Progress in the Conversion of Ginsenoside Rb1 into Minor Ginsenosides Using β-Glucosidases
Abstract
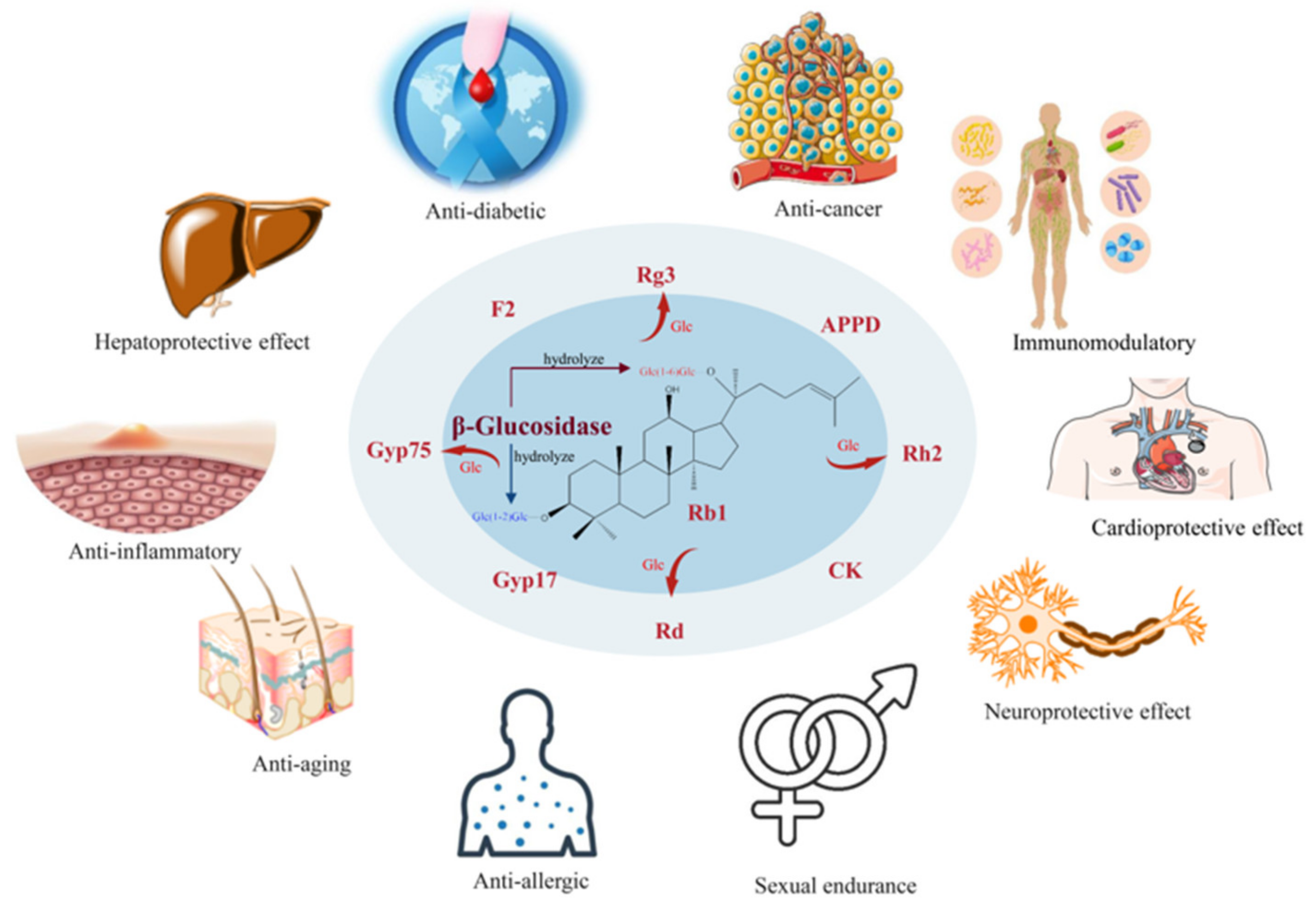
1. Introduction
2. Chemical Structure of PPD-Type Ginsenosides
3. Classification of Ginsenosidases
4. Enzymatic Conversion Pathway of Ginsenoside Rb1
5. Sources and Enzymatic Properties of β-Glucosidases That Convert Ginsenoside Rb1
5.1. Conversion of Ginsenoside Rb1 by Bacterial β-Glucosidases
5.2. Conversion of Ginsenoside Rb1 by Fungal β-Glucosidases
5.3. Conversion of Ginsenoside Rb1 by β-Glucosidase from Other Sources
| Taxonomy | GH No. | Organism | Designation | Molecular Mass (kDa) | Concentration of Rb1 (g/L) | Reaction Conditions | Reaction Time (h) | Hydrolysis Pattern and Products | Productivity (g/L/h) | Molar Conversion | Km (mM) | Vmax (U/mg) | GenBank Accession No. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | GH1 | Arthrobacter chlorophenolicus [51] | BglAch | 45.8 | 1.0 | pH 6.0 37 °C | 12.0 | C20: Rd, F2 | NR | NR | 3.19 | 20.1 | ACL38420.1 |
| GH1 | Bifidobacterium adolescentis ATCC15703 [52] | BaBgl1A | 44.2 | 5.0 | pH 7.0 37 °C | 2 | C3: Gyp17 | NR | NR | NR | NR | BAF40068.1 | |
| GH1 | Caldicellulosiruptor bescii [53] | NR | 53.0 | 1.1 | pH 5.5 80 °C | 1.0 | C20: CK | 1.1 | 100% | NR | NR | ACM59590.1 | |
| GH1 | Paenibacillus mucilaginosus [54] | BglPm | 47.7 | 1.0 | pH 7.5 37 °C | NR | C3: F2 | NR | NR | 3.24 | 10.2 | AEI42200.1 | |
| GH1 | Paenibacillus polymyxa [55] | bglB | 52.0 | 1.0 | pH6.5 40 °C | 72 | C20: Rd, F2, CK | NR | NR | 0.743 | 31400 | AAA22264.1 | |
| GH1 | Pyrococcus furiosus [56] | NR | 55.5 | 1.0 | pH 5 95 °C | 3 | C20: CK | 0.177 | 94.4% | NR | NR | AAC25555.1 | |
| GH1 | Sphingomonas sp. 2F2 [57] | BglSp | 49.4 | 1.0 | pH 7.0 37 °C | 0.5 | C3: Gyp17 | NR | NR | 2.90 | 515.0 | ADY18331.1 | |
| GH1 | Sphingopyxis alaskensis [37] | NR | 51.0 | 8.0 | pH 5.5 40 °C | 1.0 | C3: Gyp17 | 6.8 | 100% | NR | NR | ABF52736.1 | |
| GH1 | Thermus caldophilus [58] | NR | NR | 1 | pH 5.0 75 °C | 0.5 | C20: Rd | 1.60 | 93.7% | NR | NR | AAO15361.1 | |
| GH1 | T.petrophlia [34] | Tpebgl1 | 51.5 | 30.0 | pH 6.0 90 °C | 0.8 | C20: Rd | NR | 97.5% | 0.28 | 470.2 | ABQ46970.1 | |
| GH1 | T.thermarum DSM 5069T [35] | Tt-BGL | 55.0 | 36.0 | pH 4.8 90 °C | 1.0 | C20: Rd | NR | 97% | 0.59 | 142.0 | NR | |
| GH1 | Thermus thermophilus [59] | NR | NR | 1.1 | pH 6.5 90 °C | 36 | C20: Rd | NR | NR | NR | NR | WP_011229206.1 | |
| GH3 | A. mirum [28] | BglAm | 65.3 | 1.0 | pH 8.0 30 ℃ | 95.0 | C3: APPD | NR | NR | 0.33 | 5.9 | WP_015801787.1 | |
| GH3 | Bifidobacterium longum H-1 [60] | BglX | 95.0 | 1.1 | pH 7.2 37 °C | 6.0 | C20: Rd | NR | NR | 0.83 | 56.5 | NR | |
| GH3 | B. longum KACC 91,563 [61] | BlBG3 | NR | 2.0 | pH 6.0 37 °C | 0.25 | C20: Rd | NR | NR | 2.38 | NR | NR | |
| GH3 | Dictyoglomus turgidum [62] | NR | NR | 1.1 | pH 5.5 80 °C | 6.0 | C20: APPD | NR | 20% | NR | NR | WP_012582633.1 | |
| GH3 | Flavobacterium johnsoniae [63] | BglF3 | 81.8 | 1.0 | pH 6.0 37 °C | 1.50 | C20: Rd | NR | NR | 0.91 | 5.75 | ABQ03809.1 | |
| GH3 | F. johnsoniae UW101T [64] | BglBX10 | 89.3 | 1.0 | pH 6.0 37 °C | NR | C20: Rg3 | NR | NR | NR | NR | ABQ06406.1 | |
| GH3 | Gordonia terrae [38] | NR | 78.0 | 4.0 | pH 6.5 30 °C | 2.5 | C20: Rg3 | 1.13 | 100% | 14.66 | NR | GAB42172.1 | |
| GH3 | Lactobacillus brevis [65] | Bgy2 | 123.0 | 1.0 | pH 6.0 30 °C | 10.0 | C20: Rd | NR | 69% | NR | NR | BAN05876 | |
| GH3 | M. esteraromaticum [66] | Bgp1 | 87.5 | 1.0 | pH7.0 37 °C | 6.0 | C20: Rg3 | 0.074 | 71% | NR | NR | AEX88466.1 | |
