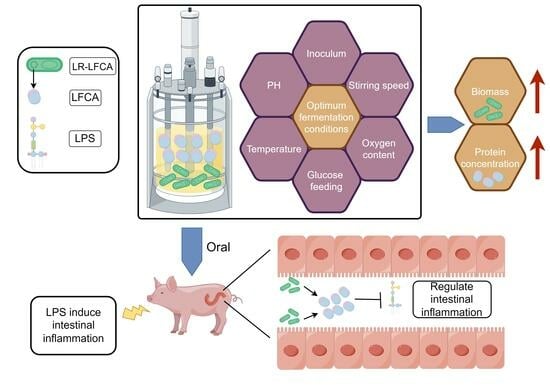
Optimum Fermentation Conditions for Bovine Lactoferricin-Lactoferrampin-Encoding LimosiLactobacillus reuteri and Regulation of Intestinal Inflammation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Determination of Growth Curve and Fermentation Culture of Recombinant LimosiLactobacillus reuteri
2.3. Western Blot Expression Identification and Quantitative Determination of LFCA Concentration
2.4. Single-Factor Screening of Inoculation Amount, Culture Temperature, Stirring Speed, Dissolved Oxygen, and ph
2.5. Response Surface Optimization Design and Model Verification
2.6. Glucose Feedback Feeding and Residual Sugar Determination in Fermentation Broth
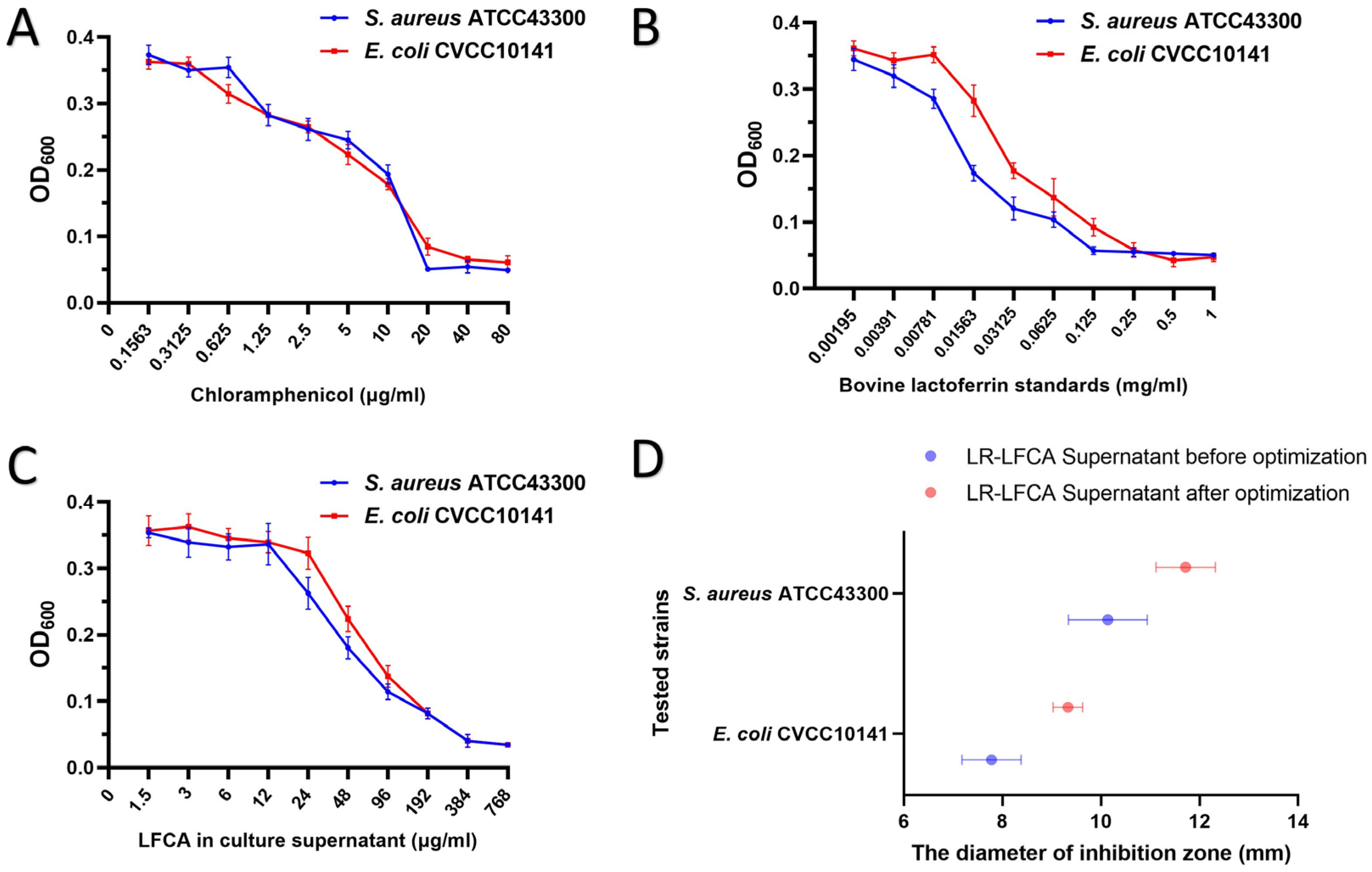
2.7. Determination of Minimum Inhibitory Concentration (MIC50) by Dilution Method with Trace Meat Broth
2.8. Oxford Cup Method for Measuring the Diameter of the Inhibition Zone
2.9. Animal Experiment Design
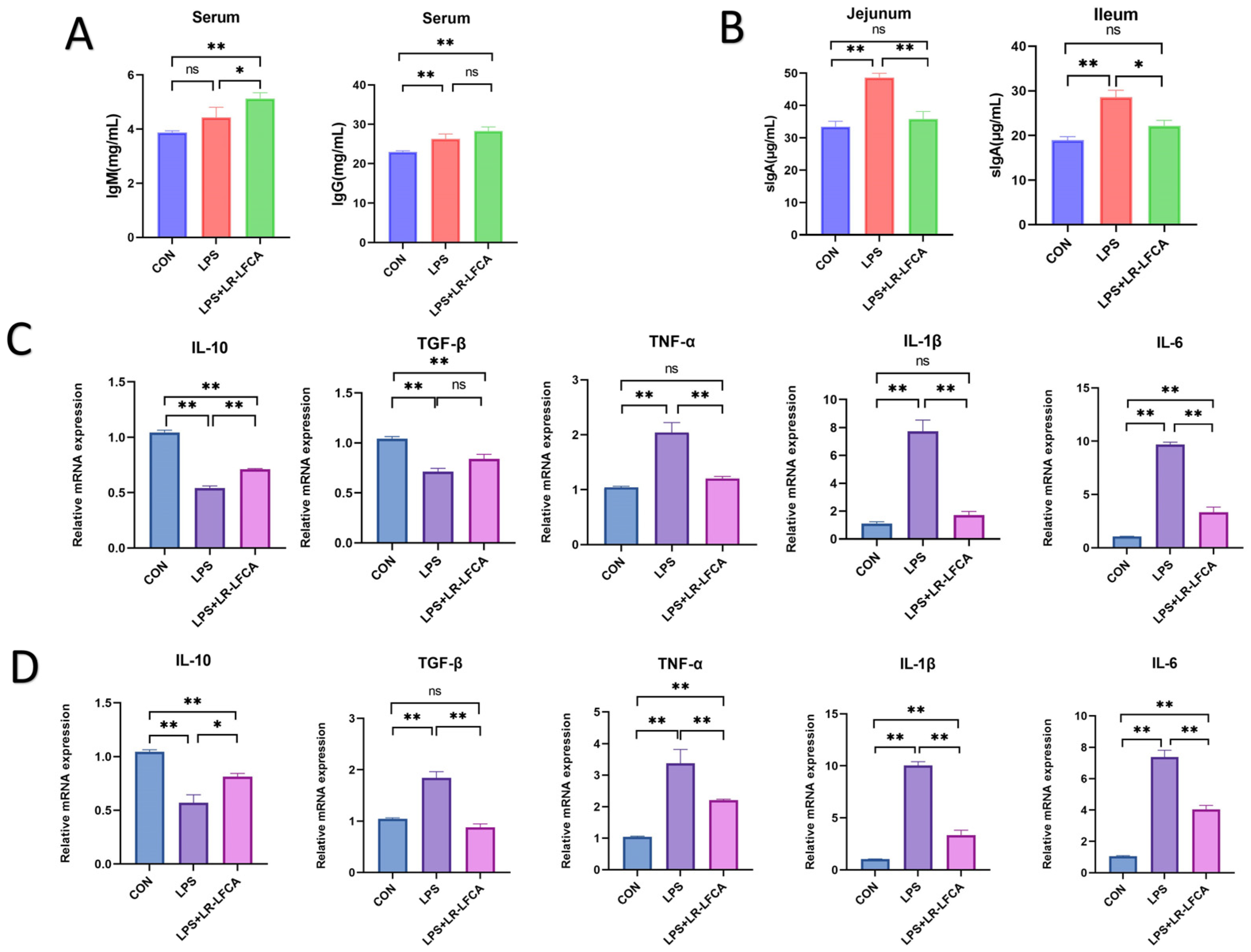
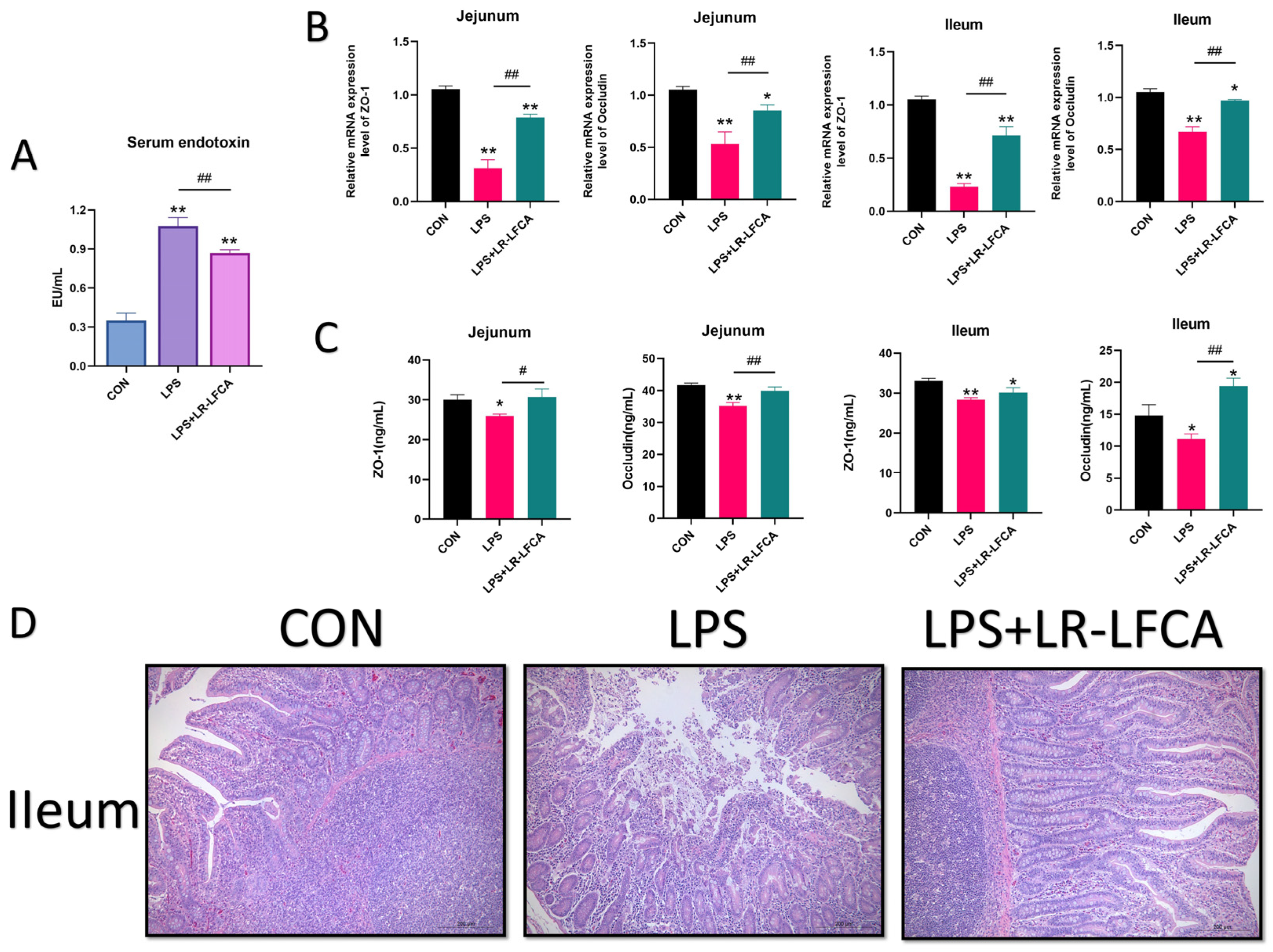
2.10. Sample Collection, Processing, and Index Determination
2.11. Determination of Tight Junction Proteins and Inflammatory Factors in Piglet Intestinal Tract
2.12. Data Processing and Statistical Analysis
3. Results
3.1. Biological Characteristics of LR-LFCA and Single-Factor Optimization of Fermentation Conditions
3.2. Screening Significant Factors Affecting LR-LFCA Fermentation by Plackett–Burman Design
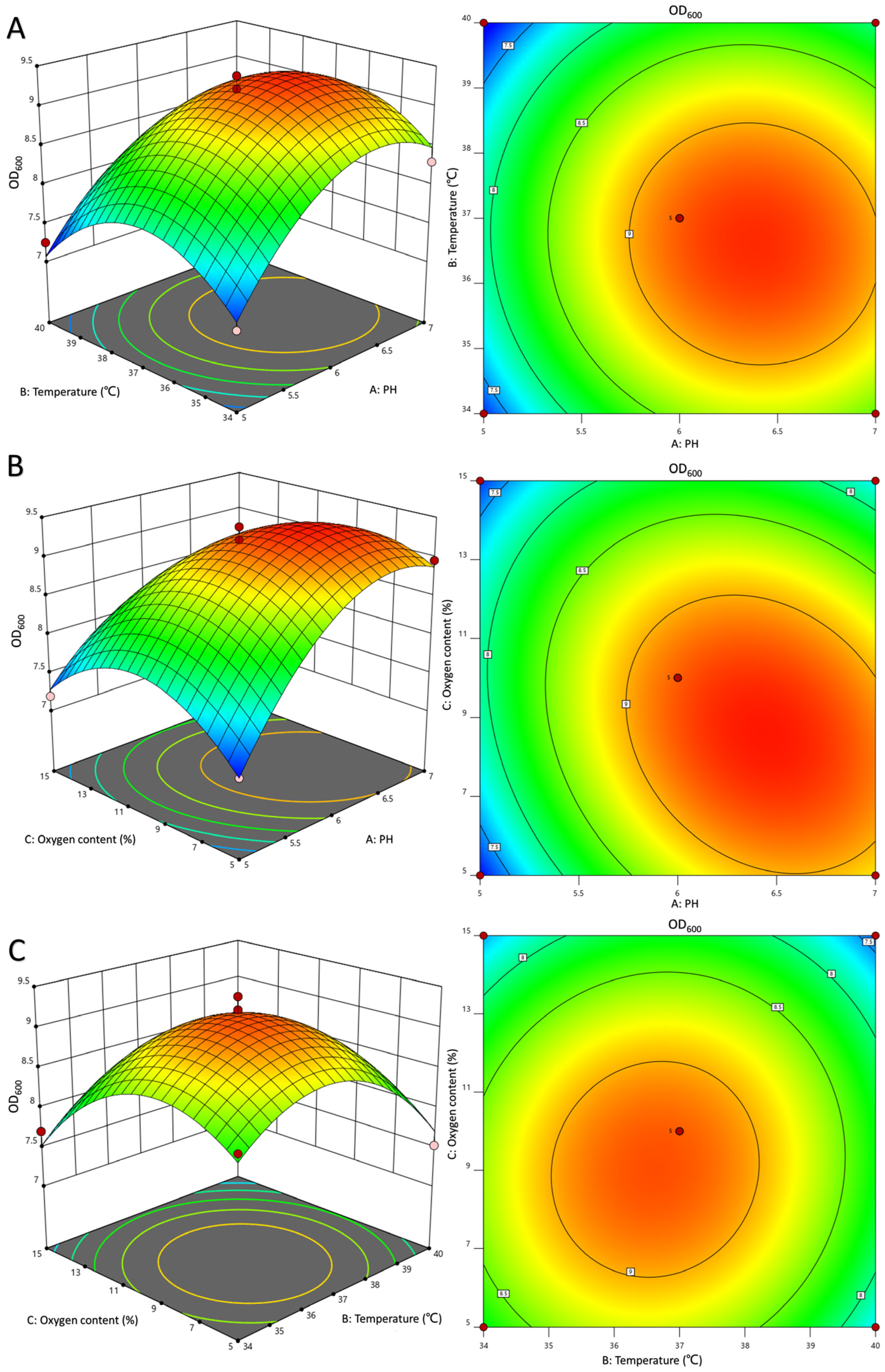
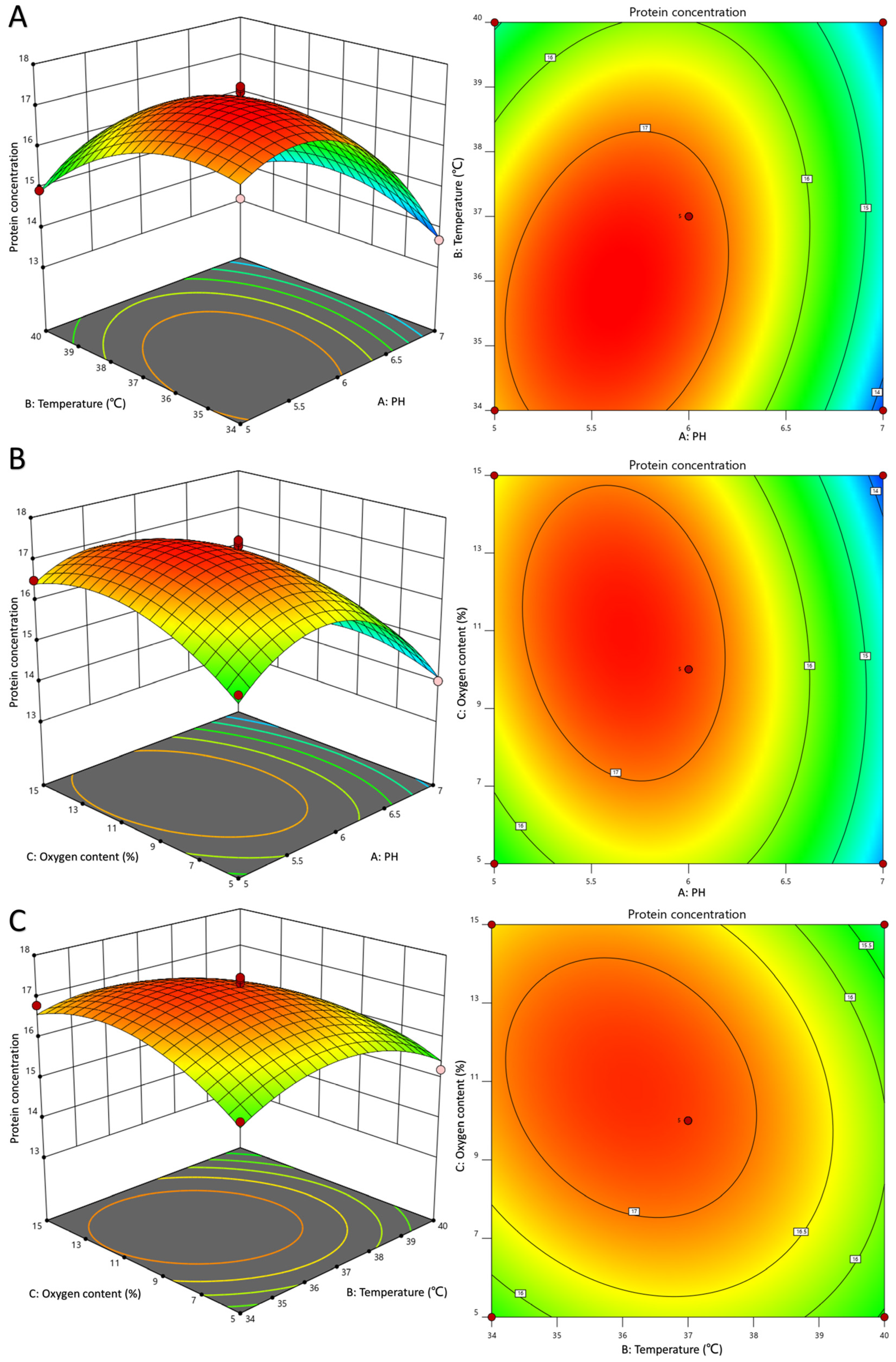
3.3. Optimization of LR-LFCA Fermentation Conditions by Response Surface Analysis
3.4. The Antibacterial Activity of LR-LFCA Fermentation Supernatant after Optimization
3.5. The Regulatory Effect of Optimized LR-LFCA on LPS-Induced Intestinal Inflammation in Piglets
3.6. The Regulatory Effects of LR-LFCA on Intestinal Barrier Function and Intestinal Tissue Morphology in LPS-Treated Piglets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, B.; Timilsena, Y.P.; Blanch, E.; Adhikari, B. Lactoferrin: Structure, function, denaturation and digestion. Crit. Rev. Food Sci. Nutr. 2019, 59, 580–596. [Google Scholar] [CrossRef]
- Flores-Villaseñor, H.; Canizalez-Román, A.; Reyes-Lopez, M.; Nazmi, K.; de la Garza, M.; Zazueta-Beltrán, J.; León-Sicairos, N.; Bolscher, J.G.M. Bactericidal effect of bovine lactoferrin, LFcin, LFampin and LFchimera on antibiotic-resistant Staphylococcus aureus and Escherichia coli. BioMetals 2010, 23, 569–578. [Google Scholar] [CrossRef]
- Shah, P.; Wu, W.-S.; Chen, C.-S. Systematical Analysis of the Protein Targets of Lactoferricin B and Histatin-5 Using Yeast Proteome Microarrays. Int. J. Mol. Sci. 2019, 20, 4218. [Google Scholar] [CrossRef]
