Identification of Key Genes Affecting Flavor Formation in Beijing-You Chicken Meat by Transcriptome and Metabolome Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Animals
2.3. RNA Extraction and Sequencing
2.4. Analysis of Gene Expression Data
2.5. Metabolite Extraction and LC-MS/MS Conditions
2.5.1. Metabolite Extraction
2.5.2. LC-MS/MS Conditions
2.6. Qualitative, Quantitative and Statistical Analysis of Metabolites
2.7. Integrative Analysis of Metabolomic and Transcriptomic Datasets
3. Results
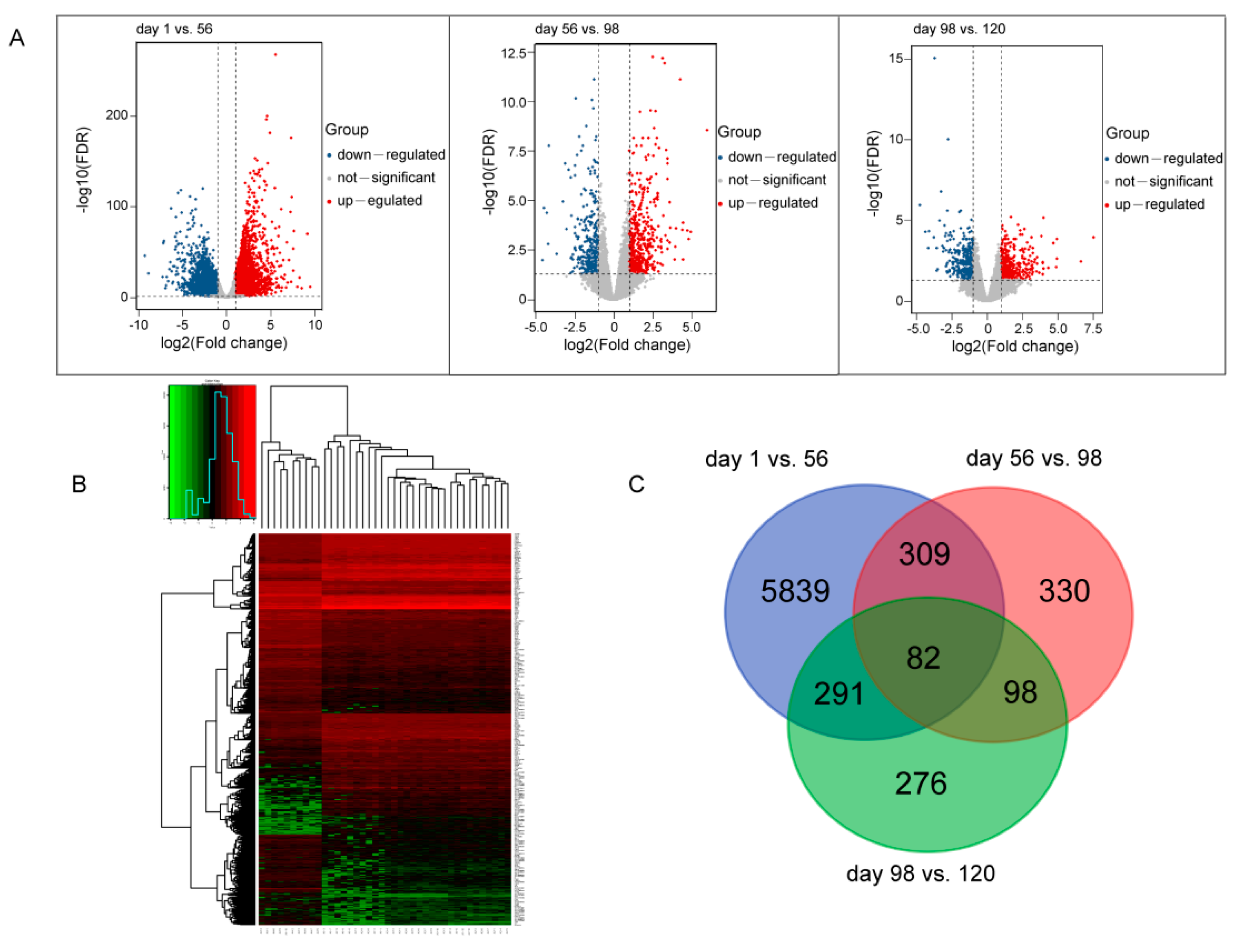
3.1. Transcriptome and DEGs Analysis
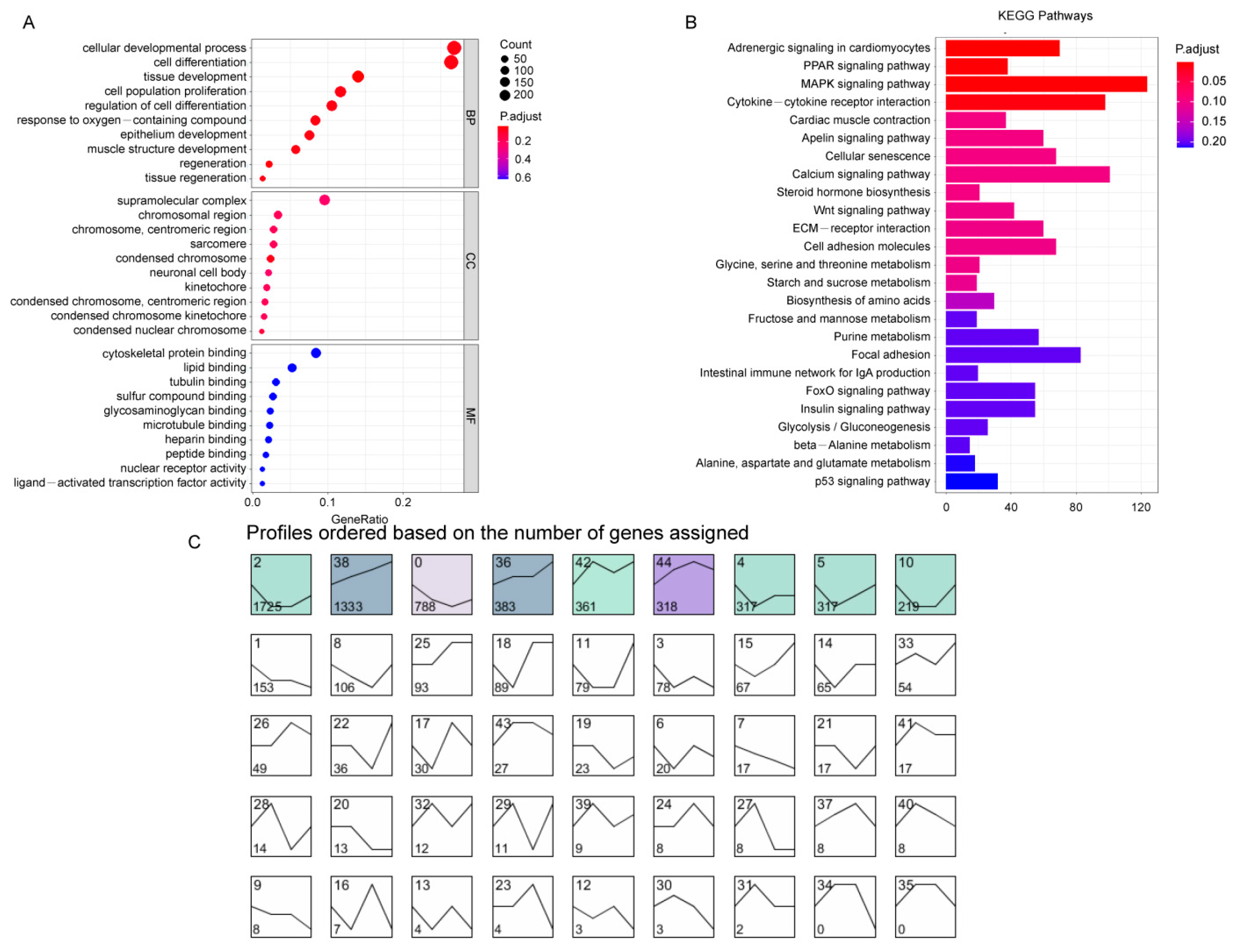
3.2. GO and KEGG Enrichment Analysis of DEGs
3.3. Time Series Analysis of DEGs
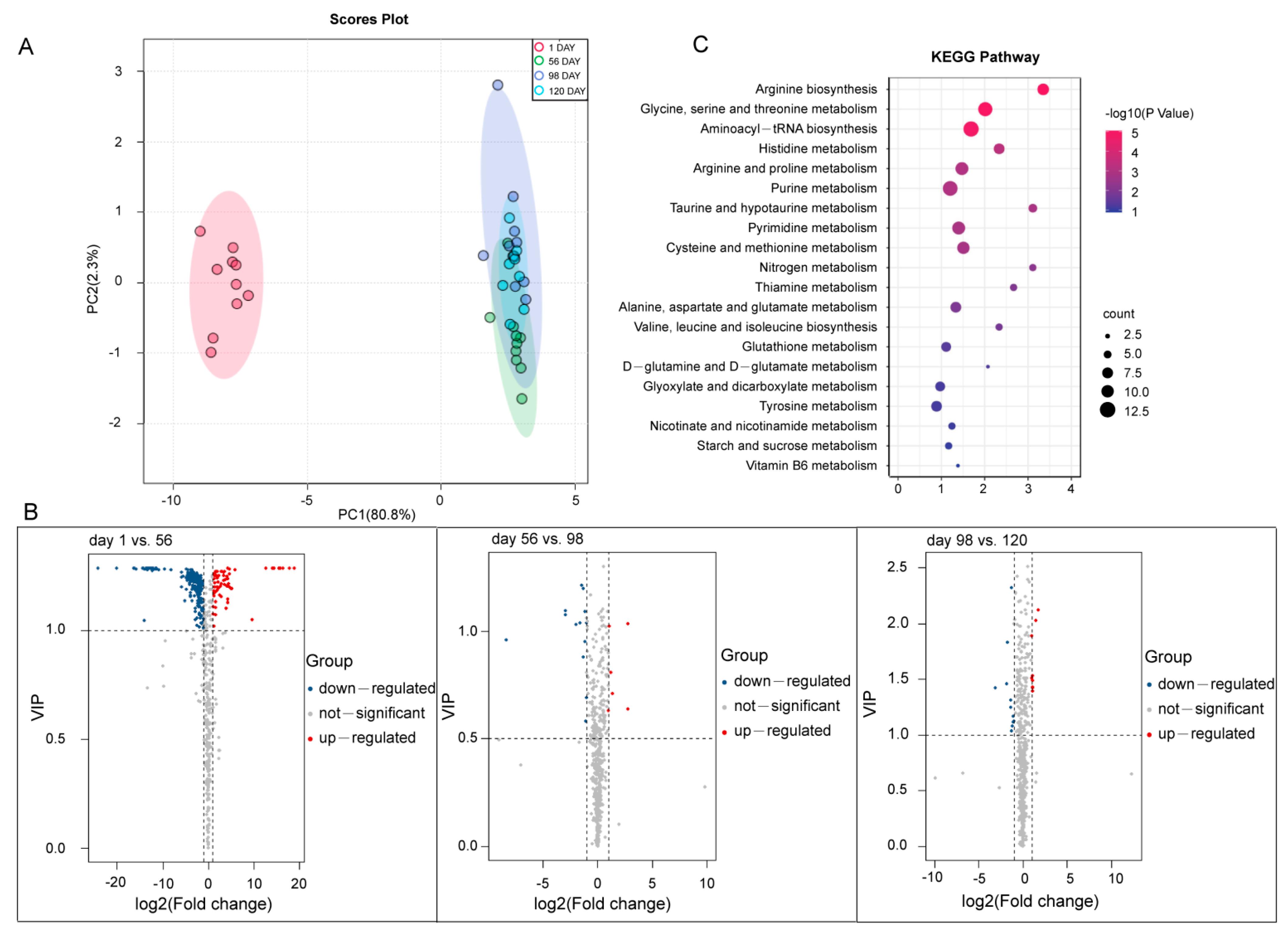
3.4. Metabolomic Data and SCMs Analysis
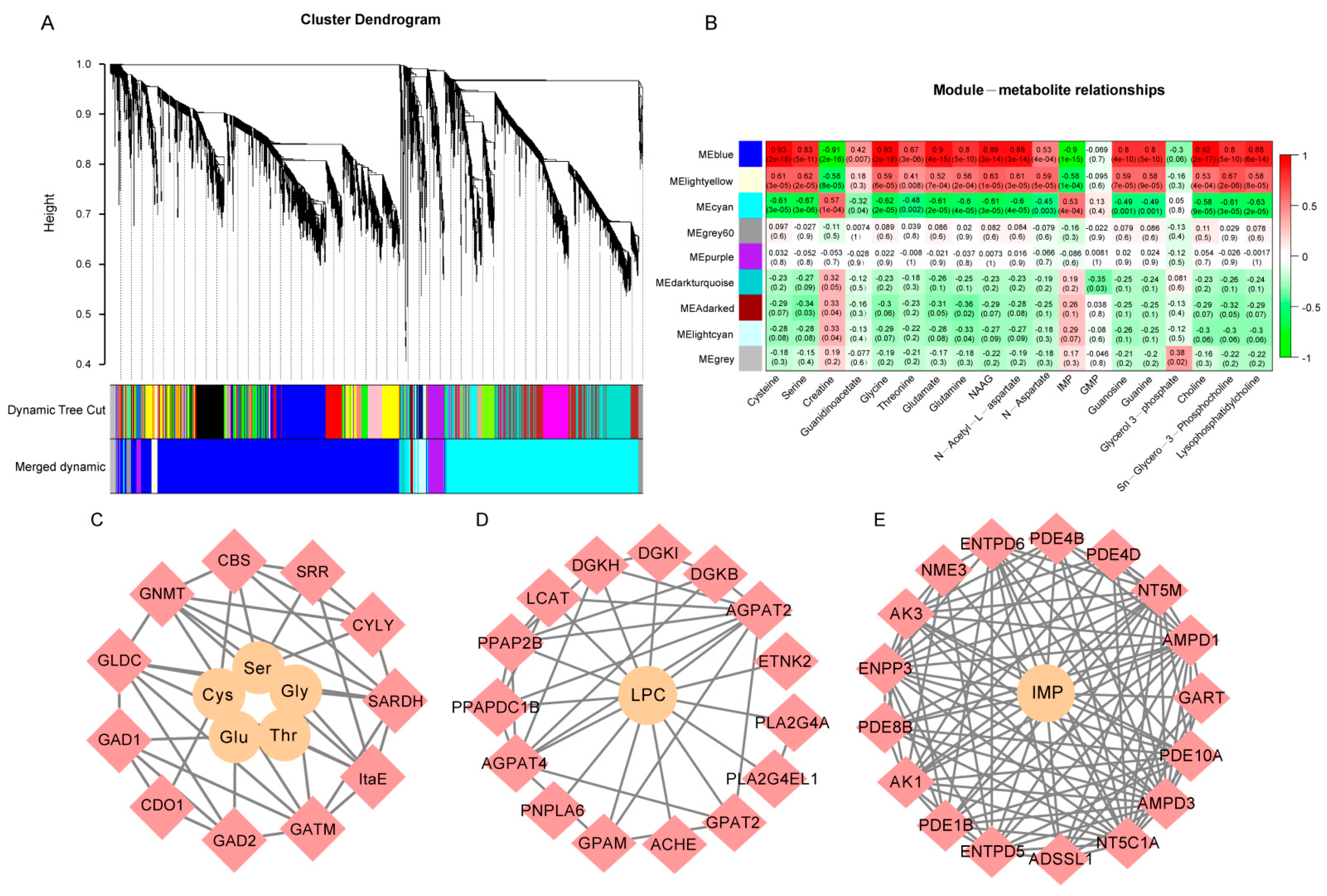
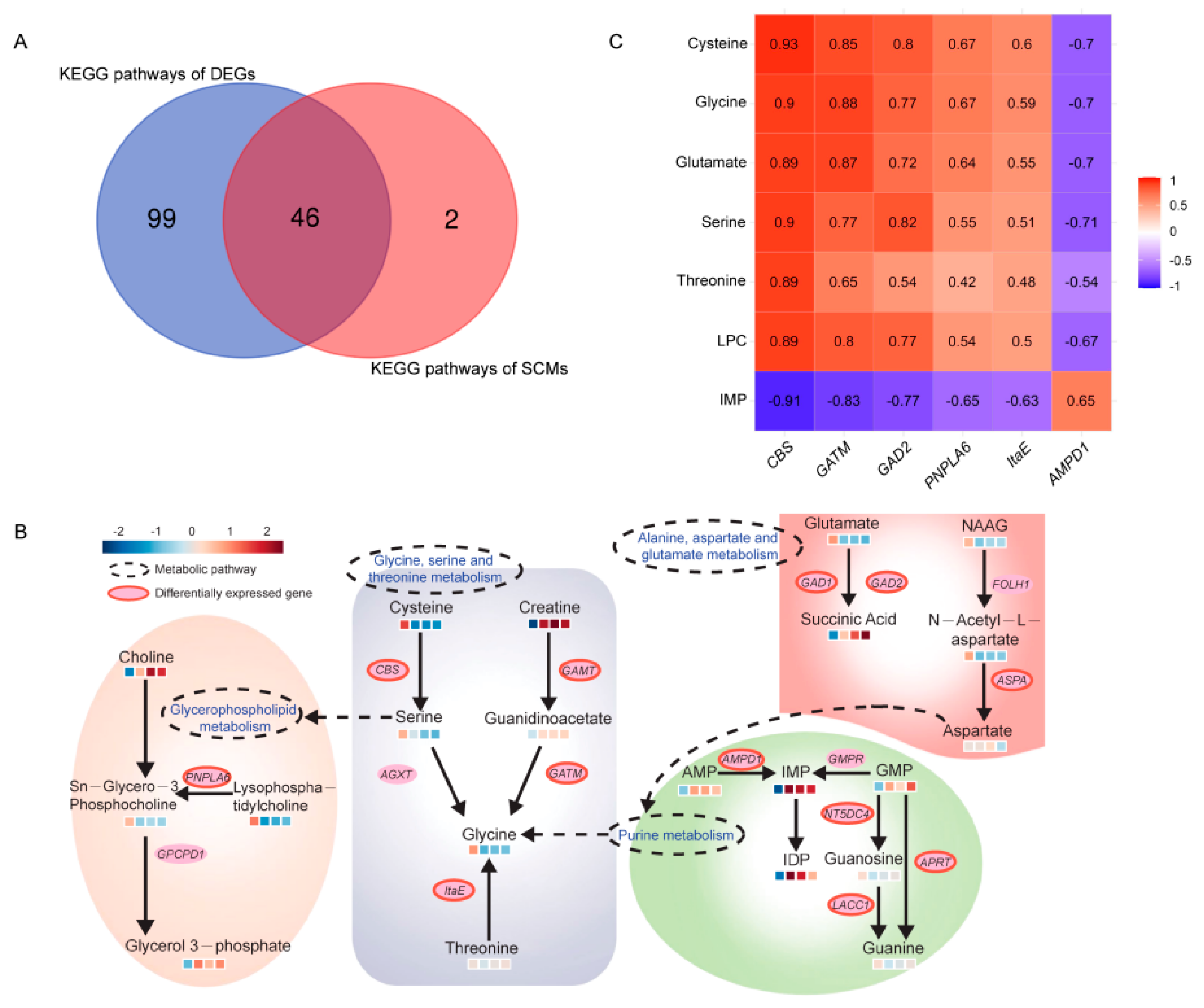
3.5. Joint Analysis of Transcriptomic and Metabolomic Data
3.6. Generation of Flavor Metabolic Regulatory Networks
3.7. Integrated Analysis of Flavor Formation during BJY Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mir, N.A.; Rafiq, A.; Kumar, F.; Singh, V.; Shukla, V. Determinants of broiler chicken meat quality and factors affecting them: A review. J. Food Sci. Technol. 2017, 54, 2997–3009. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yan, W.; Fu, Y.; Wang, L.; Lv, X.; Dai, R.; Li, X.; Jia, F. The Use of Electronic Nose in the Quality Evaluation and Adulteration Identification of Beijing-You Chicken. Foods 2022, 11, 782. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.I.; Jo, C.; Tariq, M.R. Meat flavor precursors and factors influencing flavor precursors—A systematic review. Meat Sci. 2015, 110, 278–284. [Google Scholar] [CrossRef]
- Wang, R.; Shu, P.; Zhang, C.; Zhang, J.; Chen, Y.; Zhang, Y.; Du, K.; Xie, Y.; Li, M.; Ma, T.; et al. Integrative analyses of metabolome and genome-wide transcriptome reveal the regulatory network governing flavor formation in kiwifruit (Actinidia chinensis). New Phytol. 2022, 233, 373–389. [Google Scholar] [CrossRef]
- Mottram, D.S. Flavour formation in meat and meat products: A review. Food Chem. 1998, 62, 415–424. [Google Scholar] [CrossRef]
- Jayasena, D.D.; Ahn, D.U.; Nam, K.C.; Jo, C. Flavour Chemistry of Chicken Meat: A Review. Asian Australas. J. Anim. Sci. 2013, 26, 732–742. [Google Scholar] [CrossRef]
- Sasaki, K.; Motoyama, M.; Mitsumoto, M. Changes in the amounts of water-soluble umami-related substances in porcine longissimus and biceps femoris muscles during moist heat cooking. Meat Sci. 2007, 77, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.J.; Choe, J.H.; Jung, Y.K.; Kruk, Z.A.; Lim, D.G.; Jo, C.R. Comparison of the Chemical Composition, Textural Characteristics, and Sensory Properties of North and South Korean Native Chickens and Commercial Broilers. Food Sci. Anim. Resour. 2010, 30, 171–178. [Google Scholar] [CrossRef] [Green Version]
