Brazzein and Monellin: Chemical Analysis, Food Industry Applications, Safety and Quality Control, Nutritional Profile and Health Impacts
Abstract
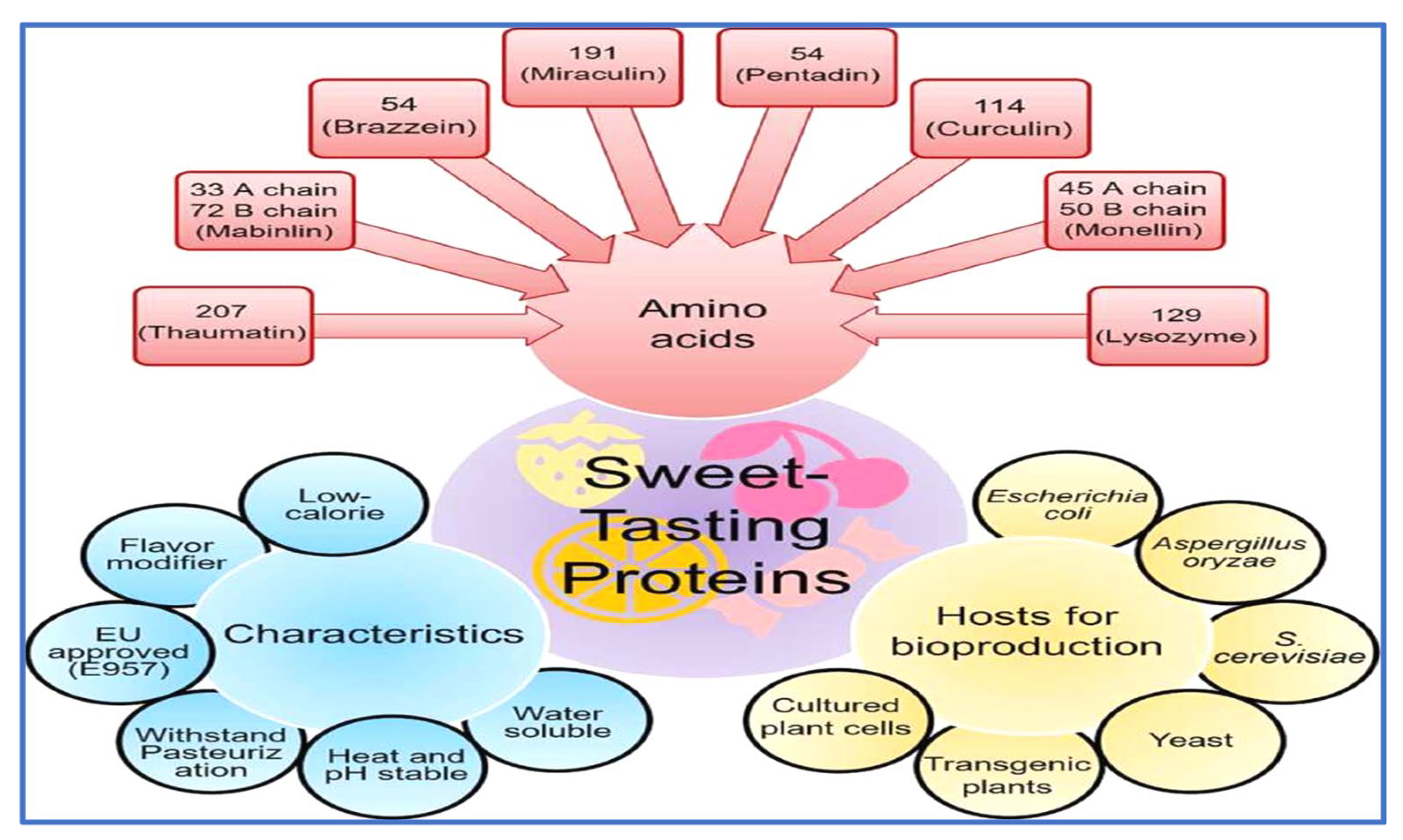
:1. Introduction
1.1. Brazzein
1.2. Monellin
2. Chemical Analysis
2.1. Brazzein
2.1.1. Isolation and Purification
2.1.2. Structural Characterization
2.2. Monellin
2.2.1. Isolation and Purification
2.2.2. Structural Characterization
3. Applications in the Food Industry and Sustainability Issues
3.1. Brazzein
3.2. Monellin
4. Safety and Quality Control
4.1. Brazzein
4.2. Monellin
5. Nutritional Profile and Health Impacts
5.1. Brazzein
5.2. Monellin
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lichtenstein, A.H.; Karpyn, A. History and development of the 2015–2020 dietary guidelines for Americans. In Food and Public Health: A Practical Introduction; Oxford University Press: Oxford, UK, 2018; pp. 31–46. [Google Scholar]
- Joseph, J.A.; Akkermans, S.; Nimmegeers, P.; Van Impe, J.F.M. Bioproduction of the recombinant sweet protein thaumatin: Current state of the art and perspectives. Front. Microbiol. 2019, 10, 695. [Google Scholar] [CrossRef] [PubMed]
- Brisbois, T.D.; Marsden, S.L.; Anderson, G.H.; Sievenpiper, J.L. Estimated intakes and sources of total and added sugars in the Canadian diet. Nutrients 2014, 6, 1899–1912. [Google Scholar] [CrossRef] [PubMed]
- You, A. Dietary Guidelines for Americans; US Department of Health and Human Services: Washington, DC, USA; US Department of Agriculture: Washington, DC, USA, 2015; p. 7.
