Application of ATR-FTIR Incorporated with Multivariate Data Analysis for Discrimination and Quantification of Urea as an Adulterant in UHT Milk
Abstract
1. Introduction
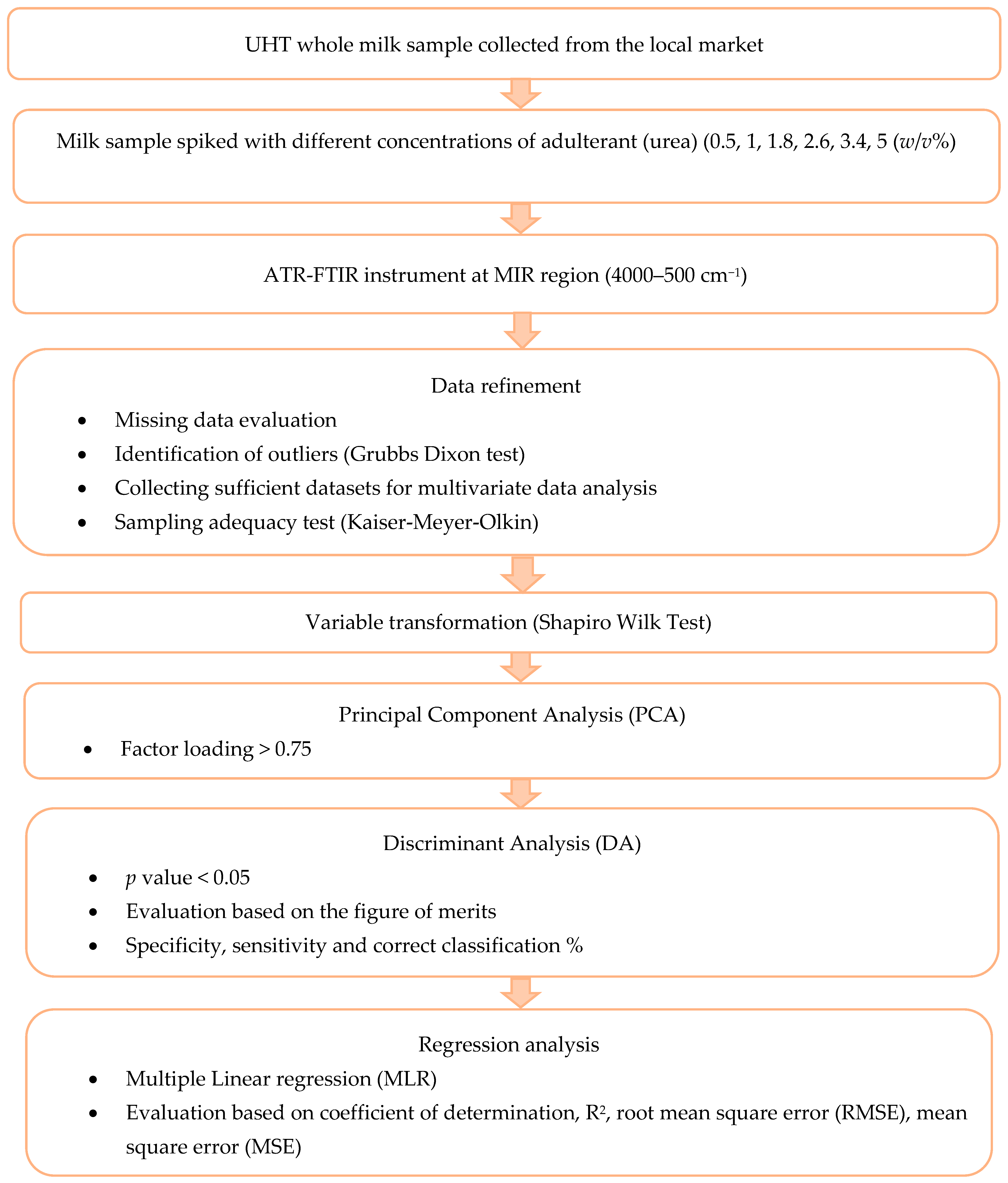
2. Materials and Method
2.1. Experimental Design
2.2. Materials and Sample Preparation
2.3. FTIR Spectral Acquisition
2.4. Dataset Refinement
2.5. Evaluation of Outlier
2.6. Variable Transformation
2.7. Principal Component Analysis (PCA)