- Kiyohara, R.; Yamaguchi, S.; Rikimaru, K.; Takahashi, H. Supplemental arachidonic acid-enriched oil improves the taste of thigh meat of Hinai-jidori chickens. Poultry Sci. 2011, 90, 1817–1822. [Google Scholar] [CrossRef]
- Choe, J.H.; Nam, K.C.; Jung, S.; Kim, B.; Yun, H.J.; Jo, C.R. Differences in the Quality Characteristics between Commercial Korean Native Chickens and Broilers. Food Sci. Anim. Resour. 2010, 30, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Mottram, D.S.; Edwards, R.A. The role of triglycerides and phospholipids in the aroma of cooked beef. J. Sci. Food Agric. 1983, 34, 517–522. [Google Scholar] [CrossRef]
- Miriam, A.; Almudena, F.; Yolanda, N.; Rita, B.; Beatriz, I.; Ana, I.F.; Ana, I.R.; Antonio, G.B.; Juan, F.M.; Ángela, C.; et al. Developmental Stage, Muscle and Genetic Type Modify Muscle Transcriptome in Pigs: Effects on Gene Expression and Regulatory Factors Involved in Growth and Metabolism. PLoS ONE. 2016, 11, e0167858. [Google Scholar]
- Zhan, H.; Xiong, Y.; Wang, Z.; Dong, W.; Zhou, Q.; Xie, S.; Li, X.; Zhao, S.; Ma, Y. Integrative analysis of transcriptomic and metabolomic profiles reveal the complex molecular regulatory network of meat quality in Enshi black pigs. Meat Sci. 2022, 183, 108642. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.P.; Cui, H.X.; Liu, R.R.; Zheng, M.Q.; Chen, J.L.; Wen, J. Comparison of breast muscle meat quality in 2 broiler breeds. Poultry Sci. 2011, 49, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.; Cui, H.; Yu, J.; Hayat, K.; Wu, X.; Zhang, X.; Ho, C.T. Characteristic flavor formation of thermally processed N-(1-deoxy-α-d-ribulos-1-yl)-glycine: Decisive role of additional amino acids and promotional effect of glyoxal. Food Chem. 2022, 371, 131–137. [Google Scholar] [CrossRef]
- Shahidi, F. Flavor of meat and meat products—An overview. In Flavor of Meat and Meat Products; Shahidi, F., Ed.; Spring: Boston, MA, USA, 1994; pp. 1–3. [Google Scholar]
- Cerny, C.; Davidek, T. Formation of Aroma Compounds from Ribose and Cysteine during the Maillard Reaction. J. Agric. Food Chem. 2003, 51, 2714–2721. [Google Scholar] [CrossRef]
- Ma, X.; Yu, M.; Liu, Z.; Deng, D.; Cui, Y.; Tian, Z.; Wang, G. Effect of amino acids and their derivatives on meat quality of finishing pigs. J. Food Sci. Technol. 2020, 57, 404–412. [Google Scholar] [CrossRef]
- Li, W.; Pierce, B.S. Steady-state substrate specificity and O2-coupling efficiency of mouse cysteine dioxygenase. Arch. Biochem. Biophys. 2015, 565, 49–56. [Google Scholar] [CrossRef]
- Tolosa, Z.A.; Maslova, P.N.; Goriachenkova, E.V. [Substrate specificity of cysteine lyase]. Biokhimiia 1975, 40, 248–256. [Google Scholar]
- Conter, C.; Fruncillo, S.; Fernández, R.C.; Martínez-Cruz, L.A.; Dominici, P.; Astegno, A. Cystathionine β-synthase is involved in cysteine biosynthesis and H2S generation in Toxoplasma gondii. Sci. Rep. 2020, 10, 14657. [Google Scholar] [CrossRef]
