Untargeted Metabolomics Analysis Revealed the Difference of Component and Geographical Indication Markers of Panax notoginseng in Different Production Areas
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Preparation
2.2. 1H-NMR Spectra Acquisition and Processing
2.3. Statistical Analysis
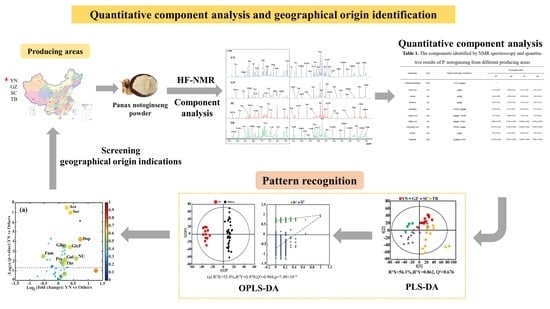
3. Results and Discussion
3.1. 1H-NMR Analysis of P. notoginseng from Different Areas
3.2. Geographical Origin Discrimination of P. notoginseng
3.3. Identification of Geographical Indication Components of P. notoginseng
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jiang, X.; Kim, S.-H.; Oh, T.-J.; Huang, L.-F.; Choi, H.-K. Proton nuclear magnetic resonance spectrometry-based metabolic characterization of Panax notoginseng roots. Anal. Lett. 2015, 48, 1341–1354. [Google Scholar] [CrossRef]
- Jee, H.S.; Chang, K.H.; Park, S.H.; Kim, K.T.; Paik, H.D. Morphological characterization, chemical components, and biofunctional activities of Panax ginseng, Panax quinquefolium, and Panax notoginseng roots: A comparative study. Food Rev. Int. 2014, 30, 91–111. [Google Scholar] [CrossRef]
- Qu, J.; Xu, N.; Zhang, J.L.; Geng, X.K.; Zhang, R.H. Panax notoginseng saponins and their applications in nervous system disorders: A narrative review. Ann. Transl. Med. 2020, 8, 1525. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Zhang, B.; Zhang, C.Y.; Sun, G.B.; Sun, X.B. Effect of Panax notoginseng saponins and major anti-obesity components on weight loss. Front. Pharmacol. 2021, 11, 601751. [Google Scholar] [CrossRef] [PubMed]
- Hawthorne, B.; Lund, K.; Freggiaro, S.; Kaga, R.; Meng, J. The mechanism of the cytotoxic effect of Panax notoginseng extracts on prostate cancer cells. Biomed. Pharmacother. 2022, 149, 112887. [Google Scholar] [CrossRef]
- Dong, T.T.; Cui, X.M.; Song, Z.H.; Zhao, K.J.; Ji, Z.N.; Lo, C.K.; Tsim, K.W. Chemical assessment of roots of Panax notoginseng in China: Regional and seasonal variations in its active constituents. J. Agric. Food Chem. 2003, 51, 4617–4623. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Xu, F.R.; Wang, Y.Z.; Zhang, J.Y. Rapid prediction study of total flavonids content in Panax notoginseng using infrared spectroscopy combined with chemometrics. Spectrosc. Spect. Anal. 2017, 37, 70–74. [Google Scholar]
- Zhou, Y.H.; Zuo, Z.T.; Xu, F.R.; Wang, Y.Z. Origin identification of Panax notoginseng by multi-sensor information fusion strategy of infrared spectra combined with random forest. Spectrochim. Acta. A Mol. Biomol. Spectrosc. 2020, 226, 117619. [Google Scholar] [CrossRef]