- Van der Kraan, M.I.A.; Groenink, J.; Nazmi, K.; Veerman, E.C.I.; Bolscher, J.G.M.; Nieuw Amerongen, A.V. Lactoferrampin: A Novel Antimicrobial Peptide in the N1-Domain of Bovine Lactoferrin. Peptides 2004, 25, 177–183. [Google Scholar] [CrossRef]
- Biasibetti, E.; Rapacioli, S.; Bruni, N.; Martello, E. Lactoferrin-derived peptides antimicrobial activity: An in vitro experiment. Nat. Prod. Res. 2020, 35, 6073–6077. [Google Scholar] [CrossRef]
- Cruz, J.; Ortiz, C.; Guzman, F.; Cardenas, C.; Fernandez-Lafuente, R.; Torres, R. Design and activity of novel lactoferrampin analogues against O157:H7 enterohemorrhagic Escherichia coli. Biopolymers 2014, 101, 319–328. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Yan, Z.B.; Meng, Y.M.; Hong, X.Y.; Shao, G.; Ma, J.J.; Cheng, X.R.; Liu, J.; Kang, J.; Fu, C.Y. Antimicrobial peptides: Mechanism of action, activity and clinical potential. Mil. Med Res. 2021, 8, 48. [Google Scholar] [CrossRef]
- Rascon-Cruz, Q.; Espinoza-Sanchez, E.A.; Siqueiros-Cendon, T.S.; Nakamura-Bencomo, S.I.; Arevalo-Gallegos, S.; Iglesias-Figueroa, B.F. Lactoferrin: A Glycoprotein Involved in Immunomodulation, Anticancer, and Antimicrobial Processes. Molecules 2021, 26, 205. [Google Scholar] [CrossRef]
- Won, J.H.; Choi, J.S.; Jun, J.-I. CCN1 interacts with integrins to regulate intestinal stem cell proliferation and differentiation. Nat. Commun. 2022, 13, 3117. [Google Scholar] [CrossRef]
- Gonzalez-Chavez, S.A.; Arevalo-Gallegos, S.; Rascon-Cruz, Q. Lactoferrin: Structure, function and applications. Int. J. Antimicrob. Agents 2009, 33, 301.e1–301.e8. [Google Scholar] [CrossRef]
- Szatraj, K.; Szczepankowska, A.K.; Chmielewska-Jeznach, M. Lactic acid bacteria—Promising vaccine vectors: Possibilities, limitations, doubts. J. Appl. Microbiol. 2017, 123, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Alexander, L.M.; Oh, J.H.; Stapleton, D.S.; Schueler, K.L.; Keller, M.P.; Attie, A.D.; van Pijkeren, J.P. Exploiting Prophage-Mediated Lysis for Biotherapeutic Release by Lactobacillus reuteri. Appl. Environ. Microbiol. 2019, 85, e02335-18. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Song, L.; Wang, X.; Xu, Y.; Liu, Z.; Zhao, D.; Wang, S.; Fan, X.; Wang, Z.; Gao, C.; et al. A bovine lactoferricin-lactoferrampin-encoding Lactobacillus reuteri CO21 regulates the intestinal mucosal immunity and enhances the protection of piglets against enterotoxigenic Escherichia coli K88 challenge. Gut Microbes 2021, 13, 1956281. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, W.; Zhang, S.; Shao, Y.; Cai, J.; Cai, L.; Wang, X.; Shan, Z.; Zhou, H.; Li, J.; et al. Effect of Microencapsulation Techniques on the Stress Resistance and Biological Activity of Bovine Lactoferricin-Lactoferrampin-Encoding Lactobacillus reuteri. Foods 2022, 11, 3169. [Google Scholar] [CrossRef]
