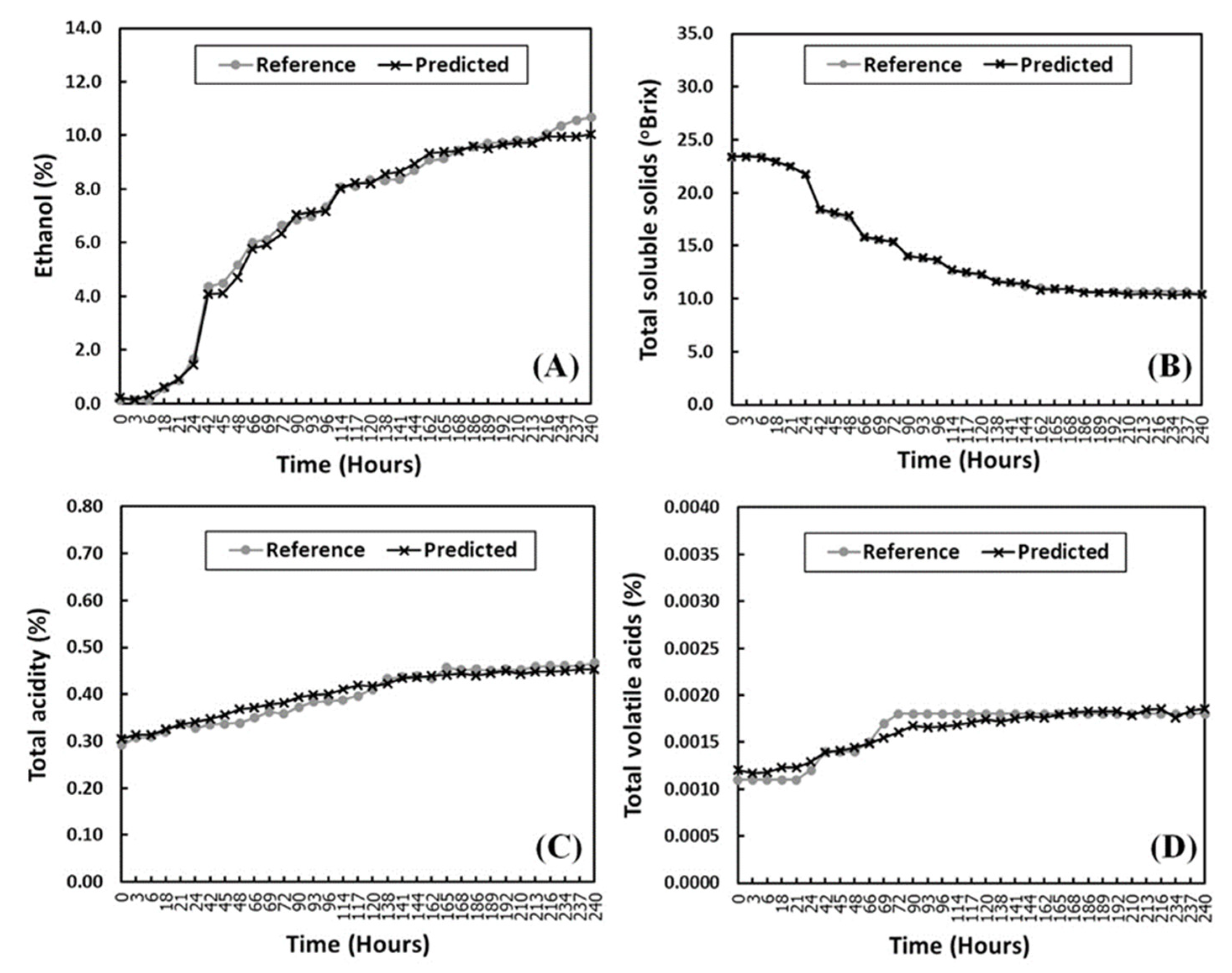
3.1. Measured Chemical Characteristics of Pineapple Wines during Fermentation by Reference Methods
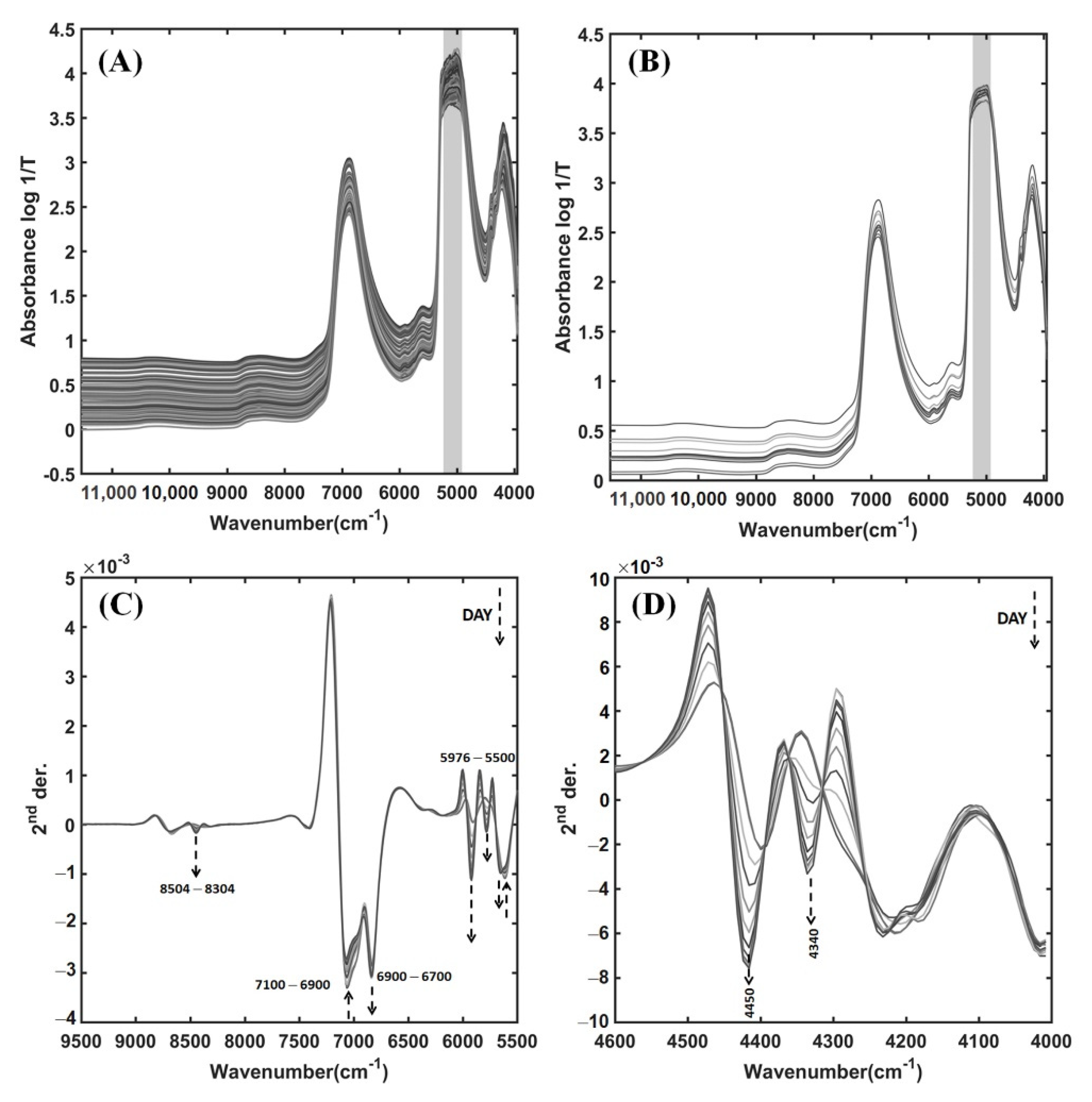
3.2. NIR Spectra of Pineapple Wines from the Fermentation Process
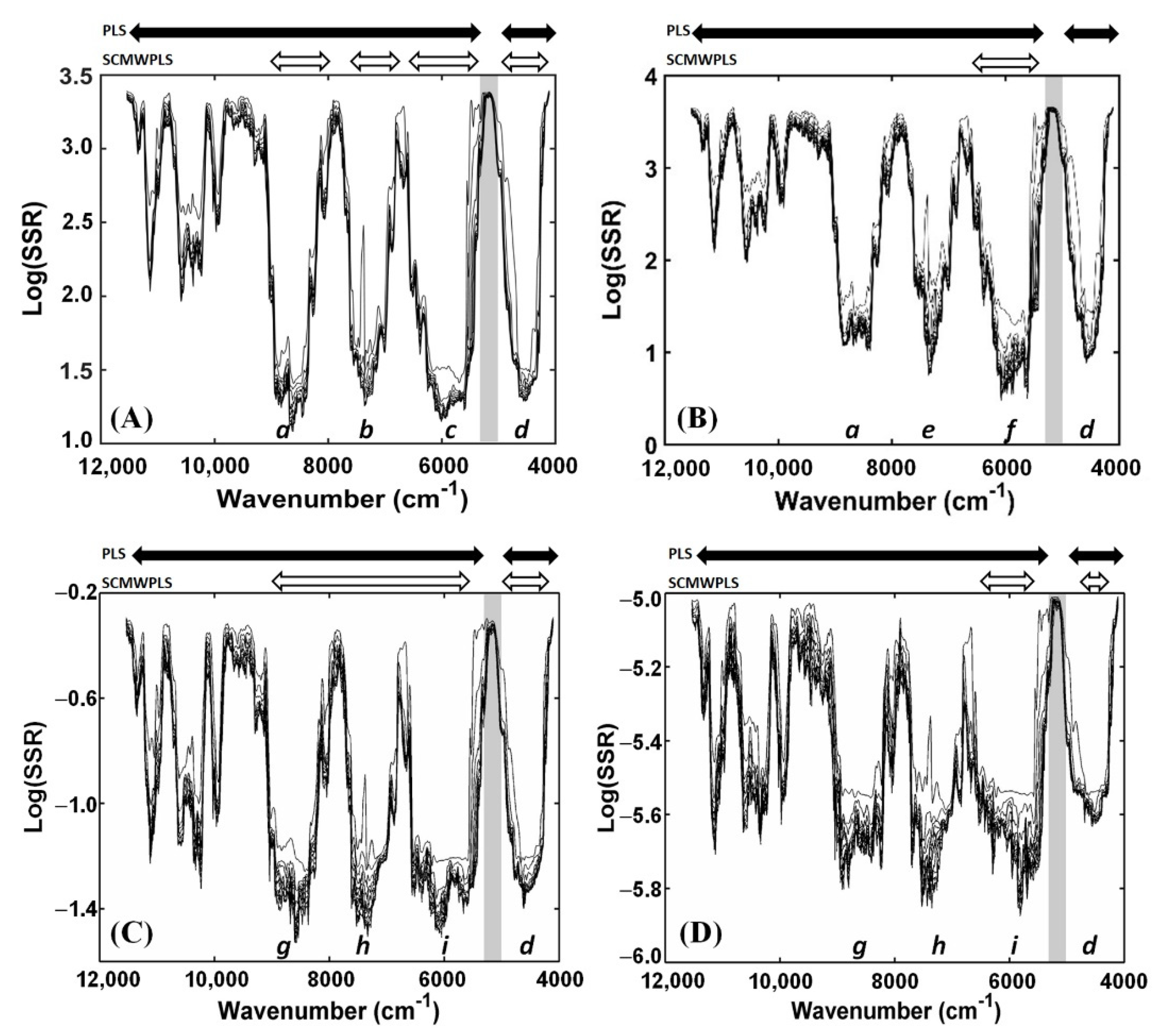
3.3. SCMWPLS Analysis
3.4. Comparison of PLS Calibration Models
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pino, J.A.; Queris, O. Analysis of volatile compounds of pineapple wine using solid-phase microextraction techniques. Food Chem. 2010, 122, 1241–1246. [Google Scholar] [CrossRef]
- Shahbandeh, M. Leading Countries in Pineapple Production Worldwide in 2019. Available online: https://www.statista.com/statistics/298517/global-pineapple-production-by-leading-countries/#statisticContainer (accessed on 13 March 2021).
- Dellacassa, E.; Trenchs, O.; Fariña, L.; Debernardis, F.; Perez, G.; Boido, E.; Carrau, F. Pineapple (Ananas comosus L. Merr.) wine production in Angola: Characterisation of volatile aroma compounds and yeast native flora. Int. J. Food Microbiol. 2017, 241, 161–167. [Google Scholar] [CrossRef]
- Lobo, M.G.; Yahia, E. Biology and postharvest physiology of pineapple. In Handbook of Pineapple Technology: Postharvest Science, Processing and Nutrition; Blackwell, W., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2016; pp. 39–61. [Google Scholar]
- Mohd-Ali, M.; Hashim, N.; Aziz, S.A.; Lasekan, O. Pineapple (Ananas comosus): A comprehensive review of nutritional values, volatile compounds, health benefits, and potential food products. Food Res. Int. 2020, 137, 109675. [Google Scholar] [CrossRef] [PubMed]
- Cannon, R.J.; Ho, C.T. Volatile sulfur compounds in tropical fruits. J. Food Drug Anal. 2018, 26, 445–468. [Google Scholar] [CrossRef] [PubMed]
- Otegbayo, B.O.; Akwa, I.M.; Tanimola, A.R. Physico-chemical properties of beetroot (Beta vulgaris l.) wine produced at varying fermentation days. Sci. Afr. 2020, 8, e00420. [Google Scholar] [CrossRef]
- Margalit, Y. Must and wine composition. In Concepts in Wine Chemistry; Wine Appreciation Guild: San Francisco, CA, USA, 2004; pp. 1–59. [Google Scholar]