| GH3 | M. esteraromaticum [24] | Bgp3 | 80.0 | 1.0 | pH 7.0 40 °C | 1.0 | C20: CK | 0.46 | 77% | NR | NR | AEX88467.1 | |
| GH3 | Microbacterium sp. Gsoil 167 [30] | BglG167b | 90.3 | 2.0 | pH 7.0 37 °C | 24.0 | C20: Rg3, APPD | NR | NR | NR | NR | AGA60132.1 | |
| GH3 | Microbacterium testaceum [29] | MT619 | 68.3 | 2.0 | pH 7.0 37 °C | NR | C3: CK, PPD | NR | NR | NR | NR | WP_013585536.1 | |
| GH3 | Mucilaginibacter sp. [67] | BglQM | 85.6 | 1.0 | pH 7.0 25 °C | NR | C20: Rd, Rg3 | NR | NR | 0.037 | 33.4 | AFS34656.1 | |
| GH3 | Pseudonocardia sp. [68] | BglPC28 | 79.0 | 2.0 | pH 7.0 37 °C | NR | C20: Rg3 | NR | NR | 6.36 | 40.0 | AGA60134.1 | |
| GH3 | Sanguibacter keddieii [69] | BglSk | NR | 1.0 | pH 8.0 25 °C | NR | C3: Gyp75, CK | NR | NR | 0.46 | 30.2 | ACZ20402.1 | |
| GH3 | T. ginsenosidimutans [70] | BgpA | 70.1 | 1.0 | pH 6.0 37 °C | NR | C3: CK | NR | NR | 4.20 | 100.6 | ACZ66247.3 | |
| GH3 | T.petrophila [36] | Tpebgl3 | 81.0 | 10.0 | pH 5.0 90 °C | 1.5 | C20: Rg3 | 4.62 | 97.9% | 1.60 | 109.0 | ABQ46916.1 | |
| Fungi | NR | A.mellea [46] | NR | NR | 1.0 | pH 4.8 37 °C | 96.0 | C20: CK | NR | NR | NR | NR | NR |
| NR | Arthrinium sp. [71] | NR | NR | 1.0 | 30 °C | 48.0 | C20: CK | NR | NR | NR | NR | NR | |
| NR | A.niger KCCM [72] | NR | NR | 1.1 | pH 5.5 30 °C | 24.0 | C20: Rd, Rg3 | NR | NR | NR | NR | NR | |
| NR | A. niger KCCM 11,239 [73] | NR | 123.0 | 0.1 | pH 4.0 50 °C | 120.0 | C20: Rd, Rg3, F2 | NR | NR | NR | NR | NR | |
| NR | A. niger XD101 [39] | NR | 110 | 6.0 | pH 4.5 50 °C | 72.0 | C20: CK | NR | 94.4% | NR | NR | NR | |
| NR | C. cladosporioides [12] | NR | NR | 0.4 | 30 °C | 168.0 | C20: CK | NR | 74.2% | NR | NR | NR | |
| NR | Esteya vermicola [74] | NR | NR | 2.0 | pH 5.0 50 °C | 30.0 | C3: Gyp75 | NR | 95.4% | NR | NR | NR | |
| NR | Fomitella fraxinea [75] | NR | NR | NR | pH 4.5 45 °C | 48.0 | C20: CK | NR | NR | NR | NR | NR | |
| GH3 | Penicillium purpurogenum [76] | NR | 110.5 | NR | pH 4.5 70 °C | NR | C3: CK | NR | NR | NR | NR | ACV87737.1 | |
| Animal | NR | A.fulica [77] | NR | 230.0 | 8.9 | pH 5.6 50 °C | 24 | C3: Rd | NR | NR | 0.338 | 0.25 | NR |
| NR | A. fulica [50] | G II | 220.0 | 4.4 | pH 5.0 50 °C | 24 | C20: Rd, F2, CK | NR | NR | 0.224 | 0.203 | NR |
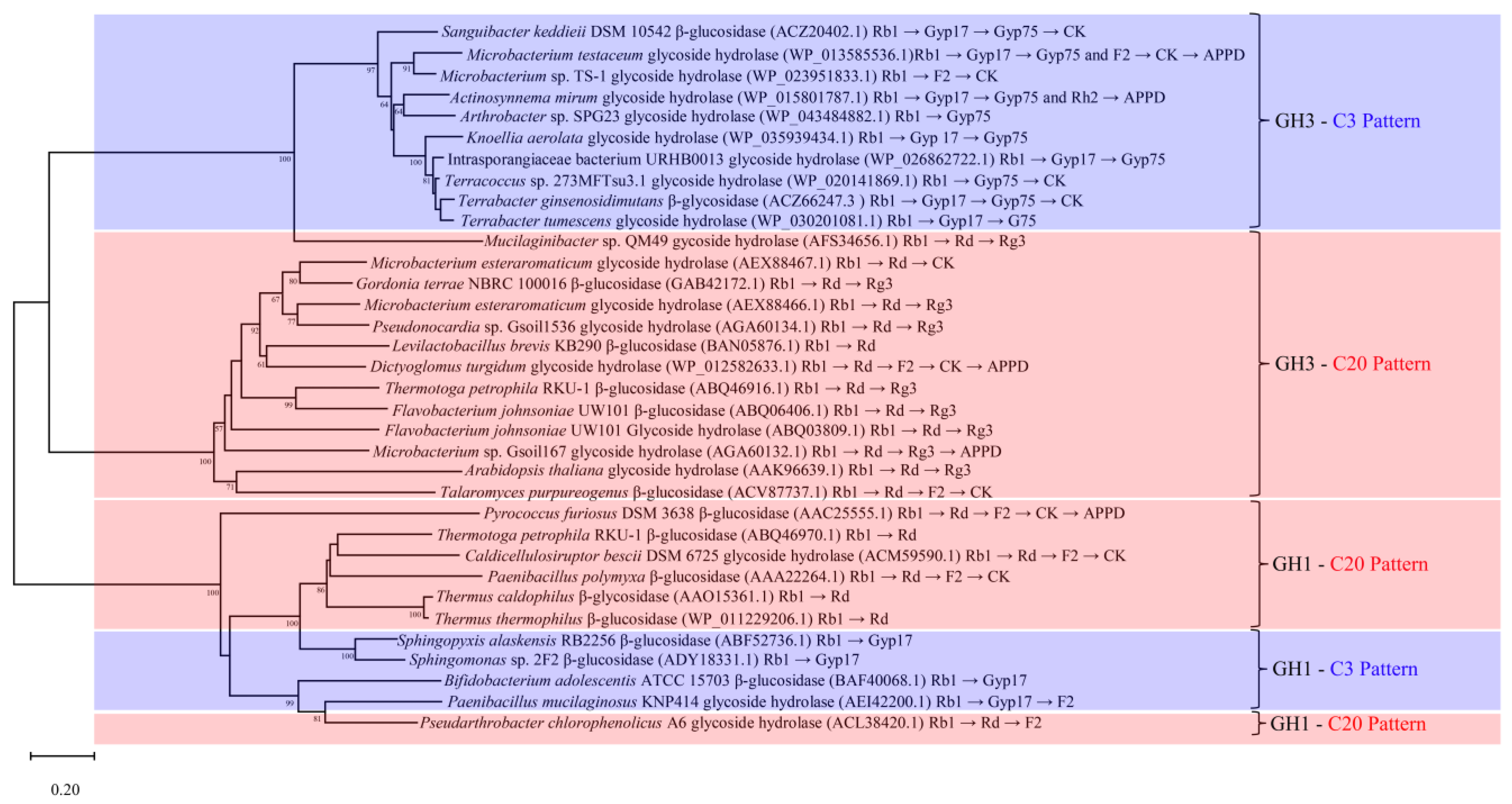
6. Phylogenetic Analysis of β-Glucosidases That Convert Ginsenoside Rb1
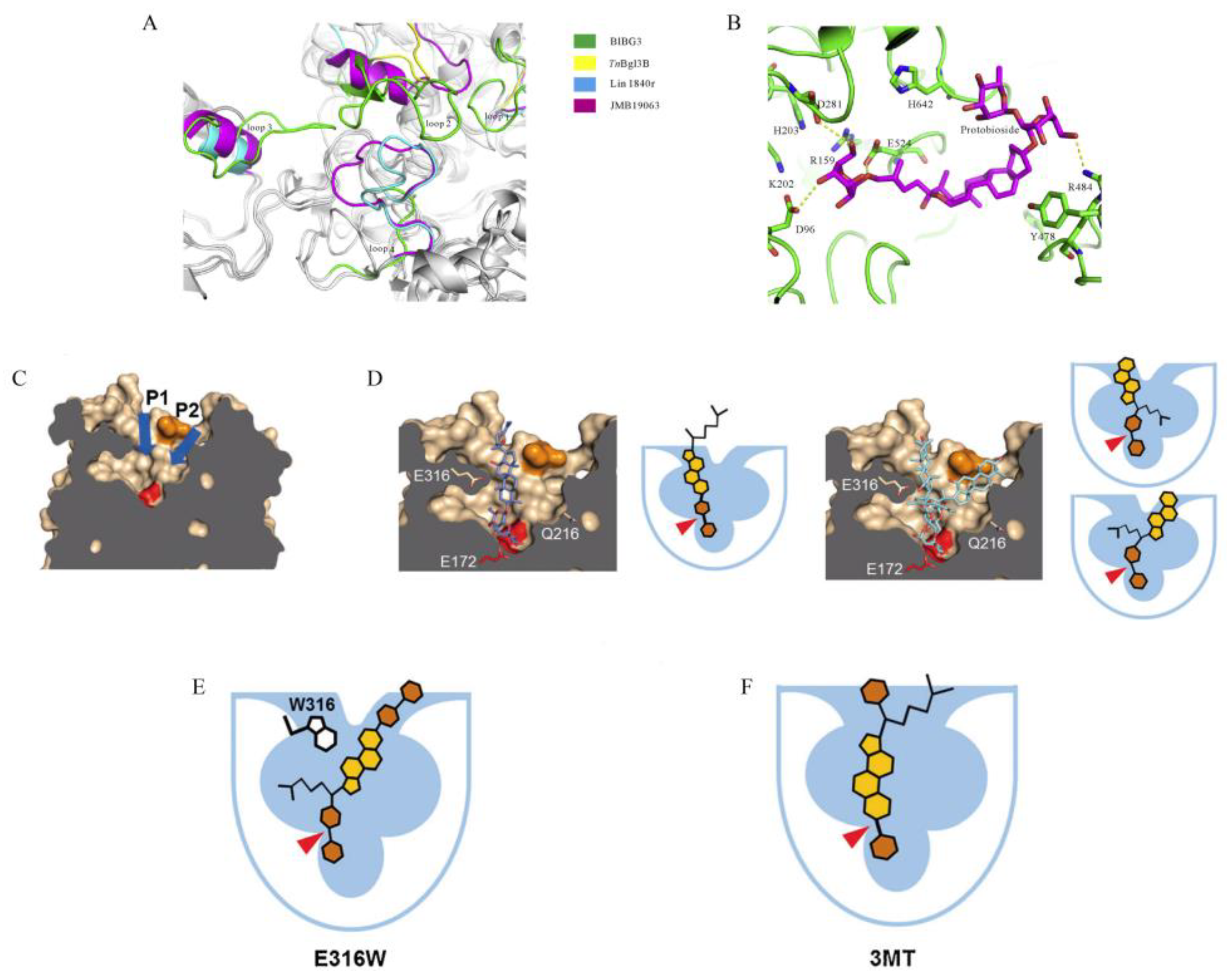
7. The Molecular Mechanism of β-Glucosidases That Convert Ginsenoside Rb1
8. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chu, L.L.; Bae, H. Bacterial endophytes from ginseng and their biotechnological application. J. Ginseng Res. 2022, 46, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.B.; Lu, X.; Hu, Y.; Fan, X.H. Chemical constituents of Panax ginseng and Panax notoginseng explain why they differ in therapeutic efficacy. Pharmacol. Res. 2020, 161, 105263. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.; Wang, R.; Zhao, S.; Wang, Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B 2021, 11, 1813–1834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Tong, Y.L.; Lu, X.; Dong, F.M.; Ma, X.X.; Yin, S.Y.; He, Y.; Liu, Y.H.; Liu, Q.C.; Fan, D.D. A derivant of ginsenoside CK and its inhibitory effect on hepatocellular carcinoma. Life Sci. 2022, 304, 120698. [Google Scholar] [CrossRef]