- Klockmann, S.; Reiner, E.; Cain, N.; Fischer, M. Food targeting: Geographical origin determination of hazelnuts (Corylus avellana) by LC-QqQ-MS/MS-based targeted metabolomics application. J. Agric. Food Chem. 2017, 65, 1456–1465. [Google Scholar] [CrossRef]
- de Rijke, E.; Fellner, C.; Westerveld, J.; Lopatka, M.; Cerli, C.; Kalbitz, K.; de Koster, C.G. Determination of n-alkanes in c-annuum (bell pepper) fruit and seed using GC-MS: Comparison of extraction methods and application to samples of different geographical origin. Anal. Bioanal. Chem. 2015, 407, 5729–5738. [Google Scholar] [CrossRef]
- Song, Y.; Xu, J.; Chen, N.; Li, M. The complete chloroplast genome of traditional Chinese medical plants Paris polyphylla var. yunnanensis. Mitochondrial DNA Part A 2017, 28, 159–160. [Google Scholar] [CrossRef]
- Bai, J.; Yue, P.; Dong, Q.; Wang, F.; He, C.Y.; Li, Y.; Guo, J.L. Identification of geographical origins of Panax notoginseng based on HPLC multi-wavelength fusion profiling combined with average linear quantitative fingerprint method. Sci. Rep. 2021, 11, 5126. [Google Scholar] [CrossRef]
- Rivera-Perez, A.; Romero-Gonzalez, R.; Frenich, A.G. A metabolomics approach based on 1H NMR fingerprinting and chemometrics for quality control and geographical discrimination of black pepper. J. Food Compost Anal. 2022, 105, 104235. [Google Scholar] [CrossRef]
- Lin, H.F.; He, C.X.; Liu, H.L.; Shen, G.P.; Xia, F.; Feng, J.H. NMR-based quantitative component analysis and geographical origin identification of China’s sweet orange. Food Control 2021, 130, 108292. [Google Scholar] [CrossRef]
- Calo, F.; Girelli, C.R.; Wang, S.C.; Fanizzi, F.P. Geographical origin assessment of extra virgin olive oil via NMR and MS combined with chemometrics as analytical approaches. Foods 2022, 11, 113. [Google Scholar] [CrossRef]
- Lalaleo, L.; Hidalgo, D.; Valle, M.; Calero-Caceres, W.; Lamuela-Raventos, R.M.; Becerra-Martinez, E. Differentiating, evaluating, and classifying three quinoa ecotypes by washing, cooking and germination treatments, using 1H NMR-based metabolomic approach. Food Chem. 2020, 331, 127351. [Google Scholar] [CrossRef]
- An, L.; Ma, J.W.; Wang, H.; Li, F.G.; Qin, D.M.; Wu, J.K.; Zhu, G.Y.; Zhang, J.F.; Yuan, Y.L.; Zhou, L.; et al. NMR-based global metabolomics approach to decipher the metabolic effects of three plant growth regulators on strawberry maturation. Food Chem. 2018, 269, 559–566. [Google Scholar] [CrossRef]
- Song, X.Y.; She, S.; Xin, M.M.; Chen, L.; Li, Y.; Vander Heyden, Y.; Rogers, K.M.; Chen, L.Z. Detection of adulteration in Chinese monofloral honey using 1H nuclear magnetic resonance and chemometrics. J. Food Compost Anal. 2020, 86, 103390. [Google Scholar] [CrossRef]
- Gougeon, L.; da Costa, G.; Guyon, F.; Richard, T. 1H NMR metabolomics applied to Bordeaux red wines. Food Chem. 2019, 301, 125257. [Google Scholar] [CrossRef]