- Agboola, D.A.; Fawibe, O.O.; Ogunyale, O.G.; Ajiboye, A.A. Botanical and protein sweeteners. J. Adv. Lab. Res. Biol. 2014, 5, 169–187. [Google Scholar]
- Kelada, K.D.; Tusé, D.; Gleba, Y.; McDonald, K.A.; Nandi, S. Process simulation and techno-economic analysis of large-scale bioproduction of sweet protein thaumatin II. Foods 2021, 10, 838. [Google Scholar] [CrossRef]
- Pearlman, M.; Obert, J.; Casey, L. The association between artificial sweeteners and obesity. Curr. Gastroenterol. Rep. 2017, 19, 64. [Google Scholar] [CrossRef]
- Whitehouse, C.R.; Boullata, J.; McCauley, L.A. The potential toxicity of artificial sweeteners. AAOHN J. 2008, 56, 251–261. [Google Scholar] [CrossRef]
- Kokotou, M.G.; Asimakopoulos, A.G.; Thomaidis, N.S. Artificial sweeteners as emerging pollutants in the environment: Analytical methodologies and environmental impact. Anal. Methods 2012, 4, 3057–3070. [Google Scholar] [CrossRef]
- Sylvetsky, A.C.; Rother, K.I. Trends in the consumption of low-calorie sweeteners. Physiol. Behav. 2016, 164, 446–450. [Google Scholar] [CrossRef]
- Kroger, M.; Meister, K.; Kava, R. Low-calorie sweeteners and other sugar substitutes: A review of the safety issues. Compr. Rev. Food Sci. Food Saf. 2006, 5, 35–47. [Google Scholar] [CrossRef]
- Etheridge, K. The sales and marketing of talin. In Thaumatin; Witty, M., Higginbotham, J.D., Eds.; CRC Press: Boca Raton, FL, USA, 1994. [Google Scholar]
- Park, S.W.; Kang, B.H.; Lee, H.M.; Lee, S.J.; Kim, H.S.; Choi, H.W.; Park, T.J.; Kong, K.H. Efficient brazzein production in yeast (Kluyveromyces lactis) using a chemically defined medium. Bioprocess Biosyst. Eng. 2021, 44, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66. [Google Scholar] [CrossRef] [PubMed]
- Kant, R. Sweet proteins—Potential replacement for artificial low calorie sweeteners. Nutr. J. 2005, 4, 5. [Google Scholar] [CrossRef]
- Faus, I. Recent developments in the characterization and biotechnological production of sweet-tasting proteins. Appl. Microbiol. Biotechnol. 2000, 53, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Kang, C.-H.; Kim, R.; Cho, J.M.; Lee, Y.-B.; Lee, T.-K. Redesigning a sweet protein: Increased stability and renaturability. Protein Eng. Des. Sel. 1989, 2, 571–575. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Ji, L.; Xu, S.; Zhang, Y.; Iqbal, H.M.N.; Cheng, H. Bioprospecting and biotechnological insights into sweet-tasting proteins by microbial hosts—A review. Bioengineered 2022, 13, 9816–9829. [Google Scholar] [CrossRef]
- Van der Wel, H.; Larson, G.; Hladik, A.; Hladik, C.M.; Hellekant, G.; Glaser, D. Isolation and characterization of pentadin, the sweet principle of Pentadiplandra brazzeana Baillon. ChemSenses 1988, 14, 75–79. [Google Scholar] [CrossRef]
- Wintjens, R.; Viet, T.M.V.N.; Mbosso, E.; Huet, J. Hypothesis/review: The structural basis of sweetness perception of sweet-tasting plant proteins can be deduced from sequence analysis. Plant Sci. 2011, 181, 347–354. [Google Scholar] [CrossRef]
- Rajan, V.; Howard, J.A. Brazzein: A Natural Sweetener 2. In Sweeteners; Springer: Cham, Switzerland, 2018; p. 17. [Google Scholar]
- Kim, M.-C.; Kim, D.-H.; Yun, C.-R.; Chung, J.-H.; Kim, H.-S.; Choi, H.-E.; Kong, K.-H. Refined Single-Interval Adjustment Matrix Yes-No Task for Estimating the Absolute Thresholds of Sweet-Tasting Molecules. J. Sens. Stud. 2017, 32, e12250. [Google Scholar] [CrossRef]
- Yusuf, E.H. An Overview of Biotransformation for the Sustainability of Sweet-Tasting Proteins as Natural Sugar Replacers. Chem. Proc. 2021, 8, 85. [Google Scholar] [CrossRef]
- Lee, Y.R.; Akter, S.; Lee, I.H.; Jung, Y.J.; Park, S.Y.; Cho, Y.-G.; Kang, K.K.; Jung, Y.J. Stable expression of brazzein protein, a new type of alternative sweetener in transgenic rice. J. Plant Biotechnol. 2018, 45, 63–70. [Google Scholar] [CrossRef]
- Jung, Y.J.; Kang, K.K. Stable expression and characterization of brazzein, thaumatin and miraculin genes related to sweet protein in transgenic lettuce. J. Plant Biotechnol. 2018, 45, 257–265. [Google Scholar] [CrossRef]
- Masuda, T.; Kitabatake, N. Developments in biotechnological production of sweet proteins. J. Biosci. Bioeng. 2006, 102, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.F.; Szczepankiewicz, O.; Thulin, E.; Linse, S.; Carey, J. Role of protein surface charge in monellin sweetness. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2009, 1794, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-B.; Kim, Y.; Lee, J.; Oh, K.-J.; Byun, M.-O.; Jeong, M.-J.; Bae, S.-C. Stable expression of the sweet protein monellin variant MNEI in tobacco chloroplasts. Plant Biotechnol. Rep. 2012, 6, 285–295. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Z.; Yu, N.; Yan, L. Expression and Secretion of a Single-Chain Sweet Protein, Monellin, in Saccharomyces cerevisiae by an α-Factor Signal Peptide. Biotechnol. Lett. 2010, 33, 721–725. [Google Scholar] [CrossRef]