- Zhou, P.; Xie, W.; He, S.; Sun, Y.; Meng, X.; Sun, G.; Sun, X. Ginsenoside Rb1 as an anti-diabetic agent and its underlying mechanism analysis. Cells 2019, 8, 204. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, L.; Zhang, Y.; Qi, W.X.; Wang, Z.Y.; Tian, L.; Zhao, D.Q.; Wu, Q.B.; Li, X.Y.; Wang, T. Pharmacological properties, molecular mechanisms and therapeutic potential of ginsenoside Rg3 as an antioxidant and anti-inflammatory agent. Front. Pharmacol. 2022, 13, 975784. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Liu, Q.P.; An, P.; Jia, M.; Luan, X.; Tang, J.Y.; Zhang, H. Ginsenoside Rd: A promising natural neuroprotective agent. Phytomedicine 2022, 95, 153883. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhu, L.; Wang, L. A narrative review of the pharmacology of ginsenoside compound K. Ann. Transl. Med. 2022, 10, 234. [Google Scholar] [CrossRef]
- Chopra, P.; Chhillar, H.; Kim, Y.J.; Jo, I.H.; Kim, S.T.; Gupta, R. Phytochemistry of ginsenosides: Recent advancements and emerging roles. Crit. Rev. Food Sci. Nutr. 2021, 1–28. [Google Scholar] [CrossRef]
- Ma, Z.; Mi, Y.; Han, X.; Li, H.; Tian, M.; Duan, Z.; Fan, D.; Ma, P. Transformation of ginsenoside via deep eutectic solvents based on choline chloride as an enzymatic reaction medium. Bioprocess Biosyst. Eng. 2020, 43, 1195–1208. [Google Scholar] [CrossRef]
- Geraldi, A.; Ni’matuzahroh, F.; Cui, C.H.; Nguyen, T.T.; Kim, S.C. Enzymatic biotransformation of ginsenoside Rb1 by recombinant β-glucosidase of bacterial isolates from Indonesia. Biocatal. Agric. Biotechnol. 2020, 23, 101449. [Google Scholar] [CrossRef]
- Wu, L.; Jin, Y.; Yin, C.; Bai, L. Co-transformation of Panax major ginsenosides Rb1 and Rg1 to minor ginsenosides C-K and F1 by Cladosporium cladosporioides. J. Ind. Microbiol. Biotechnol. 2012, 39, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Won, H.J.; Kim, H.I.; Park, T.; Kim, H.; Jo, K.; Jeon, H.; Ha, S.J.; Hyun, J.M.; Jeong, A.; Kim, J.S.; et al. Non-clinical pharmacokinetic behavior of ginsenosides. J. Ginseng Res. 2019, 43, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zou, H.; Gao, Y.; Luo, J.; Xie, X.; Meng, W.; Zhou, H.; Tan, Z. Insights into gastrointestinal microbiota-generated ginsenoside metabolites and their bioactivities. Drug Metab. Rev. 2020, 52, 125–138. [Google Scholar] [CrossRef]
- Li, W.; Jiang, Y.; Liu, Y.; Li, C.; Fan, D. Biocatalytic strategies in producing ginsenoside by glycosidase-A review. Chin. J. Biotechnol. 2019, 35, 1590–1606. [Google Scholar] [CrossRef]
- Wong, A.S.; Che, C.M.; Leung, K.W. Recent advances in ginseng as cancer therapeutics: A functional and mechanistic overview. Nat. Prod. Rep. 2015, 32, 256–272. [Google Scholar] [CrossRef]
- Li, W.N.; Fan, D.D. Biocatalytic strategies for the production of ginsenosides using glycosidase: Current state and perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 3807–3823. [Google Scholar] [CrossRef]
- Renchinkhand, G.; Magsar, U.; Bae, H.C.; Choi, S.H.; Nam, M.S. Identification of β-glucosidase activity of Lentilactobacillus buchneri URN103L and its potential to convert ginsenoside Rb1 from Panax ginseng. Foods 2022, 11, 529. [Google Scholar] [CrossRef]
- Kim, D.H. Gut microbiota-mediated pharmacokinetics of ginseng saponins. J. Ginseng Res. 2018, 42, 255–263. [Google Scholar] [CrossRef]
- Yang, X.D.; Yang, Y.Y.; Ouyang, D.S.; Yang, G.P. A review of biotransformation and pharmacology of ginsenoside compound K. Fitoterapia 2015, 100, 208–220. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell Mol. Life Sci 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Shin, K.C.; Oh, D.K. Classification of glycosidases that hydrolyze the specific positions and types of sugar moieties in ginsenosides. Crit. Rev. Biotechnol. 2016, 36, 1036–1049. [Google Scholar] [CrossRef]