2.8. Discriminant Analysis (DA)
2.9. Regression Analysis
3. Results and Discussion
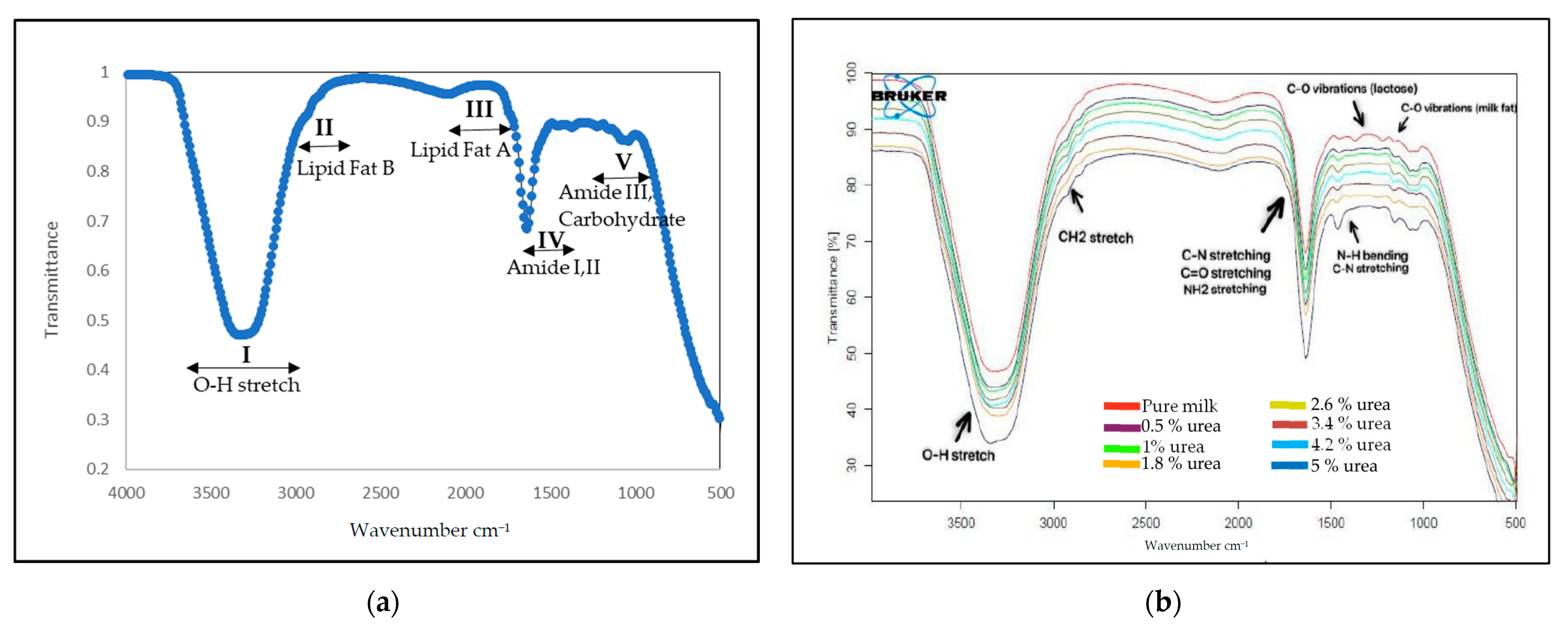
3.1. Identification of Functional Groups of Urea in UHT Milk Based on FTIR Spectra
3.2. Determination of Significant Wavenumbers for Adulterated UHT Milk via Principal Component Analysis (PCA)
3.3. Classification of Adulterated UHT Milk via Discriminant Analysis (DA)
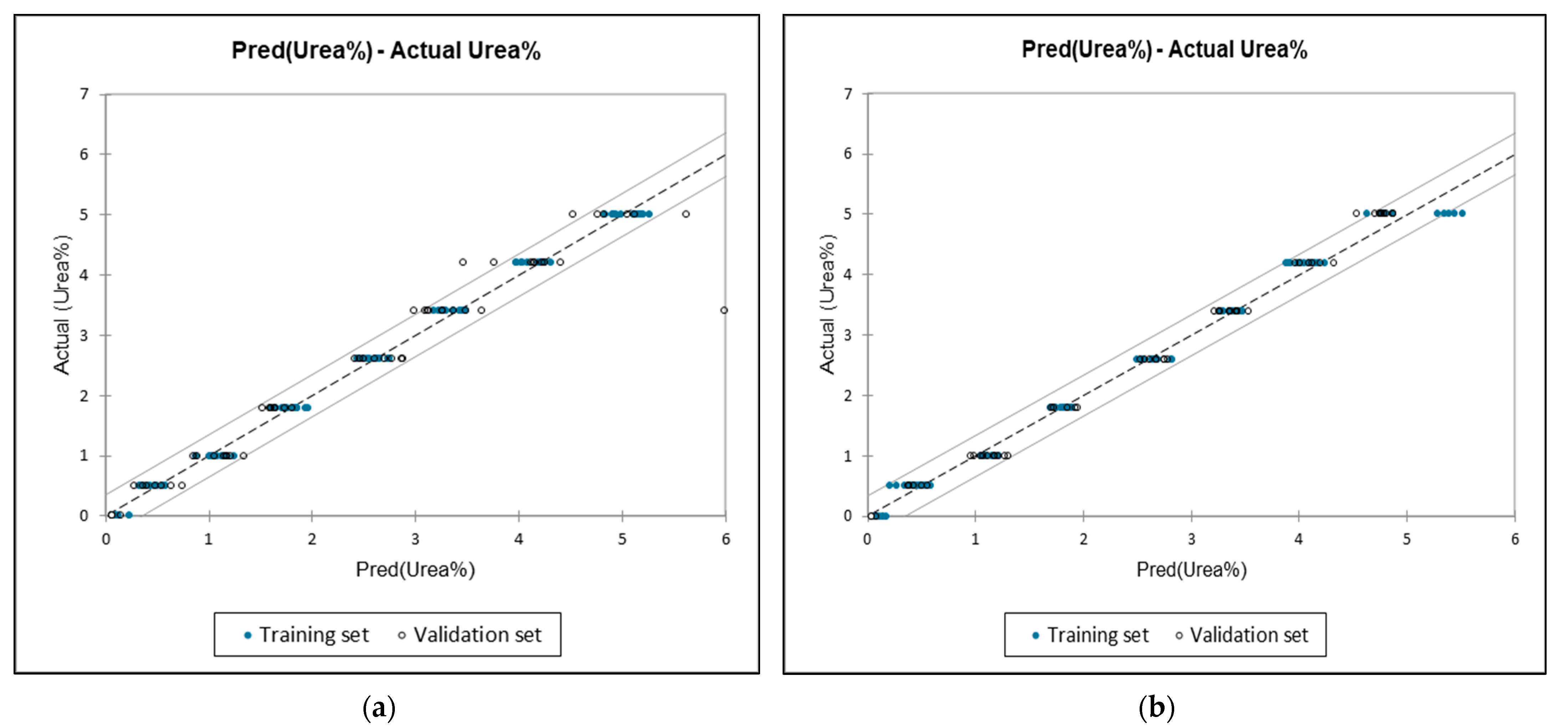
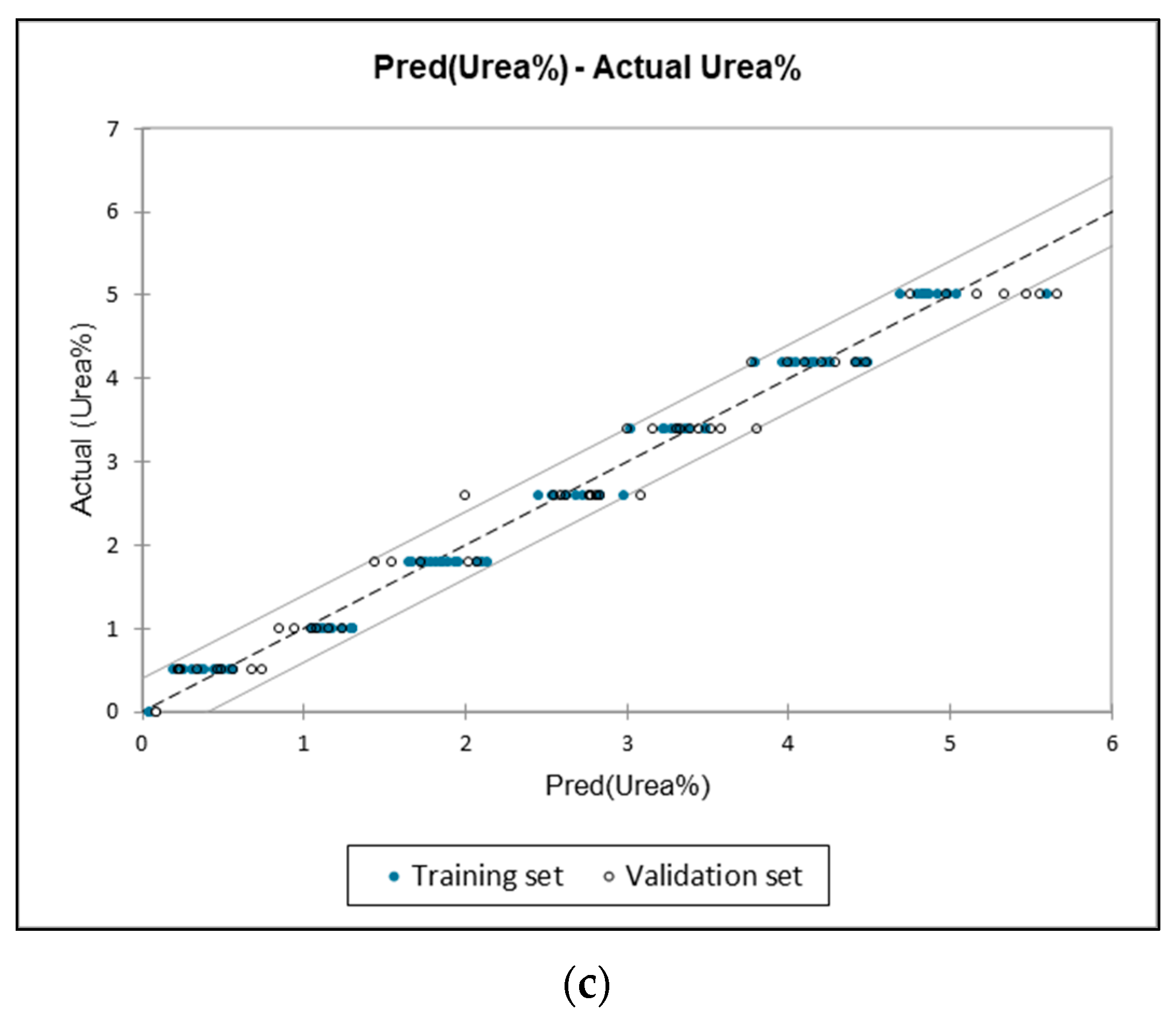
3.4. Quantification of Urea Adulteration in UHT Milk Regression Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atanasova, J.; Ivanova, I. Antibacterial peptides from goat and sheep milk proteins. Biotechnol. Biotechnol. Equip. 2010, 24, 1799–1803. [Google Scholar] [CrossRef]
- Developing a Dairy Industry State By 2025. Available online: https://www.businesstoday.com.my/2021/10/23/developing-a-dairy-industry-state-by-2025/ (accessed on 15 May 2023).