- Peñarrubia, L.; Kim, R.; Giovannoni, J.; Kim, S.-H.; Fischer, R.L. Production of the sweet protein monellin in transgetic plants. Bio/Technology 1992, 10, 561–564. [Google Scholar] [CrossRef]
- Reddy, C.S.; Vijayalakshmi, M.; Kaul, T.; Islam, T.; Reddy, M.K. Improving flavour and quality of tomatoes by expression of synthetic gene encoding sweet protein monellin. Mol. Biotechnol. 2015, 57, 448–453. [Google Scholar] [CrossRef]
- Liu, J.; Yan, D.-Z.; Zhao, S.-J. Expression of monellin in a food-grade delivery system in Saccharomyces cerevisiae. J. Sci. Food Agric. 2014, 95, 2646–2651. [Google Scholar] [CrossRef]
- Cai, C.; Li, L.; Lu, N.; Zheng, W.; Yang, L.; Liu, B. Expression of a high sweetness and heat-resistant mutant of sweet-tasting protein, monellin, in Pichia pastoris with a constitutive GAPDH promoter and modified N-terminus. Biotechnol. Lett. 2016, 38, 1941–1946. [Google Scholar] [CrossRef]
- Ming, D.; Hellekant, G. Brazzein, a new high-potency thermostable sweet protein from Pentadiplandra brazzeana B. FEBS Lett. 1994, 355, 106–108. [Google Scholar] [CrossRef]
- Neiers, F.; Naumer, C.; Krohn, M.; Briand, L. The Recent Development of a Sweet-Tasting Brazzein and its Potential Industrial Applications. Ref. Ser. Phytochem. 2018, 20, 527–546. [Google Scholar] [CrossRef]
- Choi, H.-E.; Lee, J.-I.; Jo, S.-Y.; Chae, Y.-C.; Lee, J.-H.; Sun, H.-J.; Ko, K.; Hong, S.; Kong, K.-H. Functional expression of the sweet-tasting protein brazzein in transgenic tobacco. Food Sci. Technol. 2022, 42, 40521. [Google Scholar] [CrossRef]
- Kim, T.-Y.; Woo, E.-J.; Yoon, T.-S. Binding mode of brazzein to the taste receptor based on crystal structure and docking simulation. Biochem. Biophys. Res. Commun. 2022, 592, 119–124. [Google Scholar] [CrossRef]
- Singarapu, K.K.; Tonelli, M.; Markley, J.L.; Assadi-Porter, F.M. Structure-function relationships of brazzein variants with altered interactions with the human sweet taste receptor. Protein Sci. 2016, 25, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Dittli, S.M.; Rao, H.; Tonelli, M.; Quijada, J.; Markley, J.L.; Max, M.; Assadi-Porter, F. Structural Role of the Terminal Disulfide Bond in the Sweetness of Brazzein. Chem. Senses 2011, 36, 821–830. [Google Scholar] [CrossRef]
- Jafari, S.S.; Jafarian, V.; Khalifeh, K.; Ghanavatian, P.; Shirdel, S.A. The effect of charge alteration and flexibility on the function and structural stability of sweet-tasting brazzein. RSC Adv. 2016, 6, 59834–59841. [Google Scholar] [CrossRef]
- Hung, C.-Y.; Cheng, L.-H.; Yeh, C.-M. Functional expression of recombinant sweet-tasting protein brazzein by Escherichia coli and Bacillus licheniformis. Food Biotechnol. 2019, 33, 251–271. [Google Scholar] [CrossRef]
- Berlec, A.; Jevnikar, Z.; Majhenič, A.; Rogelj, I.; Štrukelj, B. Expression of the sweet-tasting plant protein brazzein in Escherichia coli and Lactococcus lactis: A path toward sweet lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 73, 158–165. [Google Scholar] [CrossRef]
- Jo, H.-J.; Noh, J.-S.; Kong, K.-H. Efficient secretory expression of the sweet-tasting protein brazzein in the yeast Kluyveromyces lactis. Protein Expr. Purif. 2013, 90, 84–89. [Google Scholar] [CrossRef]
- Poirier, N.; Roudnitzky, N.; Brockhoff, A.; Belloir, C.; Maison, M.; Thomas-Danguin, T.; Meyerhof, W.; Briand, L. Efficient production and characterization of the sweet-tasting brazzein secreted by the yeast Pichia pastoris. J. Agric. Food Chem. 2012, 60, 9807–9814. [Google Scholar] [CrossRef]
- Yan, S.; Song, H.; Pang, D.; Zou, Q.; Li, L.; Yan, Q.; Fan, N.; Zhao, X.; Yu, H.; Li, Z.; et al. Expression of Plant Sweet Protein Brazzein in the Milk of Transgenic Mice. PLoS ONE 2013, 8, e76769. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Ota, M.; Ariyoshi, Y.; Sasaki, H.; Tanokura, M.; Ming, D.; Caldwell, J.; Abilgaad, F. Crystallization and preliminary X-ray analysis of brazzein, a new sweet protein. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 577–578. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.E.; Abildgaard, F.; Džakula, Z.; Ming, D.; Hellekant, G.; Markley, J.L. Solution structure of the thermostable sweet-tasting protein brazzein. Nat. Struct. Biol. 1998, 5, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Nagata, K.; Hongo, N.; Kameda, Y.; Yamamura, A.; Sasaki, H.; Lee, W.C.; Ishikawa, K.; Suzuki, E.-I.; Tanokura, M. The structure of brazzein, a sweet-tasting protein from the wild African plant Pentadiplandra brazzeana. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 642–647. [Google Scholar] [CrossRef]