- Yao, C.L.; Wang, J.; Li, Z.W.; Qu, H.; Pan, H.Q.; Li, J.Y.; Wei, W.L.; Zhang, J.Q.; Bi, Q.R.; Guo, D.A. Characteristic malonyl ginsenosides from the leaves of Panax notoginseng as potential quality markers for adulteration detection. J. Agric. Food Chem. 2021, 69, 4849–4857. [Google Scholar] [CrossRef]
- Manirakiza, P.; Covaci, A.; Schepens, P. Comparative study on total lipid determination using Soxhlet, Roese-Gottlieb, Bligh & Dyer, and modified Bligh & Dyer extraction methods. J. Food Compost. Anal. 2001, 14, 93–100. [Google Scholar]
- Wu, H.F.; Southam, A.D.; Hines, A.; Viant, M.R. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal. Biochem. 2008, 372, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. PCA as a practical indicator of OPLS-DA model reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Jeong, J.; Alves, A.C.; Han, S.-T.; In, G.; Kim, E.-H.; Jeong, W.-S.; Hong, Y.-S. Metabolomic understanding of intrinsic physiology in Panax ginseng during whole growing seasons. J. Ginseng Res. 2019, 43, 654–665. [Google Scholar] [CrossRef]
- Chan, P.H.; Zheng, K.Y.Z.; Tsim, K.W.K.; Lam, H. Metabonomic analysis of water extracts from Chinese and American ginsengs by 1H nuclear magnetic resonance: Identification of chemical profile for quality control. Chin. Med. 2012, 7, 25. [Google Scholar] [CrossRef]
- Cho, J.-G.; In, S.-J.; Jung, Y.-J.; Cha, B.-J.; Lee, D.-Y.; Kim, Y.-B.; Yeom, M.; Baek, N.-I. Re-evaluation of physicochemical and NMR data of triol ginsenosides Re, Rf, Rg2, and 20-gluco-Rf from Panax ginseng roots. J. Ginseng Res. 2014, 38, 116–122. [Google Scholar] [CrossRef]
- Teng, R.W.; Li, H.Z.; Chen, J.T.; Wang, D.Z.; He, Y.N.; Yang, C.R. Complete assignment of 1H and 13C NMR data for nine protopanaxatriol glycosides. Magn. Reson. Chem. 2002, 40, 483–488. [Google Scholar] [CrossRef]
- Yamashita, H. Biological function of acetic acid-improvement in obesity and glucose tolerance by acetic acid in type 2 diabetic rats. Crit. Rev. Food Sci. Nutr. 2016, 56, S171–S175. [Google Scholar] [CrossRef]
- Tabatabaie, L.; Klomp, L.W.; Berger, R.; de Koning, T.J. L-Serine synthesis in the central nervous system: A review on serine deficiency disorders. Mol. Genet. Metab. 2010, 99, 256–262. [Google Scholar] [CrossRef]
- Hussain, T.; Lokhandwala, M.F. Renal dopamine receptors and hypertension. Exp. Biol. Med. 2003, 228, 134–142. [Google Scholar] [CrossRef]
- Marsden, C.A. Dopamine: The rewarding years (vol 147, pg S136, 2006). Br. J. Pharmacol. 2021, 178, 4772. [Google Scholar]