- Xiao, H.; Wang, J.; Barwick, S.R.; Yoon, Y.; Smith, S.B. Effect of long-term chronic hyperhomocysteinemia on retinal structure and function in the cystathionine-β-synthase mutant mouse. Exp. Eye Res. 2022, 214, 108894. [Google Scholar] [CrossRef] [PubMed]
- Shaposhnikov, M.V.; Zakluta, A.S.; Zemskaya, N.V.; Guvatova, Z.G.; Shilova, V.Y.; Yakovleva, D.V.; Gorbunova, A.A.; Koval, L.A.; Ulyasheva, N.S.; Evgen’ev, M.B.; et al. Deletions of the cystathionine-β-synthase (CBS) and cystathionine-γ-lyase (CSE) genes, involved in the control of hydrogen sulfide biosynthesis, significantly affect lifespan and fitness components of Drosophila melanogaster. Mech. Ageing Dev. 2022, 203, 111656. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.J.; Ho, N.A.T.; Pack, S.P. Buffer-free production of gamma-aminobutyric acid using an engineered glutamate decarboxylase from Escherichia coli. Enzyme Microb. Technol. 2013, 53, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Tian, L.; Zhang, C.; Yuan, K.; Xu, S. Genetic diversity and natural selection footprints of the glycine amidinotransferase gene in various human populations. Sci. Rep. 2016, 6, 18755. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zhang, M.; Chen, X.; Zhang, W.; Ding, W.; Zhang, Q. Evolution of Threonine Aldolases, a Diverse Family Involved in the Second Pathway of Glycine Biosynthesis. J. Mol. Evol. 2015, 80, 102–107. [Google Scholar] [CrossRef]