- Holland, J.W.; Gupta, R.; Deeth, H.C.; Alewood, P.F. Proteomic analysis of temperature-dependent changes in stored UHT milk. J. Agric. Food Chem. 2011, 59, 1837–1846. [Google Scholar] [CrossRef]
- Godden, S.M.; Lissemore, K.D.; Kelton, D.F.; Leslie, K.E.; Walton, J.S.; Lumsden, J.H. Factors associated with milk urea concentrations in Ontario dairy cows. J. Dairy Sci. 2001, 84, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, M.I.M.; Marouf, H.A.; Bazalou, M.S.; Dawoud, A.S. Determination of urea in COW’S milk sold in damietta GOVERNORATE. J. Egypt Vet. Med. Assoc. 2008, 68, 1–7. [Google Scholar]
- Nagraik, R.; Sharma, A.; Kumar, D.; Chawla, P.; Kumar, A.P. Milk adulterant detection: Conventional and biosensor based approaches: A review. Sens. Bio-Sens. Res. 2021, 33, 100433. [Google Scholar] [CrossRef]
- Sharma, R.; Rajput, Y.S.; Barui, A.K.; Laxmana, N.N. Detection of adulterants in milk-A laboratory manual. Karnal India NDRI Publ. Natl. Dairy Res. Inst. 2012, 4, 104. [Google Scholar]
- Francis, A.; Dhiman, T.; Mounya, K.S. Adulteration of milk: A review. J. Sci. Technol. 2020, 5, 37–41. [Google Scholar] [CrossRef]
- Xie, W.Q.; Yu, K.X.; Gong, Y.X. Rapid and quantitative determination of urea in milk by reaction headspace gas chromatography. Microchem. J. 2019, 147, 838–841. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, Y.; Li, M.; Fang, X.; Li, X.; Li, H.; Xu, B. Determination of urea in milk by liquid chromatography-isotope dilution mass spectrometry. Anal. Lett. 2012, 45, 1557–1565. [Google Scholar] [CrossRef]
- Ahmad, S.A.; Ahmed, M.; Qadir, M.A.; Shafiq, M.I.; Batool, N.; Nosheen, N.; Ahmad, M.; Mahmood, R.K.; Khokhar, Z.U. Quantitation and risk assessment of chemical adulterants in milk using UHPLC coupled to photodiode array and differential refractive index detectors. Food Anal. Methods 2016, 9, 3367–3376. [Google Scholar] [CrossRef]
- Dutta, S.J.; Chakraborty, G.; Chauhan, V.; Singh, L.; Sharanagat, V.S.; Gahlawat, V.K. Development of a predictive model for determination of urea in milk using silver nanoparticles and UV–Vis spectroscopy. LWT 2022, 168, 113893. [Google Scholar] [CrossRef]
- Domingo, E.; Tirelli, A.A.; Nunes, C.A.; Guerreiro, M.C.; Pinto, S.M. Melamine detection in milk using vibrational spectroscopy and chemometrics analysis: A review. Food Res. Int. 2014, 60, 131–139. [Google Scholar] [CrossRef]
- Balan, B.; Dhaulaniya, A.S.; Jamwal, R.; Yadav, A.; Kelly, S.; Cannavan, A.; Singh, D.K. Rapid detection and quantification of sucrose adulteration in cow milk using Attenuated total reflectance-Fourier transform infrared spectroscopy coupled with multivariate analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 240, 118628. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, M.K.; Singuluri, H. Milk Adulteration in Hyderabad, India –A Comparative Study on the Levels of Different Adulterants Present in Milk. 2014. Available online: https://epubs.icar.org.in/index.php/IJDS/article/view/44300 (accessed on 7 May 2023).
- Feng, L.; Zhu, S.; Chen, S.; Bao, Y.; He, Y. Combining Fourier transform mid-infrared spectroscopy with chemometric methods to detect adulterations in milk powder. Sensors 2019, 19, 2934. [Google Scholar] [CrossRef]
- Gorla, G.; Mestres, M.; Boque, R.; Riu, J.; Spanu, D.; Giussani, B. ATR-MIR spectroscopy to predict commercial milk major components: A comparison between a handheld and a benchtop instrument. Chemom. Intell. Lab. Syst. 2020, 200, 103995. [Google Scholar] [CrossRef]
- Kamboj, U.; Kaushal, N.; Jabeen, S. Near Infrared Spectroscopy as an efficient tool for the Qualitative and Quantitative Determination of Sugar Adulteration in Milk. J. Phys. Conf. Ser. 2020, 1531, 012024. [Google Scholar] [CrossRef]
- Kamboj, U.; Kaushal, N.; Mishra, S.; Munjal, N. Application of selective near infrared spectroscopy for qualitative and quantitative prediction of water adulteration in milk. Mater. Today Proc. 2020, 24, 2449–2456. [Google Scholar] [CrossRef]