- Waterhouse, A.L.; Sacks, G.L.; Jeffery, D.W. Introduction. In Understanding Wine Chemistry; John Wiley & Sons: Hoboken, NJ, USA, 2016; pp. xvii–xxiv. [Google Scholar]
- Alañón, M.E.; Pérez-Coello, M.S.; Marina, M.L. Wine science in the metabolomics era. Trends Anal. Chem. 2015, 74, 1–20. [Google Scholar] [CrossRef]
- Teixeira dos Santos, C.A.; Páscoa, R.N.M.J.; Lopes, J.A. A review on the application of vibrational spectroscopy in the wine industry: From soil to bottle. Trends Anal. Chem. 2017, 88, 100–118. [Google Scholar] [CrossRef]
- Cozzolino, D. Sample presentation, sources of error and future perspectives on the application of vibrational spectroscopy in the wine industry. J. Sci. Food Agric. 2015, 95, 861–868. [Google Scholar] [CrossRef]
- Cozzolino, D.; Kwiatkowski, M.J.; Parker, M.; Cynkar, W.U.; Dambergs, R.G.; Gishen, M.; Herderich, M.J. Prediction of phenolic compounds in red wine fermentations by visible and near infrared spectroscopy. Anal. Chim. Acta 2004, 513, 73–80. [Google Scholar] [CrossRef]
- Martelo-Vidal, M.J.; Vázquez, M. Determination of polyphenolic compounds of red wines by UV–VIS–NIR spectroscopy and chemometrics tools. Food Chem. 2014, 158, 28–34. [Google Scholar] [CrossRef]
- Ozaki, Y.; Huck, C.; Tsuchikawa, S.; Engelsen, S.B. Near-Infrared Spectroscopy Theory, Spectral Analysis, Instrumentation, and Applications; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Burns, D.A.; Ciurczak, E.W. Handbook of Near-Infrared Analysis; Marcel Dekker: New York, NY, USA, 1992. [Google Scholar]
- Siesler, H.W.; Ozaki, Y.; Kawata, S.; Heise, H.M. Near-Infrared Spectroscopy Principles, Instruments, Applications; WILEY–VCH: Weinheim, Germany, 2002. [Google Scholar]
- Cozzolino, D. State-of-art advantages and drawbacks on the application of vibrational spectroscopy to monitor alcoholic fermentation (beer and wine). Appl. Spectrosc. Rev. 2016, 51, 302–317. [Google Scholar] [CrossRef]
- Wu, Z.; Xu, E.; Wang, F.; Long, J.; Xu, X.; Jiao, A.; Jin, Z. Rapid determination of process variables of Chinese rice wine using FT-NIR spectroscopy and Efficient Wavelengths Selection Methods. Food Anal. Methods 2015, 8, 1456–1467. [Google Scholar] [CrossRef]