- Zhang, Y.; Nie, Y.; Zhou, X.; Bi, J.; Xu, Y. Enhancement of pullulanase production from recombinant Bacillus subtilis by optimization of feeding strategy and fermentation conditions. AMB Express 2020, 10, 11. [Google Scholar] [CrossRef]
- Zhang, Q.; Jia, K.Z.; Xia, S.T.; Xu, Y.H.; Liu, R.S.; Li, H.M.; Tang, Y.J. Regulating ehrlich and demethiolation pathways for alcohols production by the expression of ubiquitin-protein ligase gene HUWE1. Sci. Rep. 2016, 6, 20828. [Google Scholar] [CrossRef]
- Juntarachot, N.; Kantachote, D.; Peerajan, S.; Sirilun, S.; Chaiyasut, C. Optimization of Fungal Dextranase Production and Its Antibiofilm Activity, Encapsulation and Stability in Toothpaste. Molecules 2020, 25, 4784. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, Y.; Bohu, T.; Wu, S.; Bai, Z.; Zhuang, X. Nitrogen Removal Characteristics and Constraints of an Alphaproteobacteria with Potential for High Nitrogen Content Heterotrophic Nitrification-Aerobic Denitrification. Microorganisms 2022, 10, 235. [Google Scholar] [CrossRef]
- Kumar, M.; Singh, A.; Beniwal, V.; Salar, R.K. Improved production of tannase by Klebsiella pneumoniae using Indian gooseberry leaves under submerged fermentation using Taguchi approach. AMB Express 2016, 6, 46. [Google Scholar] [CrossRef]
- Gunny, A.A.; Arbain, D.; Jamal, P.; Gumba, R.E. Improvement of halophilic cellulase production from locally isolated fungal strain. Saudi J. Biol. Sci. 2015, 22, 476–483. [Google Scholar] [CrossRef]
- Brito, H.O.; Radulski, D.; Wilhelms, D.B.; Stojakovic, A.; Brito, L.M.O.; Gil da Costa, R.M.; Trindade, E.; Engblom, D.; Franco, C.R.C.; Zampronio, A.R. Immune-mediated febrile response in female rats: Role of central hypothalamic mediators. Sci. Rep. 2020, 10, 4073. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.; Cheng, S.; Li, Y.; Wen, Z.; Ma, X.; Jiang, X.; Wang, Y.; Han, X. Faecal Microbiota Transplantation Reduces Susceptibility to Epithelial Injury and Modulates Tryptophan Metabolism of the Microbial Community in a Piglet Model. J. Crohn’s Colitis 2018, 12, 1359–1374. [Google Scholar] [CrossRef]
- Qiu, Y.; Yang, X.; Wang, L.; Gao, K.; Jiang, Z. L-Arginine Inhibited Inflammatory Response and Oxidative Stress Induced by Lipopolysaccharide via Arginase-1 Signaling in IPEC-J2 Cells. Int. J. Mol. Sci. 2019, 20, 1800. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tang, C.; Zhao, Q.; Fan, S.; Yang, P.; Zhang, J. Butyrate Mitigates Lipopolysaccharide-Induced Intestinal Morphological Changes in Weanling Piglets by Regulating the Microbiota and Energy Metabolism, and Alleviating Inflammation and Apoptosis. Microorganisms 2022, 10, 2001. [Google Scholar] [CrossRef]