- Zheng, M.M.; Xu, F.X.; Li, Y.J.; Xi, X.Z.; Cui, X.W.; Han, C.C.; Zhang, X.L. Study on transformation of ginsenosides in different methods. Biomed Res. Int. 2017, 2017, 8601027. [Google Scholar] [CrossRef]
- Quan, L.H.; Min, J.W.; Jin, Y.; Wang, C.; Kim, Y.J.; Yang, D.C. Enzymatic biotransformation of ginsenoside Rb1 to compound K by recombinant β-glucosidase from Microbacterium esteraromaticum. J. Agric. Food Chem. 2012, 60, 3776–3781. [Google Scholar] [CrossRef]
- Yu, H.; Liu, Q.; Zhang, C.; Lu, M.; Fu, Y.; Im, W.T.; Lee, S.T.; Jin, F. A new ginsenosidase from Aspergillus strain hydrolyzing 20-O-multi-glycoside of PPD ginsenoside. Process Biochem. 2009, 44, 772–775. [Google Scholar] [CrossRef]
- Jin, X.F.; Yu, H.S.; Wang, D.M.; Liu, T.Q.; Liu, C.Y.; An, D.S.; Im, W.T.; Kim, S.G.; Jin, F.X. Kinetics of a cloned special ginsenosidase hydrolyzing 3-O-glucoside of multi-protopanaxadiol-type ginsenosides, named ginsenosidase type III. J. Microbiol. Biotechnol. 2012, 22, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.M.; Yu, H.S.; Song, J.G.; Xu, Y.F.; Liu, C.Y.; Jin, F.X. A novel ginsenosidase from an Aspergillus strain hydrolyzing 6-O-multi-glycosides of protopanaxatriol-type ginsenosides, named ginsenosidase type IV. J. Microbiol. Biotechnol. 2011, 21, 1057–1063. [Google Scholar] [CrossRef]
- Cui, C.H.; Kim, S.C.; Im, W.T. Characterization of the ginsenoside-transforming recombinant β-glucosidase from Actinosynnema mirum and bioconversion of major ginsenosides into minor ginsenosides. Appl. Microbiol. Biotechnol. 2013, 97, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.H.; Jeon, B.M.; Fu, Y.; Im, W.T.; Kim, S.C. High-density immobilization of a ginsenoside-transforming β-glucosidase for enhanced food-grade production of minor ginsenosides. Appl. Microbiol. Biotechnol. 2019, 103, 7003–7015. [Google Scholar] [CrossRef]
- Cui, C.H.; Kim, D.J.; Jung, S.C.; Kim, S.C.; Im, W.T. Enhanced production of gypenoside LXXV using a novel ginsenoside-transforming β-glucosidase from ginseng-cultivating soil bacteria and its anti-cancer property. Molecules 2017, 22, 844. [Google Scholar] [CrossRef]
- Chi, H.; Ji, G.E. Transformation of ginsenosides Rb1 and Re from Panax ginseng by food microorganisms. Biotechnol. Lett. 2005, 27, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Seo, M.J.; Shin, K.C.; Lee, K.W.; Oh, D.K. Synergistic production of 20(S)-protopanaxadiol from protopanaxadiol-type ginsenosides by β-glycosidases from Dictyoglomus turgidum and Caldicellulosiruptor bescii. AMB Express 2017, 7, 219. [Google Scholar] [CrossRef] [PubMed]
- Salgado, J.C.S.; Meleiro, L.P.; Carli, S.; Ward, R.J. Glucose tolerant and glucose stimulated β-glucosidases—A review. Bioresour. Technol. 2018, 267, 704–713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Luo, J.; Xie, J.; Wang, Z.; Xiao, W.; Zhao, L. Cooperated biotransformation of ginsenoside extracts into ginsenoside 20(S)-Rg3 by three thermostable glycosidases. J. Appl. Microbiol. 2020, 128, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Xie, J.; Zhang, X.; Cao, F.; Pei, J. Overexpression and characterization of a glucose-tolerant β-glucosidase from Thermotoga thermarum DSM 5069T with high catalytic efficiency of ginsenoside Rb1 to Rd. J. Mol. Catal. B Enzym. 2013, 95, 62–69. [Google Scholar] [CrossRef]