- Coitinho, T.B.; Cassoli, L.D.; Cerqueira, P.H.R.; da Silva, H.K.; Coitinho, J.B.; Machado, P.F. Adulteration identification in raw milk using Fourier transform infrared spectroscopy. J. Food Sci. Technol. 2017, 54, 2394–2402. [Google Scholar] [CrossRef]
- Grassi, S.; Tarapoulouzi, M.; D’Alessandro, A.; Agriopoulou, S.; Strani, L.; Varzakas, T. How Chemometrics Can Fight Milk Adulteration. Foods 2023, 12, 139. [Google Scholar] [CrossRef] [PubMed]
- Mabood, F.; Ali, L.; Boque, R.; Abbas, G.; Jabeen, F.; Haq, Q.M.I.; Hussain, J.; Hamaed, A.M.; Naureen, Z.; Al-Nabhani, M.; et al. Robust Fourier transformed infrared spectroscopy coupled with multivariate methods for detection and quantification of urea adulteration in fresh milk samples. Food Sci. Nutr. 2020, 8, 5249–5258. [Google Scholar] [CrossRef] [PubMed]
- Amsaraj, R.; Ambade, N.D.; Mutturi, S. Variable selection coupled to PLS2, ANN and SVM for simultaneous detection of multiple adulterants in milk using spectral data. Int. Dairy J. 2021, 123, 105172. [Google Scholar] [CrossRef]
- Conceição, D.G.; Gonçalves, B.H.R.; Hora, F.F.D.; Faleiro, A.S.; Santos, L.S.; Ferrão, S.P. Use of FTIR-ATR spectroscopy combined with multivariate analysis as a screening tool to identify adulterants in raw milk. J. Braz. Chem. Soc. 2019, 30, 780–785. [Google Scholar] [CrossRef]
- Iqbal, R.; Yasmin, I.; Tehseen, S.; Khaliq, A.; Chughtai, M.F.J.; Ahsan, S.; Khan, W.A.; Nadeem, M.; Hleba, L.; Rebezov, M.; et al. Safety assessment of milk and indigenous milk products from different areas of Faisalabad. J. Microbiol. Biotechnol. Food Sci. 2020, 9, 1197–1203. Available online: https://office2.jmbfs.org/index.php/JMBFS/article/view/4516 (accessed on 20 May 2023).
- Jha, S.N.; Jaiswal, P.; Borah, A.; Gautam, A.K.; Srivastava, N. Detection and quantification of urea in milk using attenuated total reflectance-Fourier transform infrared spectroscopy. Food Bioprocess Technol. 2015, 8, 926–933. [Google Scholar] [CrossRef]
- Souza, S.S.; Cruz, A.G.; Walter, E.H.; Faria, J.A.; Celeghini, R.M.; Ferreira, M.M.; Granato, D.; Sant’Ana, A.D.S. Monitoring the authenticity of Brazilian UHT milk: A chemometric approach. Food Chem. 2011, 124, 692–695. [Google Scholar] [CrossRef]
- Jeyaletchumi, P.; Christina, R.; Norlida, B.T.; Chang, S.H.; Nurhaida, A.M.; Chennie, S.W. Safety of UHT Milk Along Supply Chain in Sabah, Malaysia. Available online: https://apcph.cphm.my/wp-content/uploads/2022/07/APCPH2022-O-47.pdf (accessed on 2 June 2023).
- Troise, A.D.; Dathan, N.A.; Fiore, A.; Roviello, G.; Di Fiore, A.; Caira, S.; Cuollo, M.; De Simone, G.; Fogliano, V.; Monti, S.M. Faox enzymes inhibited Maillard reaction development during storage both in protein glucose model system and low lactose UHT milk. Amino Acids 2014, 46, 279–288. [Google Scholar] [CrossRef]
- Grewal, M.K.; Chandrapala, J.; Donkor, O.; Apostolopoulos, V.; Stojanovska, L.; Vasiljevic, T. Fourier transform infrared spectroscopy analysis of physicochemical changes in UHT milk during accelerated storage. Int. Dairy J. 2017, 66, 99–107. [Google Scholar] [CrossRef]
- Azad, T.; Ahmed, S. Common milk adulteration and their detection techniques. Int. J. Food Contam. 2016, 3, 22. [Google Scholar] [CrossRef]
- Pramono, T.B.; Islamy, R.A.; Putra, J.J.; Suparyanto, T.; Purwandari, K.; Cenggoro, T.W.; Pardamean, B. A Model of Visual Intelligent System for Genus Identification of Fish in the Siluriformes Order. IOP Conf. Ser. Earth Environ. Sci. 2021, 794, 012114. [Google Scholar] [CrossRef]
- Yang, Y.; Shih, F.Y.; Roshan, U. Defense against adversarial attacks based on stochastic descent sign activation networks on medical images. Int. J. Pattern Recognit. Artif. Intell. 2022, 36, 2254005. [Google Scholar] [CrossRef]
- Andrada, M.F.; Vega-Hissi, E.G.; Estrada, M.R.; Martinez, J.C.G. Application of k-means clustering, linear discriminant analysis and multivariate linear regression for the development of a predictive QSAR model on 5-lipoxygenase inhibitors. Chemom. Intell. Lab. Syst. 2015, 143, 122–129. [Google Scholar] [CrossRef]