- Ye, M.; Yue, T.; Yuan, Y.; Li, Z. Application of FT-NIR spectroscopy to apple wine for rapid simultaneous determination of soluble solid content, pH, total acidity, and total ester content. Food Bioprocess Technol. 2014, 7, 3055–3062. [Google Scholar] [CrossRef]
- Peng, B.; Ge, N.; Cui, L.; Zhao, H. Monitoring of alcohol strength and titratable acidity of apple wine during fermentation using near-infrared spectroscopy. LWT-Food Sci. Technol. 2016, 66, 86–92. [Google Scholar] [CrossRef]
- Li, G.; Yan, N.; Yuan, L.; Wu, J.; Du, J.; Gao, Y.; Peng, Y. Rapid analysis of alcohol content during the green jujube wine fermentation by FT-NIR. E3S Web Conf. IAECST 2019 2020, 145, 01037. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Novales, J.; María-Isabel, L.; María-Teresa, S.; José-Antonio, G.; José, M. A feasibility study on the use of a miniature fiber optic NIR spectrometer for the prediction of volumic mass and reducing sugars in white wine fermentations. J. Food Eng. 2008, 89, 325–329. [Google Scholar] [CrossRef]
- Du, Y.P.; Liang, Y.Z.; Jiang, J.H.; Berry, R.J.; Ozaki, Y. Spectral regions selection to improve prediction ability of PLS models by changeable size moving window partial least squares and searching combination moving window partial least squares. Anal. Chim. Acta 2004, 501, 183–191. [Google Scholar] [CrossRef]
- Kasemsumran, S.; Du, Y.P.; Murayama, K.; Huehne, M.; Ozaki, Y. Near-infrared spectroscopic determination of human serum albumin, γ-globumin, and glucose in a control serum solution with searching combination moving window partial least squares. Anal. Chim. Acta 2004, 512, 223–230. [Google Scholar] [CrossRef]
- Kasemsumran, S.; Du, Y.P.; Maruo, K.; Ozaki, Y. Improvement of partial least squares models for in vitro and in vivo glucose quantifications by using near-infrared spectroscopy and searching combination moving window partial least squares. Chemometr. Intell. Lab. Syst. 2006, 82, 97–103. [Google Scholar] [CrossRef]