- Niamah, A.K.; Mohammed, A.A.; Alhelf, N.A. Antibacterial activity and identification of produced reuterin from local Lactobacillus reuteri LBIQ1 isolate. J. Microbiol. Biotechnol. Food Sci. 2023, 12, e4701. [Google Scholar] [CrossRef]
- Saroha, T.; Sharma, S.; Choksket, S.; Korpole, S.; Patil, P.B. LimosiLactobacillus walteri sp. nov., a novel probiotic antimicrobial lipopeptide-producing bacterium. FEMS Microbiol. Lett. 2023, 370, fnad004. [Google Scholar] [CrossRef]
- Nandakumar, M.P.; Shen, J.; Raman, B.; Marten, M.R. Solubilization of trichloroacetic acid (TCA) precipitated microbial proteins via naOH for two-dimensional electrophoresis. J. Proteome Res. 2003, 2, 89–93. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, H.; Ma, M.; Mu, T.; Zhao, G.; Lwin, M.M. The Sustainability of Sweet Potato Residues from Starch Processing By-Products: Preparation with Lacticaseibacillus rhamnosus and Pediococcus pentosaceus, Characterization, and Application. Foods 2022, 12, 128. [Google Scholar] [CrossRef]
- Kurabachew, M.; Lu, S.H.J.; Krastel, P.; Schmitt, E.K.; Suresh, B.L.; Goh, A.; Knox, J.E.; Ma, N.L.; Jiricek, J.; Beer, D.; et al. Lipiarmycin targets RNA polymerase and has good activity against multidrug-resistant strains of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2008, 62, 713–719. [Google Scholar] [CrossRef]
- Pastorelli, G.; Serra, V.; Turin, L.; Redaelli, V.; Luzi, F.; Barbieri, S. Tranquillizing Effect of Passiflora incarnata Extract: Outcome on Behavioral and Physiological Indicators in Weaning Pigs with Intact Tails. Animals 2022, 12, 203. [Google Scholar] [CrossRef]
- Usui, N.; Togawa, S.; Sumi, T.; Kobayashi, Y.; Koyama, Y.; Nakamura, Y.; Kondo, M.; Shinoda, K.; Kobayashi, H.; Shimada, S. Si-Based Hydrogen-Producing Nanoagent Protects Fetuses from Miscarriage Caused by Mother-to-Child Transmission. Front. Med Technol. 2021, 3, 665506. [Google Scholar] [CrossRef] [PubMed]
- Mao, R.; Teng, D.; Wang, X.; Zhang, Y.; Jiao, J.; Cao, X.; Wang, J. Optimization of expression conditions for a novel NZ2114-derived antimicrobial peptide-MP1102 under the control of the GAP promoter in Pichia pastoris X-33. Microbiol. 2015, 15, 57. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Nguyen, H.M.; Geiger, B.; Mathiesen, G.; Eijsink, V.G.; Peterbauer, C.K.; Haltrich, D.; Nguyen, T.H. Heterologous expression of a recombinant lactobacillal beta-galactosidase in Lactobacillus plantarum: Effect of different parameters on the sakacin P-based expression system. Microb. Cell Factories 2015, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Ichinose, S.; Shibata, N.; Kakeshita, H.; Kodama, H.; Igarashi, K.; Takimura, Y. Inducer-free cellulase production system based on the constitutive expression of mutated XYR1 and ACE3 in the industrial fungus Trichoderma reesei. Sci. Rep. 2022, 12, 19445. [Google Scholar] [CrossRef]