- Lau, A.-J.; Toh, D.-F.; Chua, T.-K.; Pang, Y.-K.; Woo, S.-O.; Koh, H.-L. Antiplatelet and anticoagulant effects of Panax notoginseng: Comparison of raw and steamed Panax notoginseng with Panax ginseng and Panax quinquefolium. J. Ethnopharmacol. 2009, 125, 380–386. [Google Scholar] [CrossRef]
- Fiume, Z. Final report on the safety assessment of malic acid and sodium malate. Int. J. Toxicol. 2001, 20 (Suppl. S1), 47–55. [Google Scholar]
- Owjfard, M.; Bigdeli, M.R.; Safari, A.; Haghani, M.; Namavar, M.R. Effect of dimethyl fumarate on the motor function and Spatial arrangement of primary motor cortical neurons in the sub-acutephase of stroke in a rat model. J. Stroke Cerebrovasc. 2021, 30, 105630. [Google Scholar] [CrossRef]
- Evans, M.L.; Hopkins, D.; Macdonald, I.A.; Amiel, S.A. Alanine infusion during hypoglycaemia partly supports cognitive performance in healthy human subjects. Diabet. Med. 2004, 21, 440–446. [Google Scholar] [CrossRef]
- Richerson, G.B.; Macdonald, R.L.; Mody, I.; Kullmann, D.M. The role of tonic GABA inhibition in control of brain excitability. In Proceedings of the Annual Meeting of the American Epilepsy Society, Boston, MA, USA, 5–10 December 2003. [Google Scholar]
- Smart, T.G.; Stephenson, F.A. A half century of gamma-aminobutyric acid. Brain Neurosci. Adv. 2019, 3, 2398212819858249. [Google Scholar] [CrossRef]



| Components | Abbr. | Chemical Shift (ppm) (Multiplicity) | Concentration (mM) | |||
|---|---|---|---|---|---|---|
| YN a | GZ | SC | TB | |||
| 1-Methylnicotinamide | MNA | 8.21 (t b), 9.34 (s) c | / d | / | / | / |
| Acetic acid | Ace | 1.94 (s) | 5.25 ± 0.02 e | 3.96 ± 0.02 | 4.37 ± 0.05 | 4.97 ± 0.02 |
| Alanine | Ala | 1.47 (d) | 4.54 ± 0.03 | 4.08 ± 0.04 | 3.90 ± 0.05 | 5.56 ± 0.04 |
| Arabinose | Ara | 4.48 (d) | 6.44 ± 0.03 | 5.78 ± 0.04 | 5.35 ± 0.10 | 5.85 ± 0.13 |
| Asparagine | Asn | 2.85 (dd), 2.95 (dd) | 0.58 ± 0.02 | 0.57 ± 0.01 | 0.62 ± 0.01 | 0.74 ± 0.02 |
| Aspartic acid | Asp | 2.58 (dd), 2.68 (dd) | 1.07 ± 0.01 | 0.89 ± 0.02 | 1.19 ± 0.02 | 1.30 ± 0.02 |
| Caffeic acid | Caf | 6.92 (d), 7.04 (m) | 0.089 ± 0.001 | 0.103 ± 0.003 | 0.068 ± 0.002 | 0.106 ± 0.002 |
| Chlorogenic acid | Chl | 6.95 (m), 7.11 (m) | 0.075 ± 0.001 | 0.104 ± 0.003 | 0.069 ± 0.003 | 0.100 ± 0.002 |
| Choline | Cho | 3.19 (s) | 6.45 ± 0.02 | 5.24 ± 0.02 | 4.60 ± 0.07 | 7.65 ± 0.03 |
| Dopamine | Dop | 6.74 (dd), 6.84 (d) | 0.156 ± 0.003 | 0.104 ± 0.002 | 0.084 ± 0.004 | 0.093 ± 0.003 |
| Ethanolamine | EA | 3.15 (t) | 6.11 ± 0.02 | 5.75 ± 0.03 | 5.19 ± 0.04 | 6.38 ± 0.03 |
| Ferulic acid | Fer | 6.82 (d), 7.15 (d) | 0.077 ± 0.001 | 0.101 ± 0.002 | 0.070 ± 0.002 | 0.100 ± 0.002 |
| Formic acid | For | 8.44 (s) | 0.034 ± 0.001 | 0.031 ± 0.001 | 0.049 ± 0.003 | 0.061 ± 0.003 |
| Fructose | Fru | 3.99 (m), 4.12 (d), 3.70 (m) | 3.21 ± 0.03 | 3.17 ± 0.11 | 3.39 ± 0.08 | 4.19 ± 0.04 |