- Rongtong, B.; Suwonsichon, T.; Ritthiruangdej, P.; Kasemsumran, S. Determination of water activity, total soluble solids and moisture, sucrose, glucose and fructose contents in osmotically dehydratedpapaya using near-infrared spectroscopy. Agric. Nat. Resour. 2018, 52, 557–564. [Google Scholar]
- International Federation of Fruit Juice Producers (IFU). Determination of Titratable Acidity; IFU Analysis No.3; IFU: Paris, France, 1996. [Google Scholar]
- International Federation of Fruit Juice Producers (IFU). Determination of Total Volatile Acids; IFU Analysis No.5; IFU: Paris, France, 1968. [Google Scholar]
- Jiang, J.H.; Berry, R.J.; Siesler, H.W.; Ozaki, Y. Wavelength interval selection in multicomponent spectral analysis by moving window partial least-squares regression with applications to mid-infrared and near-infrared spectroscopic data. Anal. Chem. 2002, 74, 3555–3565. [Google Scholar] [CrossRef]
- Kasemsumran, S.; Du, Y.; Li, B.; Maruo, K.; Ozaki, Y. Moving window cross validation: A new cross validation method for the selection of a rational number of components in a partial least squares calibration model. Analyst 2006, 131, 529–537. [Google Scholar] [CrossRef]
- ISO 12099:2017(E); Animal Feeding Stuffs, Cereals and Milled Cereal Products–Guidelines for Application of Near Infrared Spectrometer. ISO: Geneva, Switzerland, 2017; pp. 5–12.
- Singpoonga, N.; Rittiron, R.; Seang-on, B.; Chaiprasart, P.; Bantadjan, Y. Determination of Adenosine and Cordycepin Concentrations in Cordyceps militaris Fruiting Bodies Using Near-Infrared Spectroscopy. ACS Omega 2020, 5, 27235–27244. [Google Scholar] [CrossRef]
- Sankom, A.; Mahakarnchanakul, W.; Rittiron, R.; Sajjaanantakul, T.; Thongket, T. Detection of profenofos in Chinese kale, cabbage, and chili spur pepper using fourier transform near-infrared and fourier transform mid-infrared spectroscopies. ACS Omega 2021, 40, 26404–26415. [Google Scholar] [CrossRef]
- Marques, W.L.; Raghavendran, V.; Stambuk, B.U.; Gomber, A.K. Sucrose and Saccharomyces cerevisiae: A relationship most sweet. FEMS Yeast Res. 2016, 16, fov107. [Google Scholar] [CrossRef] [Green Version]