- Kim, D.W.; Kim, J.H.; Park, M.; Yeom, J.H.; Go, H.; Kim, S.; Han, M.S.; Lee, K.; Bae, J. Modulation of biological processes in the nucleus by delivery of DNA oligonucleotides conjugated with gold nanoparticles. Biomaterials 2011, 32, 2593–2604. [Google Scholar] [CrossRef]
- Deng, B.; Yue, Y.; Yang, J.; Yang, M.; Xing, Q.; Peng, H.; Wang, F.; Li, M.; Ma, L.; Zhai, C. Improving the activity and thermostability of PETase from Ideonella sakaiensis through modulating its post-translational glycan modification. Commun. Biol. 2023, 6, 39. [Google Scholar] [CrossRef]
- Cheng, Q.; Tao, J.; Li, Y.; Li, W.; Li, D.; Liu, Y.; Shi, X.; Liu, X.; Zhang, X.; Tong, Y.; et al. Production of nisin and lactic acid from the starch of sweet potato by simultaneous saccharification and fermentation with two stage pH adjustment. 3 Biotech 2021, 11, 320. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Lapthorne, S.; Pereira-Fantini, P.M.; Fouhy, F.; Wilson, G.; Thomas, S.L.; Dellios, N.L.; Scurr, M.; O’sullivan, O.; Ross, R.P.; Stanton, C.; et al. Gut microbial diversity is reduced and is associated with colonic inflammation in a piglet model of short bowel syndrome. Gut Microbes 2013, 4, 212–221. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The pig: A model for human infectious diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Song, L.; Qiao, X.; Zhao, D.; Xie, W.; Bukhari, S.M.; Meng, Q.; Wang, L.; Cui, W.; Jiang, Y.; Zhou, H.; et al. Effects of Lactococcus lactis MG1363 producing fusion proteins of bovine lactoferricin–lactoferrampin on growth, intestinal morphology and immune function in weaned piglet. J. Appl. Microbiol. 2019, 127, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Spinler, J.K.; Taweechotipatr, M.; Rognerud, C.L.; Ou, C.N.; Tumwasorn, S.; Versalovic, J. Human-derived probiotic Lactobacillus reuteri demonstrate antimicrobial activities targeting diverse enteric bacterial pathogens. Anaerobe 2008, 14, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Valeur, N.; Engel, P.; Carbajal, N.; Connolly, E.; Ladefoged, K. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 2004, 70, 1176–1181. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, K.; Zuo, P.; Liu, Y.; Zhang, M.; Xie, S.; Zhang, H.; Chen, X.; Liu, C. Early decrease in blood platelet count is associated with poor prognosis in COVID-19 patients-indications for predictive, preventive, and personalized medical approach. EPMA J. 2020, 11, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Liu, J.; Li, B.; Wei, X.; Qi, Y.; Zhang, B.; Liu, H.; Xiao, P. Comparison of blood tonic efficacy and chemical constituents of Kadsura interior A.C. Smith and its closely related species. Chin. Med. 2022, 17, 14. [Google Scholar] [CrossRef]
- Van De Bruaene, A.; Delcroix, M.; Pasquet, A.; De Backer, J.; De Pauw, M.; Naeije, R.; Vachiery, J.L.; Paelinck, B.; Morissens, M.; Budts, W. Iron deficiency is associated with adverse outcome in Eisenmenger patients. Eur. Heart J. 2011, 32, 2790–2799. [Google Scholar] [CrossRef]
- Zhou, J.; Yue, J.; Yao, Y.; Hou, P.; Zhang, T.; Zhang, Q.; Yi, L.; Mi, M. Dihydromyricetin Protects Intestinal Barrier Integrity by Promoting IL-22 Expression in ILC3s through the AMPK/SIRT3/STAT3 Signaling Pathway. Nutrients 2023, 15, 355. [Google Scholar] [CrossRef]
- Vergnolle, N.; Cirillo, C. Neurons and Glia in the Enteric Nervous System and Epithelial Barrier Function. Physiology 2018, 33, 269–280. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, P.; Chen, N.; Liu, Y.; Liu, D.; Guo, Y. Effects of housing systems and glucose oxidase on growth performance and intestinal health of Beijing You Chickens. Poult. Sci. 2021, 100, 100943. [Google Scholar] [CrossRef]
- Shi, C.Z.; Chen, H.Q.; Liang, Y.; Xia, Y.; Yang, Y.Z.; Yang, J.; Zhang, J.D.; Wang, S.H.; Liu, J.; Qin, H.L. Combined probiotic bacteria promotes intestinal epithelial barrier function in interleukin-10-gene-deficient mice. World J. Gastroenterol. 2014, 20, 4636–4647. [Google Scholar] [CrossRef]
- Wardill, H.R.; Bowen, J.M.; Al-Dasooqi, N.; Sultani, M.; Bateman, E.; Stansborough, R.; Shirren, J.; Gibson, R.J. Irinotecan disrupts tight junction proteins within the gut: Implications for chemotherapy-induced gut toxicity. Cancer Biol. Ther. 2014, 15, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, J.J.; Martínez-Banaclocha, H.; Angosto-Bazarra, D.; de Torre-Minguela, C.; Baroja-Mazo, A.; Alarcón-Vila, C.; Martínez-Alarcón, L.; Amores-Iniesta, J.; Martín-Sánchez, F.; Ercole, G.A.; et al. P2X7 receptor induces mitochondrial failure in monocytes and compromises NLRP3 inflammasome activation during sepsis. Nat. Commun. 2019, 10, 2711. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Zheng, N.; Luo, Q.; Jiang, L.; He, B.; Yuan, X.; Shen, L. Probiotics Lactobacillus reuteri Abrogates Immune Checkpoint Blockade-Associated Colitis by Inhibiting Group 3 Innate Lymphoid Cells. Front. Immunol. 2019, 10, 1235. [Google Scholar] [CrossRef] [PubMed]
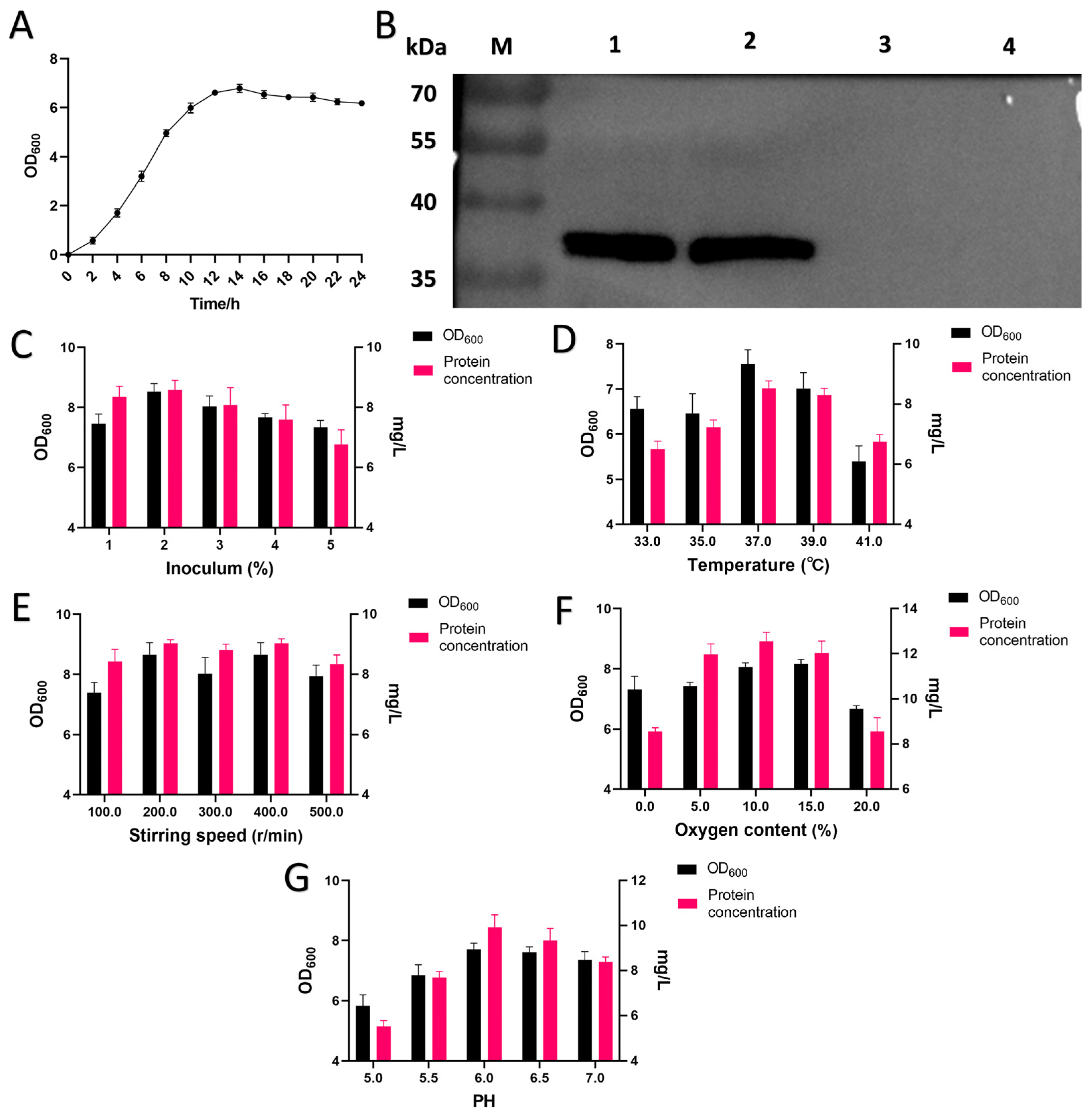