- Barbato, G.; Barini, E.M.; Genta, G.; Levi, R. Features and performance of some outlier detection methods. J. Appl. Stat. 2011, 38, 2133–2149. [Google Scholar] [CrossRef]
- Ismail, A.M.; Sani, M.S.A.; Azid, A.; Zaki, N.N.M.; Arshad, S.; Tukiran, N.A.; Abidin, S.A.S.Z.; Samsudin, M.S.; Ismail, A. Food forensics on gelatine source via ultra-high-performance liquid chromatography diode-array detector and principal component analysis. SN Appl. Sci. 2021, 3, 79. [Google Scholar] [CrossRef]
- Hosseini, E.; Ghasemi, J.B.; Daraei, B.; Asadi, G.; Adib, N. Near-infrared spectroscopy and machine learning-based classification and calibration methods in detection and measurement of anionic surfactant in milk. J. Food Compos. Anal. 2021, 104, 104170. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]
- Idris, M.H.H.; Sani, M.S.A.; Hashim, A.M.; Zaki, N.N.M.; Manaf, Y.N.A.; Desa, M.N.M.; Arshad, S.; Yuswan, M.H.; Hassan, M.S.; Yusof, Y.A.; et al. Forensic Feed Strategy: Incorporation of Multivariate and Instrumental Analyses for Authentication of Fish Feed Sources. J. Halal Ind. Serv. 2022, 5. [Google Scholar] [CrossRef]
- Idris, M.H.H.; Manaf, Y.N.; Desa, M.N.M.; Hashim, A.M.; Sani, M.S.A.; Zaki, N.N.M.; Yuswan, M.H.; Kamaruddin, M.S.; Yusof, Y.A.; Mustafa, S.; et al. A conjunction of sn-2 fatty acids and overall fatty acid composition combined with chemometric techniques increase the effectiveness of lard detection in fish feed. Chemom. Intell. Lab. Syst. 2021, 213, 104308. [Google Scholar] [CrossRef]
- Currell, G. Scientific Data Analysis; Oxford University Press: New York, NY, USA, 2015. [Google Scholar]
- Valand, R.; Tanna, S.; Lawson, G.; Bengtström, L. A review of Fourier Transform Infrared (FTIR) spectroscopy used in food adulteration and authenticity investigations. Food Addit. Contam. Part A 2020, 37, 19–38. [Google Scholar] [CrossRef]
- Ami, D.; Mereghetti, P.; Doglia, S.M. Multivariate analysis for Fourier transform infrared spectra of complex biological systems and processes. In Multivariate Analysis in Management, Engineering and the Sciences; Intechopen: London, UK, 2013; pp. 189–220. [Google Scholar] [CrossRef]
- De Souza Gondim, C.; Junqueira, R.G.; de Souza, S.V.C.; Ruisánchez, I.; Callao, M.P. Detection of several common adulterants in raw milk by MID-infrared spectroscopy and one-class and multi-class multivariate strategies. Food Chem. 2017, 230, 68–75. [Google Scholar] [CrossRef]
- Moghaddam, H.N.; Tamiji, Z.; Lakeh, M.A.; Khoshayand, M.R.; Mahmoodi, M.H. Multivariate analysis of food fraud: A review of NIR based instruments in tandem with chemometrics. J. Food Compos. Anal. 2022, 107, 104343. [Google Scholar] [CrossRef]
- Jaiswal, P.; Jha, S.N.; Kaur, J.; Borah, A. Detection and quantification of anionic detergent (lissapol) in milk using attenuated total reflectance-Fourier Transform Infrared spectroscopy. Food Chem. 2017, 221, 815–821. [Google Scholar] [CrossRef]
- Karoui, R.; Mazerolles, G.; Dufour, É. Spectroscopic techniques coupled with chemometric tools for structure and texture determinations in dairy products. Int. Dairy J. 2003, 13, 607–620. [Google Scholar] [CrossRef]
- Kher, A.; Udabage, P.; McKinnon, I.; McNaughton, D.; Augustin, M.A. FTIR investigation of spray-dried milk protein concentrate powders. Vib. Spectrosc. 2007, 44, 375–381. [Google Scholar] [CrossRef]
- Gal’perin, V.A.; Finkel’shtein, A.I. The nature of the absorption bands of urea in the range 1700–1600 cm−1. J. Appl. Spectrosc. 1968, 9, 1351–1353. [Google Scholar] [CrossRef]
- Yang, R.J.; Liu, R.; Xu, K.X. Adulteration detection of urea in milk by mid-infrared spectroscopy. Spectrosc. Spectr. Anal. 2011, 31, 2383–2385. [Google Scholar] [CrossRef]
- Sjahfirdi, L.; Nasikin, M. Protein Identification Using Fourier Transform Infrared (FTIR). 2012. Available online: www.arpapress.com/Volumes/Vol10Issue3/IJRRAS_10_3_06.pdf418 (accessed on 7 May 2023).