- Xie, J.; Zhao, D.; Zhao, L.; Pei, J.; Xiao, W.; Ding, G.; Wang, Z. Overexpression and characterization of a Ca(2+) activated thermostable β-glucosidase with high ginsenoside Rb1 to ginsenoside 20(S)-Rg3 bioconversion productivity. J. Ind. Microbiol. Biotechnol. 2015, 42, 839–850. [Google Scholar] [CrossRef]
- Shin, K.C.; Oh, D.K. Characterization of a novel recombinant β-glucosidase from Sphingopyxis alaskensis that specifically hydrolyzes the outer glucose at the C-3 position in protopanaxadiol-type ginsenosides. J. Biotechnol. 2014, 172, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.C.; Lee, H.J.; Oh, D.K. Substrate specificity of β-glucosidase from Gordonia terrae for ginsenosides and its application in the production of ginsenosides Rg3, Rg2, and Rh1 from ginseng root extract. J. Biosci. Bioeng. 2015, 119, 497–504. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, W.; Fan, D. Biotransformation of ginsenoside Rb1 to ginsenoside CK by strain XD101: A safe bioconversion strategy. Appl. Biochem. Biotechnol. 2021, 193, 2110–2127. [Google Scholar] [CrossRef]
- Lin, F.; Guo, X.; Lu, W. Efficient biotransformation of ginsenoside Rb1 to Rd by isolated Aspergillus versicolor, excreting β-glucosidase in the spore production phase of solid culture. Antonie Van Leeuwenhoek 2015, 108, 1117–1127. [Google Scholar] [CrossRef]
- Kumar, P.; Mishra, S.; Kumar, A.; Kumar, S.; Prasad, C.S. In vivo and in vitro control activity of plant essential oils against three strains of Aspergillus niger. Environ. Sci. Pollut. Res. Int. 2017, 24, 21948–21959. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Larsen, T.O.; Thrane, U.; Meijer, M.; Varga, J.; Samson, R.A.; Nielsen, K.F. Fumonisin and ochratoxin production in industrial Aspergillus niger strains. PLoS ONE 2011, 6, e23496. [Google Scholar] [CrossRef]
- Noonim, P.; Mahakarnchanakul, W.; Nielsen, K.F.; Frisvad, J.C.; Samson, R.A. Fumonisin B2 production by Aspergillus niger in Thai coffee beans. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2009, 26, 94–100. [Google Scholar] [CrossRef]
- Neergheen, V.; Kam, A.H.; Pem, Y.; Ramsaha, S.; Bahorun, T. Regulation of cancer cell signaling pathways as key events for therapeutic relevance of edible and medicinal mushrooms. Semin. Cancer Biol. 2020, 80, 145–156. [Google Scholar] [CrossRef]
- Liu, Z.; Li, J.X.; Wang, C.Z.; Zhang, D.L.; Wen, X.; Ruan, C.C.; Li, Y.; Yuan, C.S. Microbial conversion of protopanaxadiol-type ginsenosides by the edible and medicinal mushroom Schizophyllum commune: A green biotransformation strategy. ACS Omega 2019, 4, 13114–13123. [Google Scholar] [CrossRef]
- Upadhyaya, J.; Kim, M.J.; Kim, Y.H.; Ko, S.R.; Park, H.W.; Kim, M.K. Enzymatic formation of compound-K from ginsenoside Rb1 by enzyme preparation from cultured mycelia of Armillaria mellea. J. Ginseng Res. 2016, 40, 105–112. [Google Scholar] [CrossRef]
- Hsu, B.Y.; Lu, T.J.; Chen, C.H.; Wang, S.J.; Hwang, L.S. Biotransformation of ginsenoside Rd in the ginseng extraction residue by fermentation with lingzhi (Ganoderma lucidum). Food Chem. 2013, 141, 4186–4193. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Huang, Q.; Sui, X.; Xie, Y. Activities of β-glucosidase and α-L-arabinofuranosidase from Cordyceps militaris and application in the transformation of ginsenoside Rg1 and Rc. Food Sci. 2022, 1–19. [Google Scholar]
- Wang, X. Cloning and Functional Analysis of the Panax ginseng Glycoside Hydrolase Genes. Master’s Thesis, Southwest University, Chongqing, China, 2021. [Google Scholar] [CrossRef]