| Fumarate | Fum | 6.53 (s) | 0.155 ± 0.004 | 0.256 ± 0.008 | 0.258 ± 0.008 | 0.132 ± 0.003 |
| Galactose 1 phosphate | GalP | 4.18(t), 5.50 (d) | 0.234 ± 0.005 | 0.206 ± 0.005 | 0.184 ± 0.003 | 0.173 ± 0.006 |
| γ-Aminobutyric acid | GABA | 1.91 (q), 2.32 (t), 3.01 (t) | 2.46 ± 0.02 | 2.48 ± 0.04 | 2.45 ± 0.05 | 5.64 ± 0.07 |
| Ginsenoside Rb1 | GRb1 | 0.95 (s), 1.25 (s), 5.02 (d), 5.16 (br) | 19.67 ± 0.11 | 18.87 ± 0.22 | 12.45 ± 0.18 | 20.25 ± 0.16 |
| Ginsenoside Rb2 | GRb2 | 1.84 (s), 2.24 (s) | 12.91 ± 0.08 | 11.03 ± 0.07 | 10.87 ± 0.07 | 12.88 ± 0.08 |
| Ginsenoside Rc | GRc | 0.93 (s), 1.07 (br) | 30.77 ± 0.20 | 27.31 ± 0.21 | 22.93 ± 0.15 | 31.89 ± 0.11 |
| Ginsenoside Rd | GRd | 0.97 (s), 1.28 (s) | 20.11 ±0.14 | 20.60 ± 0.16 | 21.90 ± 0.41 | 22.81 ± 0.29 |
| Ginsenoside Re | GRe | 1.34 (s) | 17.24 ± 0.07 | 17.44 ± 0.10 | 14.25 ± 0.15 | 18.17 ± 0.13 |
| Ginsenoside Rg1 | GRg1 | 1.70 (s), 6.37 (br), 8.09 (s), 8.30 (s) 8.47 (s) | 23.48 ± 0.22 | 22.09 ± 0.21 | 19.48 ± 0.19 | 23.27 ± 0.24 |
| Glucosamine 6-phosphate | GlcP | 4.96 (d), 5.45 (d) | 1.57 ± 0.02 | 1.15 ± 0.01 | 1.07 ± 0.01 | 1.50 ± 0.05 |
| Guanosine | Gua | 5.85 (d), 7.99 (s) | 0.074 ± 0.001 | 0.055 ± 0.001 | 0.056 ± 0.001 | 0.093 ± 0.001 |
| Gulonolactone | Gul | 4.73 (d) | 1.84 ± 0.02 | 1.63 ± 0.02 | 1.53 ± 0.02 | 1.64 ± 0.03 |
| Inosine | Ino | 6.06 (d), 8.35 (s) | 0.122 ± 0.002 | 0.074 ± 0.001 | 0.080 ± 0.001 | 0.174 ± 0.002 |
| Malic acid | MA | 4.33 (dd) | 2.51 ± 0.03 | 3.73 ± 0.09 | 2.86 ± 0.06 | 2.03 ± 0.03 |
| Maltotriose | Mti | 4.65 (d) | 0.85 ± 0.01 | 0.405 ± 0.004 | 0.59 ± 0.02 | 0.57 ± 0.01 |
| Myo-Inositol | m-I | 3.27 (t), 3. 52 (dd), 3.61 (t) | 11.10 ± 0.03 | 9.23 ± 0.02 | 10.09 ± 0.09 | 11.78 ± 0.10 |
| N-Methylnicotinamide | NMN | 8.90 (d), 9.29 (s) | / | / | / | / |
| Notoginsenoside R1 | NR1 | 2.74 (d) | 3.20 ± 0.07 | 3.89 ± 0.06 | 2.36 ± 0.07 | 1.81 ± 0.04 |
| Notoginsenoside Re | NRe | 1.64 (br) | 41.59 ± 0.21 | 39.48 ± 0.33 | 36.22 ± 0.41 | 41.99 ± 0.28 |
| Notoginsenoside U | NU | 5.29 (s) | 1.31 ± 0.04 | 0.96 ± 0.02 | 0.95 ± 0.01 | 0.60 ± 0.02 |
| Phenylalanine | Phe | 7.32 (d), 7.37 (m), 7.42 (m) | 0.201 ± 0.001 | 0.245 ± 0.006 | 0.180 ± 0.005 | 0.361 ± 0.003 |
| Proline | Pro | 1.99 (m), 2.06 (m), 4.13 (m) | 14.78 ± 0.04 | 13.94 ± 0.07 | 12.64 ± 0.10 | 15.88 ± 0.06 |
| Pyridoxine (Vitamin B6) | VB6 | 7.67 (s) | 0.173 ± 0.002 | 0.154 ± 0.003 | 0.189 ± 0.007 | 0.166 ± 0.008 |
| Raffinose | Raf | 5.43 (d) | 0.83 ± 0.01 | 0.80 ± 0.02 | 0.93 ± 0.02 | 0.77 ± 0.01 |
| Ribonolactone | Rib | 4.42 (dd) | 2.03 ± 0.02 | 1.57 ± 0.01 | 1.82 ± 0.03 | 2.63 ± 0.05 |
| Serine | Ser | 3.95 (dd) | 6.90 ± 0.05 | 4.23 ± 0.03 | 5.60 ± 0.09 | 6.26 ± 0.06 |