- Okeke, B.C.; Agu, K.C.; Uba, P.O.; Awah, N.S.; Anaukwu, C.G.; Archibong, E.J.; Uwanta, L.I.; Ezeneche, J.N.; Ezenwa, C.U.; Orji, M.U. Wine production from mixed fruits (pineapple and watermelon) using high alcohol tolerant yeast isolated from palm wine. Univers. J. Microbiol. Res. 2015, 3, 41–44. [Google Scholar]
- Maeda, H.; Ozaki, Y.; Tanaka, M.; Hayashi, N.; Kojima, T. Near infrared spectroscopy and chemometrics studies of temperature-dependent spectral variations of water: Relationship between spectral changes and hydrogen bonds. J. Near Infrared Spectrosc. 1995, 3, 191–201. [Google Scholar] [CrossRef]
- Cozzolino, D.; Parker, M.; Dambergs, R.G.; Herderich, M.; Gishen, M. Chemometrics and visible-near infrared spectroscopic monitoring of red wine fermentation in a pilot. Biotechnol. Bioeng. 2006, 95, 1101–1107. [Google Scholar] [CrossRef]
- Dambergs, R.G.; Kambouris, A.; Francis, I.L.; Gishen, M. Rapid analysis of methanol in grape derived distillation products using near infrared transmission spectroscopy. J. Agric. Food Chem. 2002, 50, 3079–3084. [Google Scholar] [CrossRef]
- Workman, J.; Weyer, L., Jr. Practice Guide to Interpretive Near Infrared Spectroscopy; CRC Press: New York, NY, USA, 2007. [Google Scholar]
- Williams, P.; Antoniszyn, J.; Manley, M. Near Infrared Technology: Getting the Best out of Light; African Sun Media, Sun Press: Stellenbosch, South Africa, 2019. [Google Scholar]
- Beć, B.K.; Futami, Y.; Wójcik, M.; Ozaki, Y. A spectroscopic and theoretical study in the near-infrared region of low concentration aliphatic alcohols. Phys. Chem. Chem. Phys. 2016, 18, 13666–13682. [Google Scholar] [CrossRef]
- Grassia, S.; Amigo, J.M.; Lyndgaard, C.B.; Foschino, R.; Casiraghi, E. Beer fermentation: Monitoring of process parameters by FT-NIR and multivariate data analysis. Food Chem. 2014, 155, 279–286. [Google Scholar] [CrossRef] [PubMed]
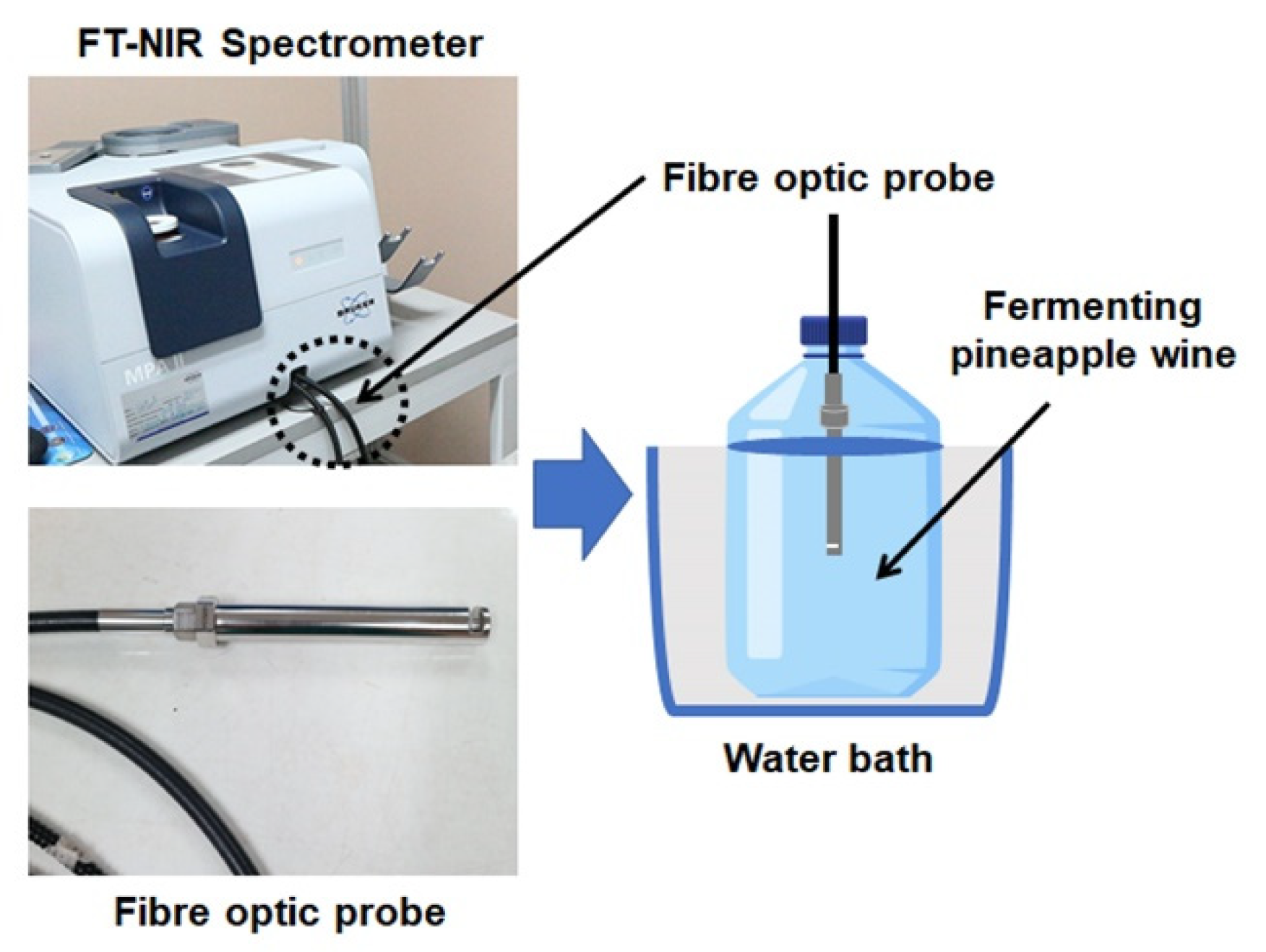
| Sample | VIS-NIR or NIR Range (cm−1) | Measurement Mode | Sample Preparation | Chemometric Method | Analyst | RMSEP or a RMSECV or b SEP or c SECV |
|---|---|---|---|---|---|---|