- Gao, G.; Dai, J.; Ding, M.; Hellekant, G.; Wang, J.; Wang, D. Studies on solution NMR structure of brazzein. Sci. China Life Sci. 1999, 42, 409–419. [Google Scholar] [CrossRef]
- Izawa, H.; Ota, M.; Kohmura, M.; Ariyoshi, Y. Synthesis and characterization of the sweet protein brazzein. Biopolymers 1998, 39, 95–101. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, C.; Zheng, Y.; Liu, B. New Insight into the Structure-Activity Relationship of Sweet-Tasting Proteins: Protein Sector and Its Role for Sweet Properties. Front. Nutr. 2021, 8, 310. [Google Scholar] [CrossRef] [PubMed]
- Assadi-Porter, F.M. Brazzein, a Small, Sweet Protein: Effects of Mutations on its Structure, Dynamics and Functional Properties. Chem. Senses 2005, 30, i90–i91. [Google Scholar] [CrossRef] [PubMed]
- Delfi, M.; Emendato, A.; Leone, S.; Lampitella, E.A.; Porcaro, P.; Cardinale, G.; Petraccone, L.; Picone, D. A Super Stable Mutant of the Plant Protein Monellin Endowed with Enhanced Sweetness. Life 2021, 11, 236. [Google Scholar] [CrossRef]
- Morris, J.A.; Martenson, R.; Deibler, G.; Cagan, R.H. Characterization of Monellin, a Protein That Tastes Sweet. J. Biol. Chem. 1973, 248, 534–539. [Google Scholar] [CrossRef]
- Izawa, K.; Amino, Y.; Kohmura, M.; Ueda, Y.; Kuroda, M. Human–Environment Interactions—Taste. Compr. Nat. Prod. II Chem. Biol. 2010, 4, 631–671. [Google Scholar] [CrossRef]
- Hobbs, J.R.; Munger, S.D.; Conn, G.L. Monellin (MNEI) at 1.15 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 162–167. [Google Scholar] [CrossRef]
- Kaul, T.; Reddy, C.S.; Pandey, S. Transgenics with Monellin. In Reference Series in Phytochemistry; Springer Science and Business: Berlin/Heidelberg, Germany, 2018; pp. 211–222. [Google Scholar] [CrossRef]
- Bohak, Z.; Li, S.-L. The structure of monellin and its relation to the sweetness of the protein. Biochim. Biophys. Acta (BBA)-Protein Struct. 1976, 427, 153–170. [Google Scholar] [CrossRef]
- Aghera, N.; Udgaonkar, J.B. Heterologous expression, purification and characterization of heterodimeric monellin. Protein Expr. Purif. 2011, 76, 248–253. [Google Scholar] [CrossRef]
- Fan, P.; Bracken, C.; Baum, J. Structural Characterization of Monellin in the Alcohol-Denatured State by NMR: Evidence for β-Sheet to α-Helix Conversion. Biochemistry 1993, 32, 1573–1582. [Google Scholar] [CrossRef]
- Kohmura, M.; Mizukoshi, T.; Nio, N.; Suzuki, E.-I.; Ariyoshi, Y. Structuretaste relationships of the sweet protein monellin. Pure Appl. Chem. 2002, 74, 1235–1242. [Google Scholar] [CrossRef]
- Bhattacharjee, R.; Udgaonkar, J.B. Structural Characterization of the Cooperativity of Unfolding of a Heterodimeric Protein using Hydrogen Exchange-Mass Spectrometry. J. Mol. Biol. 2021, 433, 167268. [Google Scholar] [CrossRef]
- Chen, Z.; Cai, H.; Lu, F.; Du, L. High-level expression of a synthetic gene encoding a sweet protein, monellin, in Escherichia coli. Biotechnol. Lett. 2005, 27, 1745–1749. [Google Scholar] [CrossRef]
- Kondo, K.; Miura, Y.; Sone, H.; Kobayashi, K.; Lijima, H. High-level expression of a sweet protein, monellin, in the food yeast Candida utilis. Nat. Biotechnol. 1997, 15, 453–457. [Google Scholar] [CrossRef]
- Xue, W.-F.; Szczepankiewicz, O.; Bauer, M.C.; Thulin, E.; Linse, S. Intra- versus Intermolecular Interactions in Monellin: Contribution of Surface Charges to Protein Assembly. J. Mol. Biol. 2006, 358, 1244–1255. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Yi, Y.; Liu, L.; He, M.; Deng, S.; Tian, H.; Yao, W.; Gao, X. Design and development of a high temperature stable sweet protein base on monellin. Process Biochem. 2020, 89, 29–36. [Google Scholar] [CrossRef]
- Tang, N.; Liu, J.; Cheng, Y. Potential improvement of the thermal stability of sweet-tasting proteins by structural calculations. Food Chem. 2020, 345, 128750. [Google Scholar] [CrossRef]
- Ghanavatian, P.; Khalifeh, K.; Jafarian, V. Structural features and activity of Brazzein and its mutants upon substitution of a surfaced exposed alanine. Biochimie 2016, 131, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-W.; Cha, J.-E.; Jo, H.-J.; Kong, K.-H. Multiple mutations of the critical amino acid residues for the sweetness of the sweet-tasting protein, brazzein. Food Chem. 2013, 138, 1370–1373. [Google Scholar] [CrossRef]
- Yoon, S.-Y.; Kong, J.-N.; Jo, D.-H.; Kong, K.-H. Residue mutations in the sweetness loops for the sweet-tasting protein brazzein. Food Chem. 2011, 129, 1327–1330. [Google Scholar] [CrossRef]