- Kienesberger, P.C.; Oberer, M.; Lass, A.; Zechner, R. Mammalian patatin domain containing proteins: A family with diverse lipolytic activities involved in multiple biological functions. J. Lipid Res. 2009, 50, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Tienhoven, M.V.; Atkins, J.; Li, Y.; Glynn, P. Human Neuropathy Target Esterase Catalyzes Hydrolysis of Membrane Lipids*. J. Biol. Chem. 2002, 277, 20942–20948. [Google Scholar] [CrossRef] [Green Version]
- Elizabeth, R.S.; Alexander, D.L.; Doris, K. Disease-Associated PNPLA6 Mutations Maintain Partial Functions When Analyzed in Drosophila. Front. Neurosci. 2019, 13, 1207. [Google Scholar]
- Ye, G.m.; Chen, C.; Huang, S.; Han, D.D.; Guo, J.H.; Wan, B.; Yu, L. Cloning and characterization a novel human 1-acyl-sn-glycerol-3-phosphate acyltransferase gene AGPAT7. DNA Seq. 2005, 16, 386–390. [Google Scholar] [CrossRef]
- Suja, S.; Barnes, R.I.; Garg, A.; Agarwal, A.K. Functional characterization of the human 1-acylglycerol-3-phosphate-O-acyltransferase isoform 10/glycerol-3-phosphate acyltransferase isoform 3. J. Mol. Endocrinol. 2009, 42, 469–478. [Google Scholar]
- Vance, J.E. Phospholipid Synthesis and Transport in Mammalian Cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef]
- Agarwal, A.K.; Garg, A. Congenital generalized lipodystrophy: Significance of triglyceride biosynthetic pathways. Trends Endocrinol. Metab. 2003, 14, 214–221. [Google Scholar] [CrossRef]