- Basak, M.; Uddin, M.N.; Jahan, R.A.; Rana, A.A.; Karim, M.M. Chemometric model for rapid detection of urea and hydrogen peroxide in milk. Bangladesh J. Sci. Ind. Res. 2021, 56, 1–8. [Google Scholar] [CrossRef]
- Granato, D.; Putnik, P.; Kovačević, D.B.; Santos, J.S.; Calado, V.; Rocha, R.S.; Cruz, A.G.D.; Jarvis, B.; Rodionova, O.Y.; Pomerantse. A Trends in chemometrics: Food authentication, microbiology, and effects of processing. Compr. Rev. Food Sci. Food Saf. 2018, 17, 663–677. [Google Scholar] [CrossRef]
- Falcó, I.; Randazzo, W.; Sánchez, G.; López-Rubio, A.; Fabra, M.J. On the use of carrageenan matrices for the development of antiviral edible coatings of interest in berries. Food Hydrocoll. 2019, 92, 74–85. [Google Scholar] [CrossRef]
- Roussel, S.; Preys, S.; Chauchard, F.; Lallemand, J. Multivariate data analysis (chemometrics). In Process Analytical Technology for the Food Industry; Springer: New York, NY, USA, 2014; pp. 7–59. [Google Scholar] [CrossRef]
- Retnam, A.; Zakaria, M.P.; Juahir, H.; Aris, A.Z.; Zali, M.A.; Kasim, M.F. Chemometric techniques in distribution, characterisation and source apportionment of polycyclic aromatic hydrocarbons (PAHS) in aquaculture sediments in Malaysia. Mar. Pollut. Bull. 2013, 69, 55–66. [Google Scholar] [CrossRef]
- Mohammadi, M.; Mianabadi, F.; Mehrad-Majd, H. Circulating visfatin levels and cancers risk: A systematic review and meta-analysis. J. Cell Physiol. 2019, 234, 5011–5022. [Google Scholar] [CrossRef]
- Data, M.C.; Salgado, C.M.; Azevedo, C.; Proença, H.; Vieira, S.M. Noise versus outliers. In Secondary Analysis of Electronic Health Records; Springer: Berlin/Heidelberg, Germany, 2016; pp. 163–183. [Google Scholar] [CrossRef]
- Perez, L.V. Principal Component Analysis to Address Multicollinearity; Whitman College: Walla Walla, WA, USA, 2017. [Google Scholar]
- Georgouli, K.; Del Rincon, J.M.; Koidis, A. Continuous statistical modelling for rapid detection of adulteration of extra virgin olive oil using mid infrared and Raman spectroscopic data. Food Chem. 2017, 217, 735–742. [Google Scholar] [CrossRef]
- Vasconcelos, M.; Coelho, L.; Barros, A.; de Almeida, J.M.M.M. Study of adulteration of extra virgin olive oil with peanut oil using FTIR spectroscopy and chemometrics. Cogent Food Agric. 2015, 1, 1018695. [Google Scholar] [CrossRef]
- Hussain, M.N.; Khir, M.F.A.; Hisham, M.H.; Yusof, Z.M. Feasibility study of detecting canola oil adulteration with palm oil using NIR spectroscopy and multivariate analysis. In Proceedings of the International Conference on Information, Communication Technology and System (ICTS) 2014, Surabaya, Indonesia, 24 September 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 111–114. [Google Scholar] [CrossRef]
- Aziz, A.A.; Abdullah, S.; Zakaria, Z.; Abu Bakar, N.K. Discrimination and authentication of lard blending with palm oil in cosmetic soap formulations. Int. J. Cosmet. Sci. 2023. early view. [Google Scholar] [CrossRef] [PubMed]
- Tharwat, A.; Gaber, T.; Ibrahim, A.; Hassanien, A.E. Linear discriminant analysis: A detailed tutorial. AI Commun. 2017, 30, 169–190. [Google Scholar] [CrossRef]
- Santos, P.M.; Pereira-Filho, E.R.; Rodriguez-Saona, L.E. Rapid detection and quantification of milk adulteration using infrared microspectroscopy and chemometrics analysis. Food Chem. 2013, 138, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Kishor, K.; Thakur, R. Analysis of milk adulteration using mid-IR spectroscopy. Int. J. Recent Innov. Trends Comput. Commun. 2015, 3, 5890–5895. Available online: https://www.academia.edu/download/53804282/1445932370_27-10-2015.pdf (accessed on 7 May 2023).
- Upadhyay, N.; Jaiswal, P.; Jha, S.N. Application of attenuated total reflectance Fourier Transform Infrared spectroscopy (ATR–FTIR) in MIR range coupled with chemometrics for detection of pig body fat in pure ghee (heat clarified milk fat). J. Mol. Struct. 2018, 1153, 275–281. [Google Scholar] [CrossRef]

| Sample Concentration | Number of Samples | Training Dataset | Testing Dataset |
|---|---|---|---|
| Control (unadulterated milk) | 25 | 20 | 5 |
| Milk with urea (0.5–5 (w/v %)) | 175 | 140 | 35 |
| Dataset | Sensitivity and Specificity % | Number of UHT Milk Samples and p-Values of Fisher Distance | Total UHT Milk Sample | |