- Zhao, M.; Xu, X.; Liu, B. Structure basis of the improved sweetness and thermostability of a unique double-sites single-chain sweet-tasting protein monellin (MNEI) mutant. Biochimie 2018, 154, 156–163. [Google Scholar] [CrossRef]
- Assadi-Porter, F.M.; Aceti, D.J.; Markley, J.L. Sweetness determinant sites of brazzein, a small, heat-stable, sweet-tasting protein. Arch. Biochem. Biophys. 2000, 376, 259–265. [Google Scholar] [CrossRef]
- Leone, S.; Pica, A.; Merlino, A.; Sannino, F.; Temussi, P.A.; Picone, D. Sweeter and stronger: Enhancing sweetness and stability of the single chain monellin MNEI through molecular design. Sci. Rep. 2016, 6, srep34045. [Google Scholar] [CrossRef]
- Hellekant, G.; Danilova, V. Brazzein a small, sweet protein: Discovery and physiological overview. Chem. Senses 2005, 30, i88. [Google Scholar] [CrossRef]
- Fake, G.; Howard, J. Brazzein: A High-Intensity Natural Sweetener. In Commercial Plant-Produced Recombinant Protein Products; Springer: Berlin/Heidelberg, Germany, 2014; Volume 68, ISBN 978-3-662-43835-0. [Google Scholar]
- Lamphear, B.J.; Barker, D.K.; Brooks, C.A.; Delaney, D.E.; Lane, J.R.; Beifuss, K.; Love, R.; Thompson, K.; Mayor, J.; Clough, R.; et al. Expression of the sweet protein brazzein in maize for production of a new commercial sweetener. Plant Biotechnol. J. 2005, 3, 103–114. [Google Scholar] [CrossRef]
- Farag, M.A.; Rezk, M.; Elashal, M.H.; El-Araby, M.; Khalifa, S.A.; El-Seedi, H.R. An updated multifaceted overview of sweet proteins and dipeptides as sugar substitutes; the chemistry, health benefits, gut interactions, and safety. Food Res. Int. 2022, 162, 111853. [Google Scholar] [CrossRef]
- Mutharasu, G.; Peddha, M.S. Identification of novel sweet protein for nutritional applications. Bioinformation 2011, 7, 112–114. [Google Scholar] [CrossRef]
- Kim, H.; Kang, J.; Hong, S.; Jo, S.; Noh, H.; Kang, B.-H.; Park, S.; Seo, Y.-J.; Kong, K.-H.; Hong, S. 3M-Brazzein as a Natural Sugar Substitute Attenuates Obesity, Metabolic Disorder, and Inflammation. J. Agric. Food Chem. 2020, 68, 2183–2192. [Google Scholar] [CrossRef]
- Sharififar, F.; Ashrafzadeh, A.; Khanaman, M.K. A Review of Natural Peptide Sweeteners. Int. J. Pept. Res. Ther. 2022, 28, 158. [Google Scholar] [CrossRef]
- Barre, A.; Caze-Subra, S.; Gironde, C.; Bienvenu, F.; Rougé, P. Allergénicité des protéines édulcorantes. Rev. Fr. D’allergologie 2015, 55, 363–371. [Google Scholar] [CrossRef]
- Chung, J.-H.; Kong, J.-N.; Choi, H.-E.; Kong, K.-H. Antioxidant, anti-inflammatory, and anti-allergic activities of the sweet-tasting protein brazzein. Food Chem. 2018, 267, 163–169. [Google Scholar] [CrossRef]
- Świąder, K.; Wegner, K.; Piotrowska, A.; Tan, F.J.; Sadowska, A. Plants as a source of natural high-intensity sweeteners: A review. J. Appl. Bot. Food Qual. 2019, 92, 160–171. [Google Scholar] [CrossRef]
- Asr, S.Y.; Yüksel, N.; Içier, S.; Türköz, B.K. Sweet Plant Proteins and Their Recombinant Production. Türk Doğa Derg. 2022, 11, 186–194. [Google Scholar] [CrossRef]
- Pradhan, R.; Ekka, M.S.; Rout, P.; Rout, M. Production of Sweet Protein Transgenics with Monellin. Int. J. Mod. Agric. 2020, 9, 771–777. [Google Scholar]
- Ruiz-Ojeda, F.J.; Plaza-Díaz, J.; Sáez-Lara, M.J.; Gil, A. Effects of Sweeteners on the Gut Microbiota: A Review of Experimental Studies and Clinical Trials. Adv. Nutr. 2019, 10, S31–S48. [Google Scholar] [CrossRef] [PubMed]
- Tomes, D.T. Methods and Compositions for Production of Plant Foodstuffs with Enhanced Sweet Component Flavor. PCT Patent WO1997042333A1, 13 November 1997. [Google Scholar]
- Hellekant, B.G.; Ming, D. Brazzein Sweetener. U.S. Patent 5346998A, 13 September 1994. [Google Scholar]
- Yun, C.-R.; Kong, J.-N.; Chung, J.-H.; Kim, M.-C.; Kong, K.-H. Improved secretory production of the sweet-tasting protein, brazzein, in Kluyveromyces lactis. J. Agric. Food Chem. 2016, 64, 6312–6316. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-E.; Park, Y.-J.; Lee, H.; Jeong, Y.-J.; Park, S.-Y. Increased brazzein expression by abiotic stress and bioreactor culture system for the production of sweet protein, brazzein. Plant Biotechnol. Rep. 2020, 14, 459–466. [Google Scholar] [CrossRef]
- Vigues, S.; Dotson, C.; Munger, S. The receptor basis of sweet taste in mammals. In Chemosensory Systems in Mammals, Fishes, and Insects; Springer: Berlin/Heidelberg, Germany, 2008; Volume 47, pp. 20–23. [Google Scholar]
- Tyo, K.; Liu, Z.; Magnusson, Y.; Petranovic, D.; Nielsen, J. Impact of protein uptake and degradation on recombinant protein secretion in yeast. Appl. Microbiol. Biotechnol. 2014, 98, 7149–7159. [Google Scholar] [CrossRef]