- Coleman, R.A.; Lee, D.P. Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 2004, 43, 134–176. [Google Scholar] [CrossRef]
- Kawai, M.; Okiyama, A.; Ueda, Y. Taste Enhancements Between Various Amino Acids and IMP. Chem. Senses 2002, 27, 739–745. [Google Scholar] [CrossRef] [Green Version]
- Kuchiba-Manabe, M.; Matoba, T.; Hasegawa, K. Sensory Changes in Umami Taste of Inosine 5′-Monophosphate Solution after Heating. J. Food Sci. 1991, 56, 1429–1432. [Google Scholar] [CrossRef]
- Katemala, S.; Molee, A.; Thumanu, K.; Yongsawatdigul, J. Meat quality and Raman spectroscopic characterization of Korat hybrid chicken obtained from various rearing periods. Poultry Sci. 2021, 100, 1248–1261. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Functional role and properties of AMP-deaminase. Biokhimiia 1996, 61, 195–211. [Google Scholar] [PubMed]
- Lane, A.N.; Fan, T.W.M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.F.; Sun, H.M.; Cheng, X.Y.; Li, Y.J.; Zhao, Y.L.; Mei, W.X.; Wei, X.; Zhou, H.R.; Du, Y.B.; Zeng, C.C. Dietary intake of fructose increases purine de novo synthesis: A crucial mechanism for hyperuricemia. Front. Nutr. 2022, 9, 2971. [Google Scholar] [CrossRef] [PubMed]
| Pathway | Pathway ID | Gene_ID | Differentially Expressed Genes |
|---|---|---|---|
| ECM–receptor interaction | gga04512 | 395210, 373987, 414837 | SPP1, THBS1, THBS2 |
| PPAR signaling pathway | gga03320 | 422345 | ACSL4 |