- Hu, Y.; Luan, H.; Hao, D.; Xiao, H.; Yang, S.; Yang, L. Purification and characterization of a novel ginsenoside-hydrolyzing β-D-glucosidase from the China white jade snail (Achatina fulica). Enzym. Microb. Technol. 2007, 40, 1358–1366. [Google Scholar] [CrossRef]
- Park, M.K.; Cui, C.H.; Park, S.C.; Park, S.K.; Kim, J.K.; Jung, M.S.; Jung, S.C.; Kim, S.C.; Im, W.T. Characterization of recombinant β-glucosidase from Arthrobacter chlorophenolicus and biotransformation of ginsenosides Rb1, Rb2, Rc, and Rd. J. Microbiol. 2014, 52, 399–406. [Google Scholar] [CrossRef]
- Hu, Y.; Zhai, L.; Hong, H.; Shi, Z.; Zhao, J.; Liu, D. Study on the biochemical characterization and selectivity of three β-glucosidases from Bifidobacterium adolescentis ATCC15703. Front. Microbiol. 2022, 13, 860014. [Google Scholar] [CrossRef]
- Shin, K.C.; Kim, T.H.; Choi, J.H.; Oh, D.K. Complete biotransformation of protopanaxadiol-type ginsenosides to 20-O-β-glucopyranosyl-20(S)-protopanaxadiol using a novel and thermostable β-glucosidase. J. Agric. Food Chem. 2018, 66, 2822–2829. [Google Scholar] [CrossRef]
- Cui, C.H.; Kim, J.K.; Kim, S.C.; Im, W.T. Characterization of a ginsenoside-transforming β-glucosidase from Paenibacillus mucilaginosus and its application for enhanced production of minor ginsenoside F2. PLoS ONE 2014, 9, e85727. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S. Study on Biotransformation of Ginsenosides by β-glucosidase and the Interaction Mechanism. Ph.D. Thesis, Jilin University, Changchun, China, 2021. [Google Scholar] [CrossRef]
- Yoo, M.H.; Yeom, S.J.; Park, C.S.; Lee, K.W.; Oh, D.K. Production of aglycon protopanaxadiol via compound K by a thermostable β-glycosidase from Pyrococcus furiosus. Appl. Microbiol. Biotechnol. 2011, 89, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Q.M.; Sung, B.H.; An, D.S.; Lee, H.G.; Kim, S.G.; Kim, S.C.; Lee, S.T.; Im, W.T. Bioconversion of ginsenosides Rb1, Rb2, Rc and Rd by novel β-glucosidase hydrolyzing outer 3-O glycoside from Sphingomonas sp. 2F2: Cloning, expression, and enzyme characterization. J. Biotechnol. 2011, 156, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Son, J.W.; Kim, H.J.; Oh, D.K. Ginsenoside Rd production from the major ginsenoside Rb1 by β-glucosidase from Thermus caldophilus. Biotechnol. Lett. 2008, 30, 713–716. [Google Scholar] [CrossRef]
- Shin, K.C.; Seo, M.J.; Oh, H.J.; Oh, D.K. Highly selective hydrolysis for the outer glucose at the C-20 position in ginsenosides by β-glucosidase from Thermus thermophilus and its application to the production of ginsenoside F2 from gypenoside XVII. Biotechnol. Lett. 2014, 36, 1287–1293. [Google Scholar] [CrossRef]
- Jung, I.H.; Lee, J.H.; Hyun, Y.J.; Kim, D.H. Metabolism of ginsenoside Rb1 by human intestinal microflora and cloning of its metabolizing β-D-glucosidase from Bifidobacterium longum H-1. Biol. Pharm. Bull. 2012, 2012. 35, 573–581. [Google Scholar] [CrossRef]
- Yan, S.; Wei, P.C.; Chen, Q.; Chen, X.; Wang, S.C.; Li, J.R.; Gao, C. Functional and structural characterization of a β-glucosidase involved in saponin metabolism from intestinal bacteria. Biochem. Biophys. Res. Commun. 2018, 496, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.W.; Kim, K.R.; Oh, D.K. Production of rare ginsenosides (compound Mc, compound Y and aglycon protopanaxadiol) by β-glucosidase from Dictyoglomus turgidum that hydrolyzes β-linked, but not α-linked, sugars in ginsenosides. Biotechnol. Lett. 2012, 34, 1679–1686. [Google Scholar] [CrossRef]