- Cancelliere, R.; Leone, S.; Gatto, C.; Mazzoli, A.; Ercole, C.; Iossa, S.; Liverini, G.; Picone, D.; Crescenzo, R. Metabolic effects of the sweet protein mnei as a sweetener in drinking water. A pilot study of a high fat dietary regimen in a rodent model. Nutrients 2019, 11, 2643. [Google Scholar] [CrossRef]
| Source | Extraction | Isolation | Purification | Characterization | Main Results | Ref. |
|---|---|---|---|---|---|---|
| P. brazzeana | 0.1 M phosphate buffer at pH 7.0 containing 5% glycerol, 0.1 mM DTT, 20 mL PMSF, 0.1 mM EDTA and 0.5% (w/v) PVP at 4 °C | Protein precipitation with ammonium sulfate 30% and 85% | Ion-exchange chromatography using a CM-Sepharose CLdB column (gradient: NaCl of 0.1 to 0.4 M in 20 mM sodium citrate at pH 3.6) | SDS- PAGE; ESI-MS; sequence determination by S-Pyridylethylation and S-carboxymethylation of brazzein and peptide fragment separation by RT-HPLC. | Brazzein is a single-chain polypeptide; the molecular weights obtained by SDS-PAGE and ESI-MS were 6.5 KDa and 6.473 KDa, respectively; C-terminal is a tyrosine; 8 cysteines out of 54 residues. | [34] |
| Nicotiana tabacum cv. Xanthi | 0.1 M phosphate buffer at pH 7.0 containing 5% glycerol, 0.1 mM DTT, 20 ml PMSF, 0.1 mM EDTA and 0.5% (w/v) PVP at 4 °C | Protein precipitation with ammonium sulfate 30% and 85% | C18 RT-HPLC | 1H NMR | The secondary structure: 1 α-helix, one short 310 -helix, two strands of antiparallel β-sheet, and probably a third strand near the N-terminal; The core of the brazzein structure is a “cysteine-stabilized alpha-beta” (CSαβ) motif; The tertiary structure stabilized by four disulfide bonds. | [35] |
| P. brazzeana | Buffer solution (40 mM Tris-HCl, 50 mM NaCl, 20 mM EDTA, 55 mM sodium citrate, and 12 mM sodium thiosulfate, pH 6.7) | Ammonium sulfate 30–80% precipitation; heat treatment (80 °C for 2 h) | DEAE-Sepharose anion-exchange chromatography (gradient: 0 to 1.0 M NaCl in 20 mM Tris-HCl at pH 8.0); CM-Sepharose cation-exchange chromatography (gradient: 50 mM sodium acetate buffer with 400 mM NaCl at pH 4.0) | SDS-PAGE; RP-HPLC; CD; N-terminal amino acid sequencing; ESI-MS/MS. | The expressed brazzein presents a molar mass of 6.5 KDa; Elution time on RT-HPLC is identical to the brazzein expressed from K. lactis; The secondary structure: 9.9% of α-helices and 19.7% of β-sheets. | [36] |
| P. brazzeana | 0.1 M phosphate buffer at pH 7.0 containing 5% glycerol, 0.1 mM DTT, 20 mL PMSF, 0.1 mM EDTA and 0.5% (w/v) PVP at 4 °C | Protein precipitation with ammonium sulfate 30% and 85% | Ion-exchange chromatography using a CM-Sepharose CLdB column (gradient: NaCl of 0.1 to 0.4 M in 20 mM sodium citrate at pH 3.6) | 1H NMR (pH 5.2; 22 °C) | Folding is due to the ‘cysteine-stabilized alpha-beta’ (CSαβ) motif stabilizing the α-helix by two disulfide bonds with the nearest β-strand; Total of four disulfide bonds responsible for protein folding and its sweetness. | [37] |
| P. brazzeana | 0.1 M phosphate buffer at pH 7.0 containing 5% glycerol, 0.1 mM DTT, 20 mL PMSF, 0.1 mM EDTA and 0.5% (w/v) PVP at 4 °C | Protein precipitation with ammonium sulfate 30% and 85% | HPLC (mobile phase composed of 0.05% TFA (A) and acetonitrile with 0.05% TFA gradient: 10% B to 18% B obtained in 55 min, 18% B to 25% B in 65 min and 25% B to 10% B in 75 min; flow rate of 10 mL/min) | X-ray Crystallography (pH 4; 293 K) to 1.8 Ă resolution | The first brazzein crystal to 1.8 Ă resolution is reported. | [38] |
| Escherichia coli (E. coli) | 0.1 M phosphate buffer at pH 7.0 containing 5% glycerol, 0.1 mM DTT, 20 mL PMSF, 0.1 mM EDTA and 0.5% (w/v) PVP at 4 °C | Protein precipitation with ammonium sulfate 30% and 85% | HPLC (mobile phase composed of 0.05% TFA (A) and acetonitrile (B) with 0.05% TFA gradient: 10% B to 18% B obtained in 55 min, 18% B to 25% B in 65 min and 25% B to 10% B in 75 min; flow rate of 10 mL/min) | X-ray Crystallography (pH 4; 293 K) to 1.8 Ă resolution | The crystal structure is composed of one short α -helix and three β-strands which form a triple-stranded antiparallel β–sheet; it also contains an additional α-helix that is absent in the brazzein solution structure; In solution, brazzein exists as a monomer; In crystal, brazzein forms a homodimer stabilized by six hydrogen bonds. | [39] |
| E. coli | - | - | Nickel-affinity chromatography (mobile phase: 0–500 mM Imidazol in PBS) Cation-exchange chromatography (SP-Sepharose column; gradient: 30–1000 mM NaCl) | X-ray Crystallography (pH 4–4.5; 291 K) | Structures of the recombinant brazzein exhibit two α -helices and three β-strands linked by four disulfide bonds with a significantly altered electrostatic distribution on the surface. | [40] |