|---|---|---|---|---|
| Pure Milk | Milk + Urea | |||
| Training dataset | Full spectra | |||
| Pure Milk | 100 | 11 (1) | 0 (<0.0001) | 11 |
| Milk + Urea | 100 | 0 (<0.0001) | 79 (1) | 79 |
| 1675–1560 cm−1 | ||||
| Pure Milk | 55.56 | 5 (1) | 4 (<0.0001) | 9 |
| Milk + Urea | 100 | 0 (<0.0001) | 81 (1) | 81 |
| 1585–1454 cm−1 | ||||
| Pure milk | 78.57 | 11 (1) | 3 (<0.0001) | 14 |
| Milk + Urea | 100 | 0 (<0.0001) | 76 (1) | 76 |
| Validation dataset | Full spectra | |||
| Pure milk | 100 | 11 (1) | 0 (<0.0001) | 11 |
| Milk + Urea | 98.73 | 1 (<0.0001) | 78 (1) | 79 |
| 1675–1560 cm−1 | ||||
| Pure milk | 44.44 | 4 (<0.0001) | 5 (1) | 9 |
| Milk +Urea | 97.53 | 2 (<0.0001) | 79 (1) | 81 |
| 1585–1454 cm−1 | ||||
| Pure milk | 78.57 | 11(1) | 3 (<0.0001) | 14 |
| Milk + Urea | 98.68 | 1 (<0.0001) | 75 (1) | 76 |
| Testing dataset | Full Spectra | |||
| Pure milk | 100 | 5 (1) | 0 (<0.0001) | 5 |
| Milk + Urea | 100 | 0 (<0.0001) | 35 (1) | 35 |
| 1675–1560 cm−1 | ||||
| Pure milk | 40 | 2 (<0.0001) | 3 (1) | 5 |
| Milk + Urea | 91.43 | 3 (<0.0001) | 32 (1) | 35 |
| 1585–1454 cm−1 | ||||
| Pure milk | 80 | 4 (1) | 1 (<0.0001) | 5 |
| Milk + urea | 94.29 | 2 (<0.0001) | 33 (1) | 35 |
| Wavenumber Range (cm−1) | Calibration | Validation | Most Significant Wavenumber | ||||
|---|---|---|---|---|---|---|---|
| R2 | RMSEC | MSE | R2 | RMSE | MSE | ||
| Full spectra | 0.996 | 0.171 | 0.029 | 0.940 | 0.941 | 0.886 | 1626.63 cm−1 |
| 1675–1560 | 0.991 | 0.170 | 0.029 | 0.993 | 0.168 | 0.028 | 1601.98 cm−1 |
| 1584–1453 | 0.988 | 0.203 | 0.041 | 0.981 | 0.303 | 0.092 | 1585.5534 cm−1 |
| Wavenumber cm−1 | Actual Urea Adulteration (%) | Determined Urea Concentration (%) ± SD | 95% Lower and Upper Bounds of Determined Urea Adulteration | t-Test Value |
|---|---|---|---|---|
| Full spectra | 0 | 0.027 ± 0.287 | −0.324–0.377 | 0.857 |
| 0.5 | 0.272 ± 0.063 | −0.066–0.610 | <0.0001 | |
| 1 | 1.038 ± 0.103 | 0.734–1.342 | 0.483 | |
| 1.8 | 1.686 ± 0.189 | 1.365–2.006 | 0.261 | |
| 2.6 | 2.499 ± 0.208 | 2.168–2.931 | 0.358 | |
| 3.4 | 3.299 ± 0.287 | 2.994–3.604 | 0.503 | |
| 4.2 | 4.199 ± 0.571 | 3.694–4.704 | 0.997 | |
| 5 | 5.691 ± 0.112 | 5.255–6.127 | <0.0001 | |
| 1675–1560 | 0 | −0.249 ± 0.100 | −0.269–(−0.146) | <0.0001 |
| 0.5 | 0.263 ± 0.024 | 0.151–0.375 | <0.0001 | |
| 1 | 1.065 ± 0.051 | 0.972–1.159 | 0.002 | |
| 1.8 | 1.842 ± 0.060 | 1.735–1.949 | 0.158 | |
| 2.6 | 2.498 ± 0.190 | 2.387–2.610 | 0.268 | |
| 3.4 | 3.201 ± 0.306 | 3.061–3.341 | 0.183 | |
| 4.2 | 4.118 ± 0.434 | 3.928–4.309 | 0.686 | |
| 5 | 5.865 ± 0.247 | 5.722–6.007 | <0.0001 | |
| 1585–1485 | 0 | −0.401 ± 0.104 | −0.573–(−0.229) | <0.0001 |
| 0.5 | 0.071 ± 0.214 | −0.145–0.286 | 0.002 | |
| 1 | 1.075 ± 0.116 | 0.918–1.231 | 0.190 | |
| 1.8 | 1.815 ± 0.186 | 1.612–2.018 | 0.860 | |
| 2.6 | 2.701 ± 0.191 | 2.541–2.861 | 0.271 | |
| 3.4 | 3.225 ± 0.569 | 3.013–3.436 | 0.510 | |
| 4.2 | 4.306 ± 0.394 | 3.967–4.645 | 0.565 | |
| 5 | 5.962 ± 0.513 | 5.747–6.176 | 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, E.; Binti Julmohammad, N.; Koh, W.Y.; Abdullah Sani, M.S.; Rasti, B. Application of ATR-FTIR Incorporated with Multivariate Data Analysis for Discrimination and Quantification of Urea as an Adulterant in UHT Milk. Foods 2023, 12, 2855. https://doi.org/10.3390/foods12152855
Tan E, Binti Julmohammad N, Koh WY, Abdullah Sani MS, Rasti B. Application of ATR-FTIR Incorporated with Multivariate Data Analysis for Discrimination and Quantification of Urea as an Adulterant in UHT Milk. Foods. 2023; 12(15):2855. https://doi.org/10.3390/foods12152855
Chicago/Turabian StyleTan, Emeline, Norliza Binti Julmohammad, Wee Yin Koh, Muhamad Shirwan Abdullah Sani, and Babak Rasti. 2023. "Application of ATR-FTIR Incorporated with Multivariate Data Analysis for Discrimination and Quantification of Urea as an Adulterant in UHT Milk" Foods 12, no. 15: 2855. https://doi.org/10.3390/foods12152855
APA StyleTan, E., Binti Julmohammad, N., Koh, W. Y., Abdullah Sani, M. S., & Rasti, B. (2023). Application of ATR-FTIR Incorporated with Multivariate Data Analysis for Discrimination and Quantification of Urea as an Adulterant in UHT Milk. Foods, 12(15), 2855. https://doi.org/10.3390/foods12152855