| Run | Temperature (°C)—X1 | PH—X2 | Inoculum (%)—X3 | Oxygen Content (%)—X4 | Stirring Speed (r/min)—X5 | Biomass (OD600)—Y1 | Protein Concentration (mg/L)—Y2 |
|---|---|---|---|---|---|---|---|
| 1 | 39 | 7 | 3 | 5 | 100 | 7.0081 | 12.224 |
| 2 | 35 | 5 | 1 | 5 | 100 | 5.3086 | 7.296 |
| 3 | 39 | 7 | 1 | 5 | 100 | 6.7272 | 11.942 |
| 4 | 39 | 7 | 1 | 15 | 300 | 8.9154 | 14.769 |
| 5 | 39 | 5 | 1 | 5 | 300 | 5.2155 | 9.927 |
| 6 | 39 | 5 | 3 | 15 | 300 | 6.0287 | 10.951 |
| 7 | 35 | 5 | 3 | 5 | 300 | 5.8037 | 7.884 |
| 8 | 35 | 5 | 1 | 15 | 100 | 6.0858 | 9.516 |
| 9 | 35 | 7 | 1 | 15 | 300 | 8.7379 | 11.984 |
| 10 | 35 | 7 | 3 | 5 | 300 | 8.0638 | 10.748 |
| 11 | 35 | 7 | 3 | 15 | 100 | 9.5285 | 11.804 |
| 12 | 39 | 5 | 3 | 15 | 100 | 5.9826 | 10.968 |
| Source (Biomass) | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | Remark |
|---|---|---|---|---|---|---|
| Model | 22.38 | 5 | 4.48 | 38.04 | 0.0002 | significant |
| X1 | 0.94 | 1 | 0.94 | 7.95 | 0.0304 | * |
| X2 | 16.94 | 1 | 16.94 | 143.93 | <0.0001 | ** |
| X3 | 0.11 | 1 | 0.11 | 0.90 | 0.3803 | |
| X4 | 3.91 | 1 | 3.91 | 33.25 | 0.0012 | ** |
| X5 | 0.49 | 1 | 0.49 | 4.16 | 0.0874 | |
| Pure error | 0.71 | 6 | 0.12 | |||
| Total | 23.09 | 11 | ||||
| Source (protein concentration) | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | Remark |
| Model | 43.87 | 5 | 8.77 | 61.47 | <0.0001 | significant |
| X1 | 11.11 | 1 | 11.11 | 77.88 | 0.0001 | ** |
| X2 | 23.88 | 1 | 23.88 | 167.33 | <0.0001 | ** |
| X3 | 0.061 | 1 | 0.061 | 0.43 | 0.5378 | |
| X4 | 8.29 | 1 | 8.29 | 58.05 | 0.0003 | ** |
| X5 | 0.53 | 1 | 0.53 | 3.69 | 0.1032 | |
| Pure error | 0.86 | 6 | 0.14 | |||
| Total | 44.73 | 11 |
| Run | PH—A | Temperature (°C)—B | Oxygen Content (%)—C | Biomass (OD600)—R1 | Protein Concentration (mg/L)—R2 |
|---|---|---|---|---|---|
| 1 | 6 | 34 | 5 | 8.335 | 15.732 |
| 2 | 7 | 34 | 10 | 8.3025 | 13.696 |
| 3 | 5 | 37 | 15 | 7.1925 | 16.506 |
| 4 | 7 | 40 | 10 | 8.0911 | 14.238 |
| 5 | 6 | 34 | 15 | 7.7075 | 16.801 |
| 6 | 5 | 35 | 10 | 7.1625 | 16.448 |
| 7 | 6 | 37 | 10 | 9.3807 | 17.05 |
| 8 | 6 | 40 | 15 | 7.2675 | 14.968 |
| 9 | 6 | 37 | 10 | 9.0942 | 16.923 |
| 10 | 6 | 40 | 5 | 7.5275 | 15.227 |
| 11 | 6 | 37 | 10 | 9.0732 | 17.128 |
| 12 | 7 | 37 | 15 | 7.7175 | 13.49 |
| 13 | 5 | 37 | 5 | 7.1702 | 15.529 |
| 14 | 5 | 40 | 10 | 7.251 | 14.946 |
| 15 | 7 | 37 | 5 | 8.9599 | 14.023 |
| 16 | 6 | 37 | 10 | 9.2178 | 16.936 |
| 17 | 6 | 37 | 10 | 9.1236 | 17.172 |
| Source (R1) | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | Remark |
|---|---|---|---|---|---|---|
| Model | 11.02 | 9 | 1.22 | 34.36 | <0.0001 | significant |
| A | 2.31 | 1 | 2.31 | 64.68 | <0.0001 | ** |