| E. coli | Tris–HCl buffer 50 mM (pH 8.0, with 2 mM EDTA) | - | CM-cellulose ion-exchange chromatography (mobile phase: 50 mM Tris-HCl with 0.6 M NaCl, pH 7.6); RT-HPLC | NMR (25 °C) | The recombinant protein adopts a cysteine-stabilized αβ (CSαβ) fold stabilized by 17 inter-strand α-helical hydrogen bonds and four disulfide bridges, that together contribute to the marked heat (100 °C) and cold (216 °C) stability of brazzein within a pH range of 2.5–11.0. | [41] |
| E. coli | Tris-HCl buffer 50 mM (pH 8.0, with 2 mM EDTA) | - | CM-cellulose ion-exchange chromatography (mobile phase: 50 mM Tris-HCl with 0.6 M NaCl, pH 7.6); RT-HPLC | RT-HPLC; NMR (pH 5.2; 37 °C). | Compared to the wild-type protein, the mutated brazzein displays an extended β-structure due to the terminal β-sheets and increased dynamics. | [42] |
| Kluyveromyces lactis | - | - | Nickel-affinity chromatography | CD (pH 7.6, 25 °C); Intrinsic fluorescence; ANS fluorescence. | Recombinant proteins (E9K and E9G) presented a molar mass of 6.5 KDa; The secondary structure of E9K brazzein is stabler than E9K and the wild-type brazzein; Brazzein has 6 tyrosine residues at positions 8, 11, 24, 39, 51 and 54, and a phenylalanine residue at position 38; The tertiary structure of the recombinant proteins is more compact than the wild-type brazzein; The local tertiary structure around tyrosine residues in the wild-type brazzein and E9G brazzein is more exposed to a polar environment; | [43] |
| Pichia pastoris | - | - | CM-Sepharose cation-exchange chromatography (gradient: 100 to 1000 mM NaCl IN 50 mM sodium acetate, at pH 4.0; flow rate of 1 mL/min) | SDS-PAGE; RT-HPLC (mobile phase: 0.1% TFA and 70% acetonitrile with 0.1% TFA); CD (25 °C). | 104 mg/L can be obtained from the recombinant brazzein; The molar mass of recombinant protein is 6.5 KDa with an elution time in RT-HPLC of 9 ± 0.5 min; Compared to the wild-type brazzein, no significant alterations in the secondary structure of recombinant brazzein are observed by CD analysis. | [44] |
| Bacillus licheniformis | - | - | Cation-exchange chromatography (SP-Sepharose column; gradient: 0 to 1 M NaCl in 50 mM sodium acetate buffer pH 4 in 50 min; flow rate of 1 mL/min) | SDS-PAGE; ESI-MS; NMR. | Recombinant proteins are correctly folded; | [45] |
| E. coli | - | - | Cation-exchange chromatography (Q-Sepharose column; gradient: 300 to 1000 mM NaCl in 20 mM Tris buffer; flow rate of 1 mL/min) | SDS-PAGE; LC-MS/MS; CD (25 °C). | 85% purity; formation of disulfide bonds is confirmed by LC-MS/MS; The secondary structure of the recombinant protein is similar to the wild-type brazzein. | [46] |
| E. coli | - | - | Nickel-affinity chromatography | SDS-PAGE; LC-MS/MS; MALDI-TOF. | Formation of disulfide bonds is confirmed by LC-MS/MS; The secondary structure of the recombinant protein is similar to the wild-type brazzein | [46] |
| Lactococcus lactis | Talon-affinity chromatography; RT-HPLC. | Edman degradation; SDS-PAGE. | The primary structure of the recombinant brazzein is similar to the wild-type brazzein. | [47] | ||
| Talon-affinity chromatography; RT-HPLC. | Edman degradation; SDS-PAGE. | The primary structure of the recombinant brazzein is similar to the wild-type brazzein. | [47] |
| Source | Extraction | Isolation | Purification | Characterization | Main Results | Ref. |
|---|---|---|---|---|---|---|
| D. cumminsii | - | Salt precipitation | Ion-exchange chromatography | SDS- PAGE; Gel filtration; Fluorescence spectroscopy. | The molecular weight of monellin obtained by SDS-PAGE and gel filtration is 10.5 KDa and 10.0 KDa, respectively. The primary structure has 91 amino acids with single residues of tryptophan, methionine, and cysteine | [54] |
| D. cumminsii | - | Salt precipitation; Gel filtration on Sephadex G-50 (mobile phase: 1% aqueous acetic acid). | Ion-exchange chromatography; Affinity chromatography (mobile phase: 6 M guanidine. HCl, 0.1 M sodium phosphate buffer, pH 7.4, containing 0.2 M dithiothreitol). | SDS-PAGE; Edman degradation. | Monellin is composed of two chains of similar length linked by non-covalent bonds. However, the subunits, devoid of sweetness, are not identical. By abolishing the thiol group, monellin loses its sweet taste | [58] |
| Standard | - | - | Ion-exchange chromatography using a Sephadex-CM 25-gel (mobile phase: 100 mM NaCl, 10 mM phosphate buffer); RT-HPLC (gradient: 30% methanol, 0.1% TFA to 70% methanol, 0.1% TFA). | N-terminal amino acid sequencing; ESI-MS/MS; Size-exclusion chromatography; NMR; CD | The secondary structure: in the native state, the chain A of monellin consists of β-structure, and chain B contains both α and β-structures; Addition of 50% ethanol or TFE denatures the protein. | [60] |