| Succinic acid | Suc | 2.51 (s) | 2.16 ± 0.04 | 2.86 ± 0.11 | 1.37 ± 0.05 | 2.00 ± 0.05 |
| Sucrose | Sur | 3.46 (t), 3.55(dd), 3.67 (s), 3.75 (s), 3.82 (m), 3.87 (dd), 4.04 (t), 4.21 (d), 5.40 (d) | 19.04 ± 0.08 | 26.29 ± 0.12 | 24.66 ± 0.33 | 16.21 ± 0.09 |
| Threonine | Thr | 1.32 (d), 4.27 (m) | 7.61 ± 0.04 | 7.53 ± 0.10 | 5.38 ± 0.15 | 7.74 ± 0.04 |
| Trigonelline | Tri | 8.12 (m), 8.94 (t), 9.19 (m) | / | / | / | / |
| Tryptophan | Trp | 7.27 (m), 7.52 (d), 7.72 (d) | 0.042 ± 0.001 | 0.039 ± 0.001 | 0.030 ± 0.001 | 0.085 ± 0.001 |
| Tyrosine | Tyr | 6.89 (d), 7.19 (d) | 0.197 ± 0.001 | 0.244 ± 0.004 | 0.210 ± 0.006 | 0.331 ± 0.004 |
| Unknown1 | U1 | 5.79 (d) | / | / | / | / |
| Unknown2 | U2 | 7.89 (d) | / | / | / | / |
| Unknown3 | U3 | 8.61 (s) | / | / | / | / |
| Uridine | Ud | 5.89 (d), 5.91 (d), 7.87 (d) | 0.129 ± 0.002 | 0.094 ± 0.03 | 0.093 ± 0.04 | 0.178 ± 0.03 |
| α-Glucose | α-Glc | 3.42 (t), 5.22 (d) | 1.52 ± 0.02 | 0.98 ± 0.03 | 1.88 ± 0.04 | 1.75 ± 0.03 |
| β-Glucose | β-Glc | 3.23 (dd), 3.41 (t), 4.63 (d) | 2.28 ± 0.03 | 1.88 ± 0.05 | 2.75 ± 0.07 | 3.06 ± 0.06 |
| Components | YN-Others | GZ-Others | SC-Others | TB-Others | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r a | FC b | p c | VIP d | r | FC | p | VIP | r | FC | p | VIP | r | FC | p | VIP | |
| Acetic acid | −0.66 | 1.20 | 3.3 × 10−8 | 1.80 | 0.81 | 0.82 | 6.4 × 10−10 | 0.96 | / e | / | / | / | / | / | / | / |
| Alanine | / | / | / | / | 0.63 | 0.90 | 0.02 | 1.23 | / | / | / | / | −0.72 | 1.33 | 2.7 × 10−6 | 1.56 |
| Choline | / | / | / | / | / | / | / | / | / | / | / | / | −0.51 | 1.41 | 5.7 × 10−11 | 2.00 |
| Dopamine | −0.70 | 1.66 | 5.9 × 10−5 | 1.90 | / | / | / | / | / | / | / | / | / | / | / | |
| Ethanolamine | / | / | / | / | / | / | / | / | 0.83 | 0.86 | 4.1 × 10−5 | 2.21 | / | / | / | / |
| Fumarate | −0.55 | 0.69 | 3.2 × 10−3 | 1.49 | / | / | / | / | −0.80 | 1.38 | 0.04 | 2.18 | / | / | / | / |
| γ-Aminobutyric acid | / | / | / | / | / | / | / | / | / | / | / | / | −0.82 | 2.29 | 1.2 × 10−5 | 2.49 |
| Ginsenoside Rb1 | / | / | / | / | / | / | / | / | 0.69 | 0.69 | 5.1 × 10−8 | 1.94 | / | / | / | / |
| Ginsenoside Rb2 | / | / | / | 0.64 | 0.91 | 0.003 | 1.29 | 0.79 | 0.89 | 2.1 × 10−4 | 2.16 | −0.57 | 1.11 | 0.006 | 1.28 | |
| Ginsenoside Rc | −0.53 | 1.15 | 2.9 × 10−4 | 1.53 | / | / | / | / | 0.89 | 0.77 | 8.2 × 10−11 | 2.40 | / | / | 6.11 × 10−7 | 2.70 |
| Ginsenoside Re | / | / | / | / | / | / | / | / | 0.65 | 0.81 | 3. 5 × 10−5 | 1.82 | −0.55 | 1.11 | 0.01 | 3.09 |
| Ginsenoside Rg1 | / | / | / | / | / | / | / | / | 0.56 | 0.85 | 3.1 × 10−4 | 1.63 | / | / | / | / |
| Glucosamine-6-phosphate | −0.59 | 1.30 | 3.6 × 10−4 | 1.68 | 0.65 | 0.84 | 0.01 | 1.39 | / | / | / | / | / | / | / | / |