| Focal adhesion | gga04510 | 395210, 373987, 414837 | SPP1, THBS1, THBS2 |
| Melanogenesis | gga04916 | 408082, 395703 | EDNRB2, WNT11B |
| Calcium signaling pathway | gga04020 | 408082, 395971, 428149 | EDNRB2, ADRB1, TBXA2R |
| Fatty acid biosynthesis | gga00061 | 422345 | ACSL4 |
| Apelin signaling pathway | gga04371 | 395210 | SPP1 |
| Phagosome | gga04145 | 373987, 414837 | THBS1, THBS2 |
| DNA replication | gga03030 | 423688 | DNA2 |
| Fatty acid degradation | gga00071 | 422345 | ACSL4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gai, K.; Ge, Y.; Liu, D.; Zhang, H.; Cong, B.; Guo, S.; Liu, Y.; Xing, K.; Qi, X.; Wang, X.; et al. Identification of Key Genes Affecting Flavor Formation in Beijing-You Chicken Meat by Transcriptome and Metabolome Analyses. Foods 2023, 12, 1025. https://doi.org/10.3390/foods12051025
Gai K, Ge Y, Liu D, Zhang H, Cong B, Guo S, Liu Y, Xing K, Qi X, Wang X, et al. Identification of Key Genes Affecting Flavor Formation in Beijing-You Chicken Meat by Transcriptome and Metabolome Analyses. Foods. 2023; 12(5):1025. https://doi.org/10.3390/foods12051025
Chicago/Turabian StyleGai, Kai, Yu Ge, Dapeng Liu, He Zhang, Bailin Cong, Shihao Guo, Yizheng Liu, Kai Xing, Xiaolong Qi, Xiangguo Wang, and et al. 2023. "Identification of Key Genes Affecting Flavor Formation in Beijing-You Chicken Meat by Transcriptome and Metabolome Analyses" Foods 12, no. 5: 1025. https://doi.org/10.3390/foods12051025