- Hong, H.; Cui, C.H.; Kim, J.K.; Jin, F.X.; Kim, S.C.; Im, W.T. Enzymatic biotransformation of ginsenoside Rb1 and gypenoside XVII into ginsenosides Rd and F2 by recombinant β-glucosidase from Flavobacterium johnsoniae. J. Ginseng Res. 2012, 36, 418–424. [Google Scholar] [CrossRef]
- Kim, J.K.; Cui, C.H.; Liu, Q.; Yoon, M.H.; Kim, S.C.; Im, W.T. Mass production of the ginsenoside Rg3(S) through the combinative use of two glycoside hydrolases. Food Chem. 2013, 141, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.L.; Ma, R.; Jiang, M.; Dong, W.W.; Jiang, J.; Wu, S.; Li, D.; Quan, L.H. Cloning and characterization of ginsenoside-hydrolyzing β-glucosidase from Lactobacillus brevis that transforms ginsenosides Rb1 and F2 into ginsenoside Rd and compound K. J. Microbiol. Biotechnol. 2016, 26, 1661–1667. [Google Scholar] [CrossRef]
- Quan, L.H.; Min, J.W.; Yang, D.U.; Kim, Y.J.; Yang, D.C. Enzymatic biotransformation of ginsenoside Rb1 to 20(S)-Rg3 by recombinant β-glucosidase from Microbacterium esteraromaticum. Appl. Microbiol. Biotechnol. 2012, 94, 377–384. [Google Scholar] [CrossRef]
- Cui, C.H.; Liu, Q.M.; Kim, J.K.; Sung, B.H.; Kim, S.G.; Kim, S.C.; Im, W.T. Identification and characterization of a Mucilaginibacter sp. strain QM49 β-glucosidase and its use in the production of the pharmaceutically active minor ginsenosides (S)-Rh1 and (S)-Rg2. Appl. Environ. Microbiol. 2013, 79, 5788–5798. [Google Scholar] [CrossRef]
- Du, J.; Cui, C.H.; Park, S.C.; Kim, J.K.; Yu, H.S.; Jin, F.X.; Sun, C.; Kim, S.C.; Im, W.T. Identification and characterization of a ginsenoside-transforming β-glucosidase from Pseudonocardia sp. Gsoil 1536 and its application for enhanced production of minor ginsenoside Rg2(S). PLoS ONE 2014, 9, e96914. [Google Scholar] [CrossRef]
- Kim, J.K.; Cui, C.H.; Yoon, M.H.; Kim, S.C.; Im, W.T. Bioconversion of major ginsenosides Rg1 to minor ginsenoside F1 using novel recombinant ginsenoside hydrolyzing glycosidase cloned from Sanguibacter keddieii and enzyme characterization. J. Biotechnol. 2012, 161, 294–301. [Google Scholar] [CrossRef]
- An, D.S.; Cui, C.H.; Lee, H.G.; Wang, L.; Kim, S.C.; Lee, S.T.; Jin, F.; Yu, H.; Chin, Y.W.; Lee, H.K.; et al. Identification and characterization of a novel Terrabacter ginsenosidimutans sp. nov. β-glucosidase that transforms ginsenoside Rb1 into the rare gypenosides XVII and LXXV. Appl. Environ. Microbiol. 2010, 76, 5827–5836. [Google Scholar] [CrossRef]
- Fu, Y.; Yin, Z.H.; Wu, L.P.; Yin, C.R. Biotransformation of ginsenoside Rb1 to ginsenoside C-K by endophytic fungus Arthrinium sp. GE 17-18 isolated from Panax ginseng. Lett. Appl. Microbiol. 2016, 63, 196–201. [Google Scholar] [CrossRef]
- Chang, K.H.; Jo, M.N.; Kim, K.T.; Paik, H.D. Evaluation of glucosidases of Aspergillus niger strain comparing with other glucosidases in transformation of ginsenoside Rb1 to ginsenosides Rg3. J. Ginseng Res. 2014, 38, 47–51. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Jo, M.N.; Kim, K.T.; Paik, H.D. Purification and characterization of a ginsenoside Rb1-hydrolyzing β-glucosidase from Aspergillus niger KCCM 11239. Int. J. Mol. Sci. 2012, 13, 12140–12152. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.G.; Xue, J.J.; Sun, M.Q.; Wang, C.Y.; Liu, L.; Zhang, D.L.; Lee, M.R.; Gu, L.J.; Wang, C.L.; Wang, Y.B.; et al. Highly selective microbial transformation of major ginsenoside Rb1 to gypenoside LXXV by Esteya vermicola CNU120806. J. Appl. Microbiol. 