| Apple wine [20] | 12,000–4000 | Transmission; Quartz cuvette | Centrifugation and filtration | PLS | Soluble solid content | 0.60% |
| pH | 0.08 | |||||
| Total acidity | 0.02 g 100 mL−1 | |||||
| Total ester content | 0.10 g L−1 | |||||
| Apple wine [21] | 12,000–4000 | Transflection; Liquid probe | Centrifugation | PLS | Alcohol strength | 4.25 mL L−1 |
| Titratable acidity | 0.21 g L−1 | |||||
| Jujube wine [22] | 10,526–6060 | Transmission; Quartz cuvette | Filtration | PLS | Alcohol | a 0.70% |
| Red wine [13] | 25,000–4000 | Transmission; Quartz cuvette | Centrifugation | PLS | Malvidin-3-glucoside | c 17.50–31.50 mg L−1 |
| Pigmented polymers | c 3.20–26.80 mg L−1 | |||||
| Tannins | c 49.10–131.20 mg L−1 | |||||
| Rice wine [19] | 10,000–4000 | Transmission; Quartz cuvette | Centrifugation | RCA–SVM–PLS | Ethanol | 2.60 g L−1 |
| GA–SVM–PLS | Total acid | 0.10 g L−1 | ||||
| White wine [23] | 14,285–9434 | Transmission; Quartz cuvette | Filtration | PLS | Volumic mass | a 4.18 g (dm3)−1 |
| Reducing sugars | a 10.35 g L−1 |
| Statistical Terms | Computations |
|---|---|
| Coefficient of determination (R2) | |
| Root mean square error (RMSE) RMSEC in the calibration set RMSEP in the test set | |
| Standard error of prediction (SEP) | |
| Bias | |
| Residual predictive deviation (RPD) | |
| Bias confidence limits (Tb) = | |
| Unexplained error confidence limits (TUE) |
| Fermentation Day | Ethanol (%) | Total Soluble Solids (°Brix) | Total Acidity (%) | Total Volatile Acids (%) |
|---|---|---|---|---|
| 0 | 0.06 | 23.70 | 0.29 | 1.30 × 10−3 |
| 1 | 0.40 | 23.17 | 0.32 | 1.30 × 10−3 |
| 2 | 3.91 | 18.42 | 0.35 | 1.28 × 10−3 |
| 3 | 5.54 | 15.88 | 0.38 | 1.50 × 10−3 |
| 4 | 6.74 | 13.87 | 0.39 | 1.67 × 10−3 |
| 5 | 7.75 | 12.33 | 0.41 | 1.65 × 10−3 |
| 6 | 8.49 | 11.35 | 0.42 | 1.75 × 10−3 |
| 7 | 9.06 | 10.83 | 0.44 | 1.80 × 10−3 |
| 8 | 9.71 | 10.52 | 0.44 | 1.78 × 10−3 |
| 9 | 10.08 | 10.33 | 0.45 | 1.78 × 10−3 |
| 10 | 10.76 | 10.25 | 0.46 | 1.80 × 10−3 |
| Parameters | Sample Set | Minimum | Mean | Maximum | Standard Deviation |
|---|---|---|---|---|---|
| Ethanol (%) | Calibration | 0.04 | 6.61 | 11.56 | 3.60 |