| B | 0.23 | 1 | 0.23 | 6.58 | 0.0372 | * |
| C | 0.56 | 1 | 0.56 | 15.58 | 0.0056 | ** |
| AB | 0.022 | 1 | 0.02 | 0.63 | 0.4532 | |
| AC | 0.4 | 1 | 0.40 | 11.22 | 0.0123 | * |
| BC | 0.034 | 1 | 0.03 | 0.95 | 0.3629 | |
| A2 | 2.14 | 1 | 2.14 | 60 | 0.0001 | |
| B2 | 2.45 | 1 | 2.45 | 68.83 | <0.0001 | |
| C2 | 2.09 | 1 | 2.09 | 58.73 | 0.0001 | |
| Residual | 0.25 | 7 | 0.036 | |||
| Lack of Fit | 0.19 | 3 | 0.062 | 3.9 | 0.111 | not significant |
| Pure Error | 0.064 | 4 | 0.016 | |||
| Total | 11 | 16 | ||||
| R2 = 0.9779 | (C.V.%) = 2.32 | |||||
| Source (R2) | Sum of Squares | Degree of Freedom | Mean Square | F-Value | p-Value | Remark |
| Model | 29.07 | 9 | 3.23 | 35.89 | <0.0001 | significant |
| A | 7.96 | 1 | 7.96 | 88.48 | <0.0001 | ** |
| B | 1.36 | 1 | 1.36 | 15.11 | 0.006 | ** |
| C | 0.2 | 1 | 0.2 | 2.18 | 0.183 | |
| AB | 1.04 | 1 | 1.04 | 11.6 | 0.0113 | * |
| AC | 0.57 | 1 | 0.57 | 6.33 | 0.04 | * |
| BC | 0.44 | 1 | 0.44 | 4.9 | 0.0625 | |
| A2 | 10.81 | 1 | 10.81 | 120.12 | <0.0001 | |
| B2 | 2.74 | 1 | 2.74 | 30.5 | 0.0009 | |
| C2 | 2.38 | 1 | 2.38 | 26.48 | 0.0013 | |
| Residual | 0.63 | 7 | 0.09 | |||
| Lack of Fit | 0.4 | 3 | 0.13 | 2.24 | 0.2257 | not significant |
| Pure Error | 0.23 | 4 | 0.059 | |||
| Total | 30 | 16 | ||||
| R2 = 0.9788 | (C.V.%) = 1.90 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, W.; Wang, X.; Cai, J.; Bai, H.; Shao, Y.; Li, Z.; Cai, L.; Zhang, S.; Li, J.; Cui, W.; et al. Optimum Fermentation Conditions for Bovine Lactoferricin-Lactoferrampin-Encoding LimosiLactobacillus reuteri and Regulation of Intestinal Inflammation. Foods 2023, 12, 4068. https://doi.org/10.3390/foods12224068
Xie W, Wang X, Cai J, Bai H, Shao Y, Li Z, Cai L, Zhang S, Li J, Cui W, et al. Optimum Fermentation Conditions for Bovine Lactoferricin-Lactoferrampin-Encoding LimosiLactobacillus reuteri and Regulation of Intestinal Inflammation. Foods. 2023; 12(22):4068. https://doi.org/10.3390/foods12224068
Chicago/Turabian StyleXie, Weichun, Xueying Wang, Jiyao Cai, Huitao Bai, Yilan Shao, Zhuoran Li, Limeng Cai, Senhao Zhang, Jiaxuan Li, Wen Cui, and et al. 2023. "Optimum Fermentation Conditions for Bovine Lactoferricin-Lactoferrampin-Encoding LimosiLactobacillus reuteri and Regulation of Intestinal Inflammation" Foods 12, no. 22: 4068. https://doi.org/10.3390/foods12224068
APA StyleXie, W., Wang, X., Cai, J., Bai, H., Shao, Y., Li, Z., Cai, L., Zhang, S., Li, J., Cui, W., Jiang, Y., & Tang, L. (2023). Optimum Fermentation Conditions for Bovine Lactoferricin-Lactoferrampin-Encoding LimosiLactobacillus reuteri and Regulation of Intestinal Inflammation. Foods, 12(22), 4068. https://doi.org/10.3390/foods12224068