| Gulonolactone | −0.55 | 1.15 | 7.2 × 10−3 | 1.54 | / | / | / | / | / | / | / | / | / | / | / | / |
| Malic acid | / | / | / | / | −0.80 | 1.48 | 0.002 | 2.04 | / | / | / | / | / | / | / | / |
| Myo-Inositol | / | / | / | / | 0.72 | 0.85 | 1.3 × 10−9 | 1.53 | / | / | / | / | −0.59 | 1.16 | 0.005 | 1.63 |
| Notoginsenoside R1 | / | / | / | / | −0.66 | 1.53 | 2.4 × 10−5 | 1.28 | / | / | / | / | / | / | / | / |
| Notoginsenoside Re | / | / | / | / | / | / | / | / | 0.55 | 0.89 | 0.008 | 1.61 | / | / | / | / |
| Notoginsenoside U | −0.56 | 1.51 | 5.6 × 10−3 | 1.55 | / | / | / | / | / | / | / | / | / | / | / | / |
| Proline | −0.59 | 1.06 | 0.01 | 1.67 | / | / | / | / | 0.75 | 0.86 | 4.3 × 10−5 | 2.06 | −0.73 | 1.15 | 1.3 × 10−5 | 1.55 |
| Raffinose | / | / | / | / | / | / | / | / | / | / | / | / | / | / | / | / |
| Ribonolactone | / | / | / | / | 0.70 | 0.74 | 1.2 × 10−6 | 1.50 | / | / | / | / | / | / | / | / |
| Serine | −0.65 | 1.31 | 1.0 × 10−7 | 1.79 | 0.77 | 0.68 | 1.9 × 10−12 | 1.69 | / | / | / | / | / | / | / | / |
| Sucrose | / | / | / | / | / | / | / | / | −0.54 | 1.17 | 0.02 | 1.51 | / | / | / | / |
| Threonine | −0.65 | 1.12 | 0.02 | 1.79 | / | / | / | / | 0.68 | 0.71 | 0.001 | 1.92 | / | / | / | / |
| α-Glucose | / | / | / | / | 0.61 | 0.57 | 4.1 × 10−7 | 0.99 | / | / | / | / | / | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Fang, K.; Ding, Z.; Wu, J.; Lin, J.; Xu, D.; Zhong, J.; Xia, F.; Feng, J.; Shen, G. Untargeted Metabolomics Analysis Revealed the Difference of Component and Geographical Indication Markers of Panax notoginseng in Different Production Areas. Foods 2023, 12, 2377. https://doi.org/10.3390/foods12122377
Zhang S, Fang K, Ding Z, Wu J, Lin J, Xu D, Zhong J, Xia F, Feng J, Shen G. Untargeted Metabolomics Analysis Revealed the Difference of Component and Geographical Indication Markers of Panax notoginseng in Different Production Areas. Foods. 2023; 12(12):2377. https://doi.org/10.3390/foods12122377
Chicago/Turabian StyleZhang, Shijia, Kexin Fang, Zenan Ding, Jinxia Wu, Jianzhong Lin, Dunming Xu, Jinshui Zhong, Feng Xia, Jianghua Feng, and Guiping Shen. 2023. "Untargeted Metabolomics Analysis Revealed the Difference of Component and Geographical Indication Markers of Panax notoginseng in Different Production Areas" Foods 12, no. 12: 2377. https://doi.org/10.3390/foods12122377
APA StyleZhang, S., Fang, K., Ding, Z., Wu, J., Lin, J., Xu, D., Zhong, J., Xia, F., Feng, J., & Shen, G. (2023). Untargeted Metabolomics Analysis Revealed the Difference of Component and Geographical Indication Markers of Panax notoginseng in Different Production Areas. Foods, 12(12), 2377. https://doi.org/10.3390/foods12122377