2012, 113, 807–814. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, W.J.; Gebru, Y.A.; Upadhyaya, J.; Ko, S.R.; Kim, Y.H.; Kim, M.K. Production of minor ginsenosides C-K and C-Y from naturally occurring major ginsenosides using crude β-glucosidase preparation from submerged culture of Fomitella fraxinea. Molecules 2021, 26, 4820. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.W.; Yoo, M.H.; Shin, K.C.; Kim, K.R.; Kim, Y.S.; Lee, K.W.; Oh, D.K. β-Glucosidase from Penicillium aculeatum hydrolyzes exo-, 3-O-, and 6-O-β-glucosides but not 20-O-β-glucoside and other glycosides of ginsenosides. Appl. Microbiol. Biotechnol. 2013, 97, 6315–6324. [Google Scholar] [CrossRef]
- Luan, H.; Liu, X.; Qi, X.; Hu, Y.; Hao, D.; Cui, Y.; Yang, L. Purification and characterization of a novel stable ginsenoside Rb1-hydrolyzing β-D-glucosidase from China white jade snail. Process Biochem. 2006, 41, 1974–1980. [Google Scholar] [CrossRef]
- Dobson, P.D.; Cai, Y.D.; Stapley, B.J.; Doig, A.J. Prediction of protein function in the absence of significant sequence similarity. Curr. Med. Chem. 2004, 11, 2135–4212. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Mills, C.L.; Beuning, P.J.; Ondrechen, M.J. Biochemical functional predictions for protein structures of unknown or uncertain function. Comput. Struct. Biotechnol. J. 2015, 13, 182–191. [Google Scholar] [CrossRef] [PubMed]
- Samaei-Daryan, S.; Goliaei, B.; Ebrahim-Habibi, A. Characterization of surface binding sites in glycoside hydrolases: A computational study. J. Mol. Recognit. 2017, 30, e2624. [Google Scholar] [CrossRef]
- Park, S.J.; Choi, J.M.; Kyeong, H.H.; Kim, S.G.; Kim, H.S. Rational design of a β-glycosidase with high regiospecificity for triterpenoid tailoring. ChemBioChem 2015, 16, 854–860. [Google Scholar] [CrossRef]
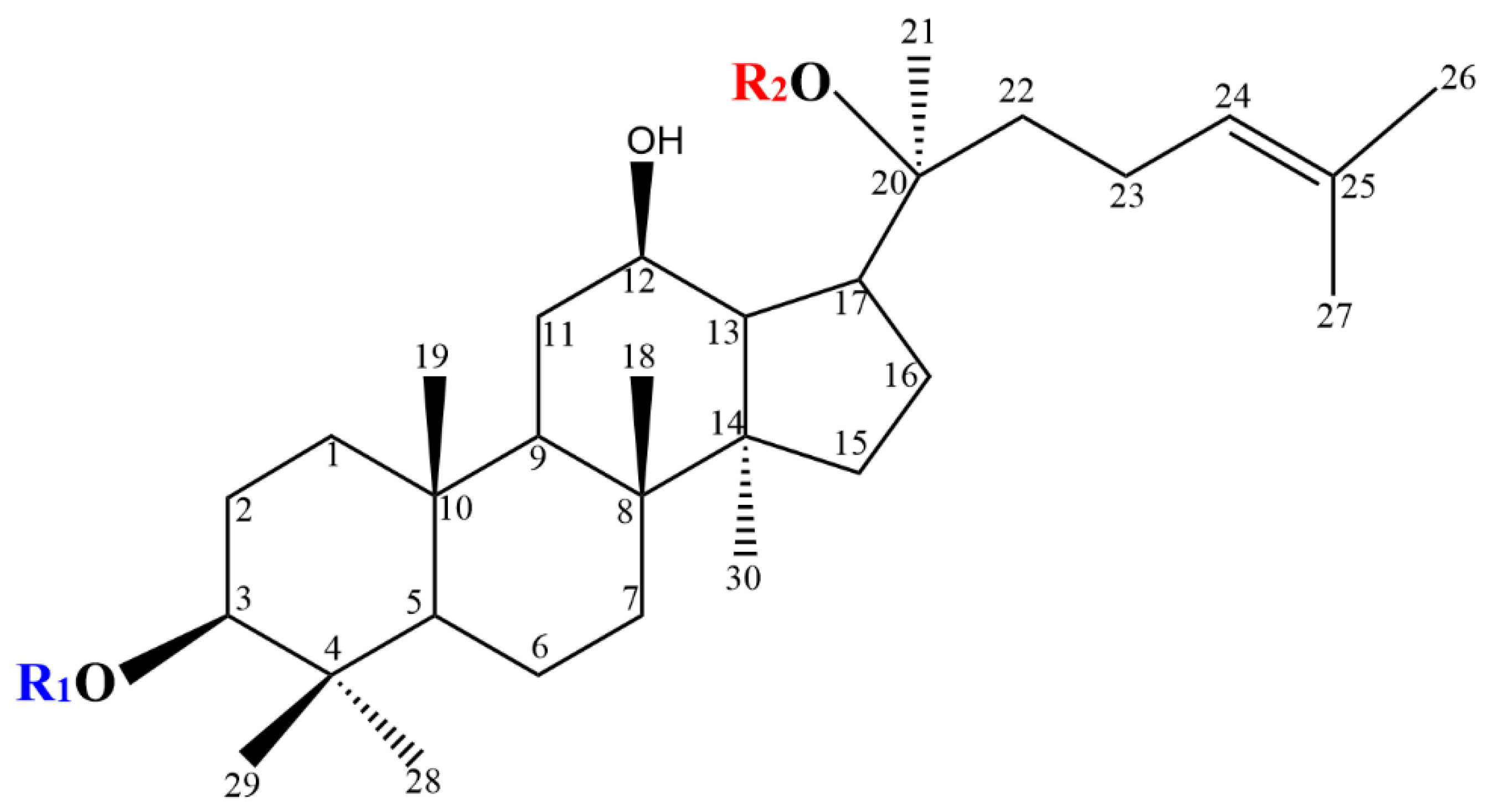
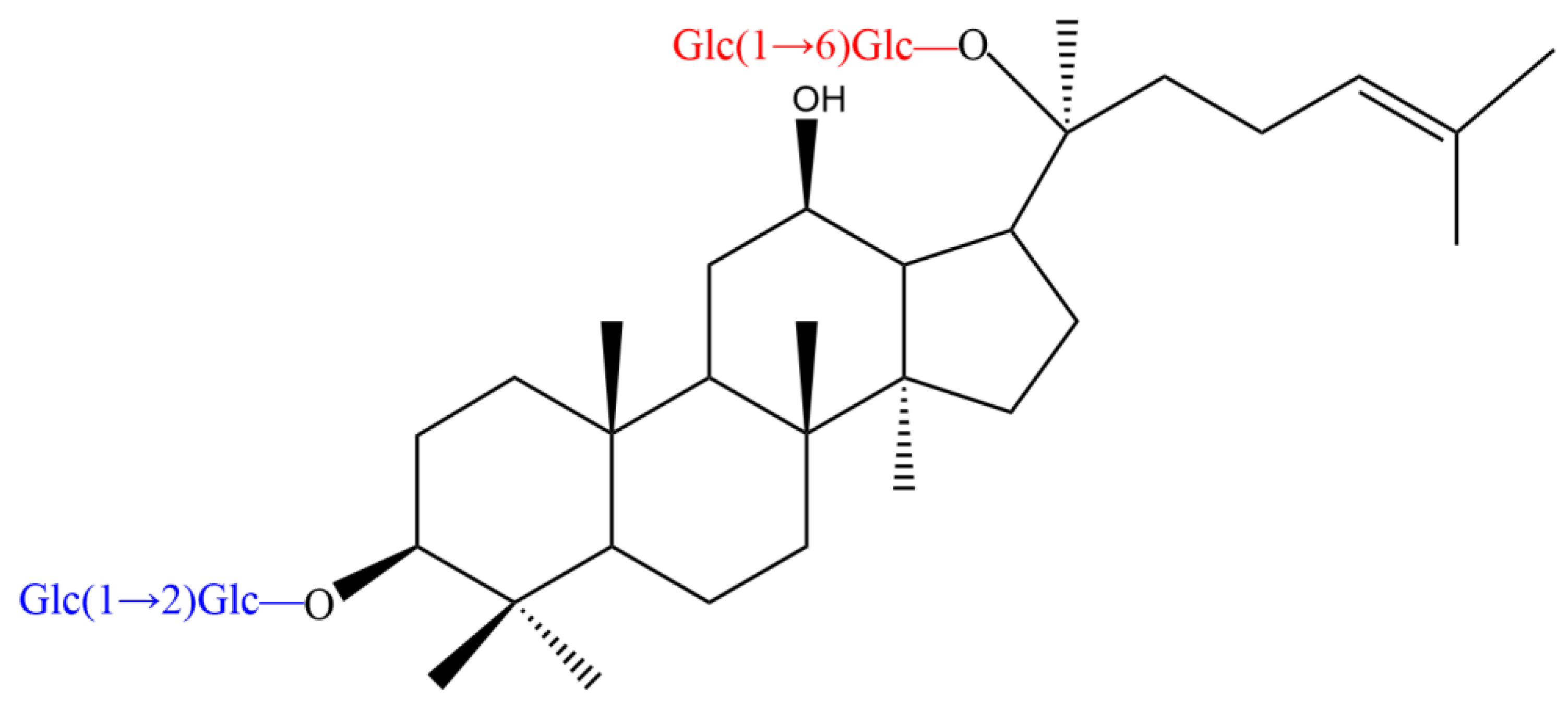
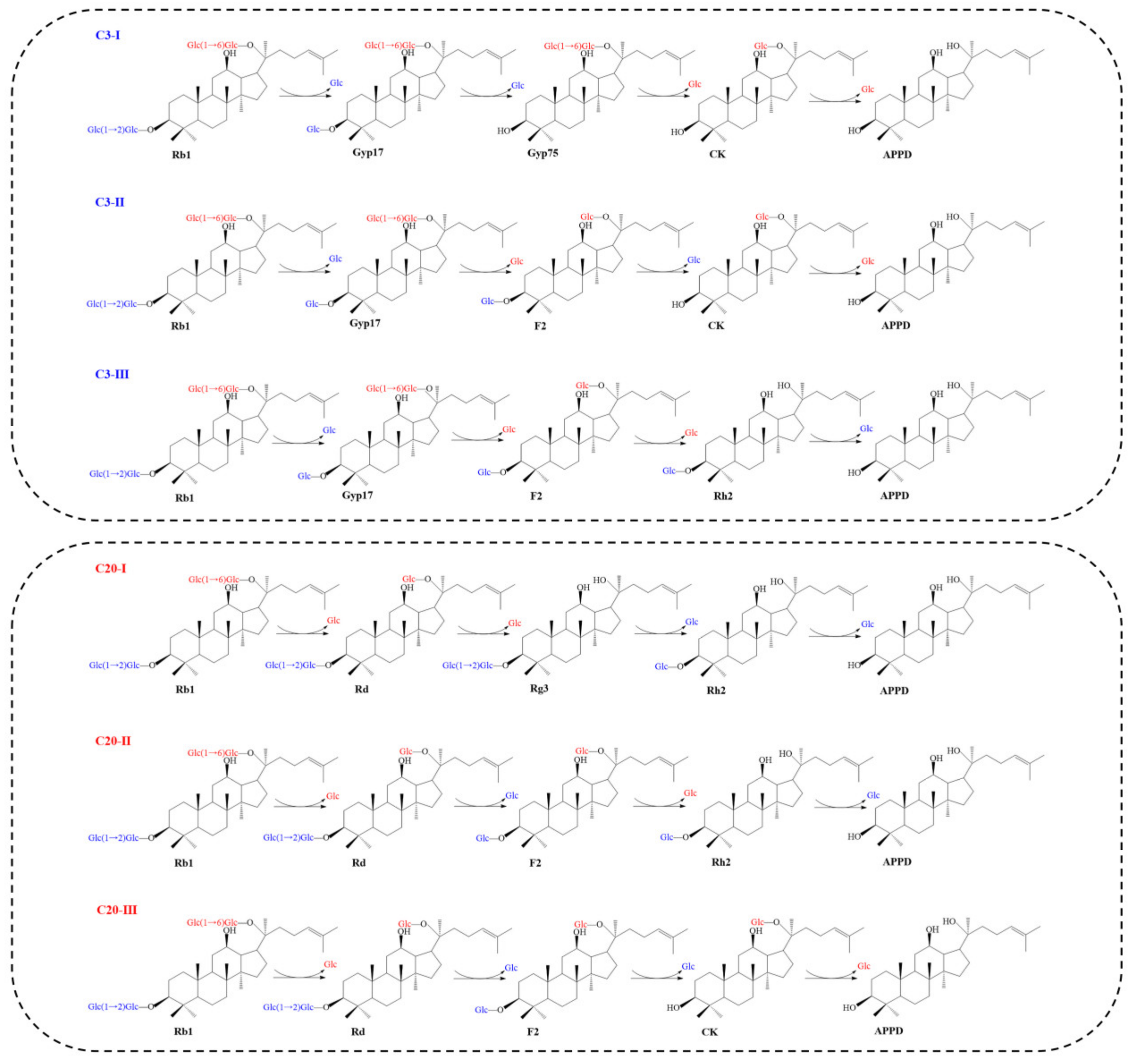
| Ginsenoside | R1 (C-3) | R2 (C-20) |
|---|---|---|
| Rb1 | Glc(1 → 2)Glc- | Glc(1 → 6)Glc- |
| Rb2 | Glc(1 → 2)Glc- | Arap(1 → 6)Glc- |
| Rb3 | Glc(1 → 2)Glc- | Xyl(1 → 6)Glc- |
| Rc | Glc(1 → 2)Glc- | Araf(1 → 6)Glc- |
| Rd | Glc(1 → 2)Glc- | Glc- |
| F2 | Glc- | Glc- |
| CMc1 | Glc- | Araf(1 → 6)Glc- |
| CMc | H- | Araf(1 → 6)Glc- |
| CMx1 | Glc- | Xyl(1 → 6)Glc- |
| CMx | H- | Xyl(1 → 6)Glc- |
| CO | Glc- | Arap(1 → 6)Glc- |
| CY | H- | Arap(1 → 6)Glc- |
| CK | H- | Glc- |
| Gyp17 | Glc- | Glc(1 → 6)Glc- |
| Gyp75 | H- | Glc(1 → 6)Glc- |
| Rg3 | Glc(1 → 2)Glc- | H- |
| Rh2 | Glc- | H- |
| APPD | H- | H- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Zhang, R.; Huang, Z.; Zhou, J. Progress in the Conversion of Ginsenoside Rb1 into Minor Ginsenosides Using β-Glucosidases. Foods 2023, 12, 397. https://doi.org/10.3390/foods12020397
Zhu H, Zhang R, Huang Z, Zhou J. Progress in the Conversion of Ginsenoside Rb1 into Minor Ginsenosides Using β-Glucosidases. Foods. 2023; 12(2):397. https://doi.org/10.3390/foods12020397
Chicago/Turabian StyleZhu, Hongrong, Rui Zhang, Zunxi Huang, and Junpei Zhou. 2023. "Progress in the Conversion of Ginsenoside Rb1 into Minor Ginsenosides Using β-Glucosidases" Foods 12, no. 2: 397. https://doi.org/10.3390/foods12020397
APA StyleZhu, H., Zhang, R., Huang, Z., & Zhou, J. (2023). Progress in the Conversion of Ginsenoside Rb1 into Minor Ginsenosides Using β-Glucosidases. Foods, 12(2), 397. https://doi.org/10.3390/foods12020397