| Test | 0.12 | 6.83 | 10.68 | 3.43 | |
| Total soluble solids (°Brix) | Calibration | 10.00 | 14.57 | 24.20 | 4.88 |
| Test | 10.53 | 14.55 | 23.50 | 4.62 | |
| Total acidity (%) | Calibration | 0.29 | 0.40 | 0.48 | 0.05 |
| Test | 0.29 | 0.40 | 0.47 | 0.06 | |
| Total volatile acids (%) | Calibration | 1.10 × 10−3 | 1.60 × 10−3 | 1.90 × 10−3 | 2.00 × 10−4 |
| Test | 1.10 × 10−3 | 1.60 × 10−3 | 1.80 × 10−3 | 3.00 × 10−4 |
| Wavenumber (cm−1) | Band Assignment | Substance [40,41] |
|---|---|---|
| 8900–8504 | O–H | Water [37] |
| 8504–8304 b | C–H stretch 2nd overtones of –CH3, –CH2 | Ethanol, Sugars, Citric acid, Acetic acid |
| 7100–6900 b | O–H stretch 1st overtones | Sugars |
| 6900–6700 b | O–H stretch 1st overtones | Ethanol (primary alcohols), Starch |
| 6896 b | C=O stretch 1st overtones from carbonyl group | Citric acid, Acetic acid |
| 6850 | O–H | Water [36] |
| 6500–6300 | O–H stretch 1st overtones | Starch |
| 5976–5500 b | C–H stretch 1st overtones of –CH3, –CH2 | Ethanol [13,38,39,42], Sugars, Citric acid, Acetic acid |
| 4504–4250 b | C–H combinations of stretch and deformation from the CH3 group | Ethanol [13,38,39,42,43] |
| 4504–4250 b | O–H stretch and C–O stretch combinations, C–H combinations of stretching and deformation | Sugars [43], Glucose, Cellulose, Starch |
| 4504–4250 b | C–H stretch and C=O stretch combinations | Citric acid, Acetic acid |
| Parameters | Methods | Preprocessing | LVs | R2 | RMSEC | RMSEP | RPD | Spectral Data Points |
|---|---|---|---|---|---|---|---|---|
| Ethanol (%) | PLS | none | 4 | 0.973 | 0.588 | 0.466 | 7.36 | 915 |
| PLS | SD | 4 | 0.985 | 0.438 | 0.406 | 8.44 | 901 | |
| SCMWPLS (cm−1) 9104–7984, 7752–6704, 6600–5256, 4976–4008 | SD | 3 | 0.984 | 0.457 | 0.393 | 8.72 | 564 | |
| TSS (°Brix) | PLS | none | 5 | 0.997 | 0.269 | 0.441 | 10.47 | 915 |
| PLS | SD | 2 | 0.995 | 0.330 | 0.219 | 21.08 | 901 | |
| SCMWPLS (cm−1) 6800–5360 | SD | 2 | 0.996 | 0.286 | 0.166 | 27.82 | 181 | |
| TA (%) | PLS | none | 2 | 0.883 | 0.174 × 10−1 | 0.182 × 10−1 | 3.15 | 915 |
| PLS | SD | 2 | 0.892 | 0.167 × 10−1 | 0.199 × 10−1 | 2.88 | 901 | |