| Synthesized | - | - | RT-HPLC; HIC. | HPLC; ESI-MS; Quantitative amino acid analysis. | Monellin contains five Aspartate residues and nine Lysine residues; Asp87 h plays an important role in monellin sweetness. | [61] |
| MNEI monellin * expressed in E. coli | - | - | Ion-exchange chromatography using a HiPrep26/60 Sephacryl 100 column (mobile phase: sodium acetate and sodium chloride); Gel filtration in a G-75 column (elution: 150 mM ammonium bicarbonate). | X-ray Crystallography (resolution of 1.15 Ă) | The crystal contains a single MNEI protein in the asymmetric unit and lacks the dimer interface observed in all the previous crystal structures of monellin and its single-chain derivatives; Four stably bound negative ions are also located and can be related to potential electrostatic interactions with the surface of the sweet taste receptor; | [56] |
| MNEI and muted MNEI monellin expression in E. coli | - | - | Ion-exchange chromatography; Size-exclusion chromatography. | SDS-PAGE; CD. | Mutated protein eluted at high salt concentrations (200 mM NaCl) relative to MNEI monellin (100–150 mM NaCl). CD spectra of the mutated monellin presents two minimums at 201 and 213 nm; At pH 2.5–6.8, the β-sheet and α-helix content exhibit minor changes, which corroborates the folding stability of the mutated monellin | [59] |
| E. coli | - | - | Ion exchange chromatography; Size-exclusion chromatography. | ESI-MS; Hydrogen exchange-mass spectrometry. | Monellin purity exceeds 95%. Chain A and B molar masses are 5.382 KDa and 5.965 KDa, respectively; Double-chain monellin (dcMN) unfolds in a barrier-limited manner in which chain B undergoes non cooperative exchange and chain A cooperatively. | [62] |
| E. coli | - | Incubation of the cell extract at 60 °C for 10 min and at pH 4 for 1 h at 4 °C | Ion-exchange chromatography using a Sephadex CM-50 column (gradient: 0 to 0.4 M NaCl); SDS-PAGE. | - | Recombinant monellin yield of 43 mg/g of dry cell wt. Purity confirmed by SDS-PAGE. | [63] |
| Candida utilis | - | - | Ion exchange chromatography using a CM-Sepharose column (gradient: 0–0.4 M NaCl); | SDS-PAGE | Molecular mass of single-chain protein is 10.0 KDa. In total soluble protein, 5% is monellin. | [64] |
| E. coli | - | - | Ion-exchange chromatography using CM-cellulose and DEAE-cellulose columns due to the nature of the different mutants proteins; RT-HPLC using a Resource RPC column (mobile phase: eluent A composed of 10% acetonitrile/water with 0.1% TFA, and eluent B of 90% acetonitrile/water with 0.1% TFA; gradient: 20–50% B). | SDS-PAGE; Amino acid analysis followed by hydrolysis; MALDI-TOF-MS; NMR-HSQC spectroscopy; Fluorescence emission spectroscopy; CD. | The intermolecular and intramolecular Coulombic interactions are involved in the stabilization of recombinant monellin and in the reconstitution of the wild-type monellin. Charge interactions may significantly modulate the folding of monellin and its binding to the sweet receptors by modifying association rates. | [65] |
| E. coli | - | - | Nickel-affinity chromatography; RT-HPLC (mobile phase: acetonitrile 5%; flow rate of 1 mL/min). | SDS-PAGE | H-Monellin shows an identical fold and a typical β-sheet-rich structure. The molar mass of H-monellin is 16.0 KDa, with 14.0 KDa for the MNEI monellin. | [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saraiva, A.; Carrascosa, C.; Ramos, F.; Raheem, D.; Pedreiro, S.; Vega, A.; Raposo, A. Brazzein and Monellin: Chemical Analysis, Food Industry Applications, Safety and Quality Control, Nutritional Profile and Health Impacts. Foods 2023, 12, 1943. https://doi.org/10.3390/foods12101943
Saraiva A, Carrascosa C, Ramos F, Raheem D, Pedreiro S, Vega A, Raposo A. Brazzein and Monellin: Chemical Analysis, Food Industry Applications, Safety and Quality Control, Nutritional Profile and Health Impacts. Foods. 2023; 12(10):1943. https://doi.org/10.3390/foods12101943
Chicago/Turabian StyleSaraiva, Ariana, Conrado Carrascosa, Fernando Ramos, Dele Raheem, Sónia Pedreiro, Angelo Vega, and António Raposo. 2023. "Brazzein and Monellin: Chemical Analysis, Food Industry Applications, Safety and Quality Control, Nutritional Profile and Health Impacts" Foods 12, no. 10: 1943. https://doi.org/10.3390/foods12101943
APA StyleSaraiva, A., Carrascosa, C., Ramos, F., Raheem, D., Pedreiro, S., Vega, A., & Raposo, A. (2023). Brazzein and Monellin: Chemical Analysis, Food Industry Applications, Safety and Quality Control, Nutritional Profile and Health Impacts. Foods, 12(10), 1943. https://doi.org/10.3390/foods12101943