| SCMWPLS (cm−1) 9200–5408, 4976–4008 | SD | 2 | 0.894 | 0.166 × 10−1 | 0.181 × 10−1 | 3.17 | 597 | |
| TVA (%) | PLS | none | 6 | 0.776 | 0.112 × 10−3 | 0.117 × 10−3 | 2.56 | 915 |
| PLS | SD | 5 | 0.753 | 0.118 × 10−3 | 0.113 × 10−3 | 2.65 | 901 | |
| SCMWPLS (cm−1) 6504–5280, 4504–4248 | SD | 6 | 0.761 | 0.116 × 10−3 | 0.105 × 10−3 | 2.86 | 187 |
| NIR Models by SCMWPLS | Statistics | Obtained Results | Criterion | Performance |
|---|---|---|---|---|
| Ethanol (%) | SEP | 0.374 | TUE = 0.554 | accepted |
| Bias | 0.128 | Tb = ±0.132 | accepted | |
| TSS (°Brix) | SEP | 0.164 | TUE = 0.346 | accepted |
| Bias | 0.029 | Tb = ±0.058 | accepted | |
| TA (%) | SEP | 0.175 × 10−1 | TUE = 0.201 × 10−1 | accepted |
| Bias | 0.005 | Tb = ±0.006 | accepted | |
| TVA (%) | SEP | 0.105 × 10−3 | TUE = 0.140 × 10−3 | accepted |
| Bias | 0.012 × 10−3 | Tb = ± 0.037 × 10−3 | accepted |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kasemsumran, S.; Boondaeng, A.; Ngowsuwan, K.; Jungtheerapanich, S.; Apiwatanapiwat, W.; Janchai, P.; Meelaksana, J.; Vaithanomsat, P. Simultaneous Monitoring of the Evolution of Chemical Parameters in the Fermentation Process of Pineapple Fruit Wine Using the Liquid Probe for Near-Infrared Coupled with Chemometrics. Foods 2022, 11, 377. https://doi.org/10.3390/foods11030377
Kasemsumran S, Boondaeng A, Ngowsuwan K, Jungtheerapanich S, Apiwatanapiwat W, Janchai P, Meelaksana J, Vaithanomsat P. Simultaneous Monitoring of the Evolution of Chemical Parameters in the Fermentation Process of Pineapple Fruit Wine Using the Liquid Probe for Near-Infrared Coupled with Chemometrics. Foods. 2022; 11(3):377. https://doi.org/10.3390/foods11030377
Chicago/Turabian StyleKasemsumran, Sumaporn, Antika Boondaeng, Kraireuk Ngowsuwan, Sunee Jungtheerapanich, Waraporn Apiwatanapiwat, Phornphimon Janchai, Jiraporn Meelaksana, and Pilanee Vaithanomsat. 2022. "Simultaneous Monitoring of the Evolution of Chemical Parameters in the Fermentation Process of Pineapple Fruit Wine Using the Liquid Probe for Near-Infrared Coupled with Chemometrics" Foods 11, no. 3: 377. https://doi.org/10.3390/foods11030377





