Glycosyltransferases Expression Changes in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 Grown on Different Carbon Sources
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Aerobic Culture Conditions
2.3. Anaerobic Culture Conditions
2.4. Biomass and pH Determination
2.5. Protein Determination
2.6. GTFs Activity Determination
2.7. Zymograms
2.8. RNA-seq Analysis
2.9. Differential Gene Expression Analysis
2.10. Operon Search
2.11. Motif Search
2.12. RT-qPCR
3. Results and Discussion
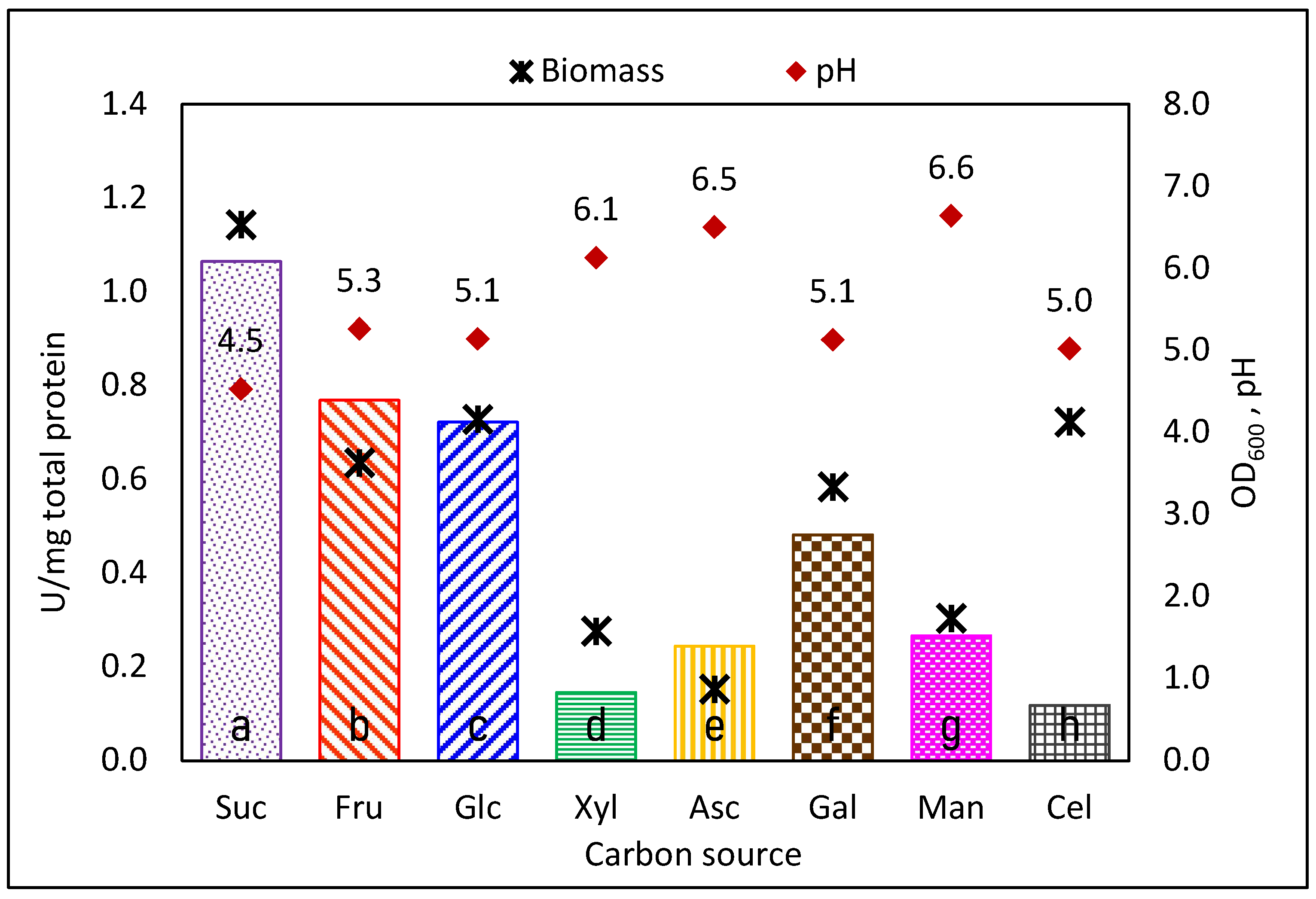
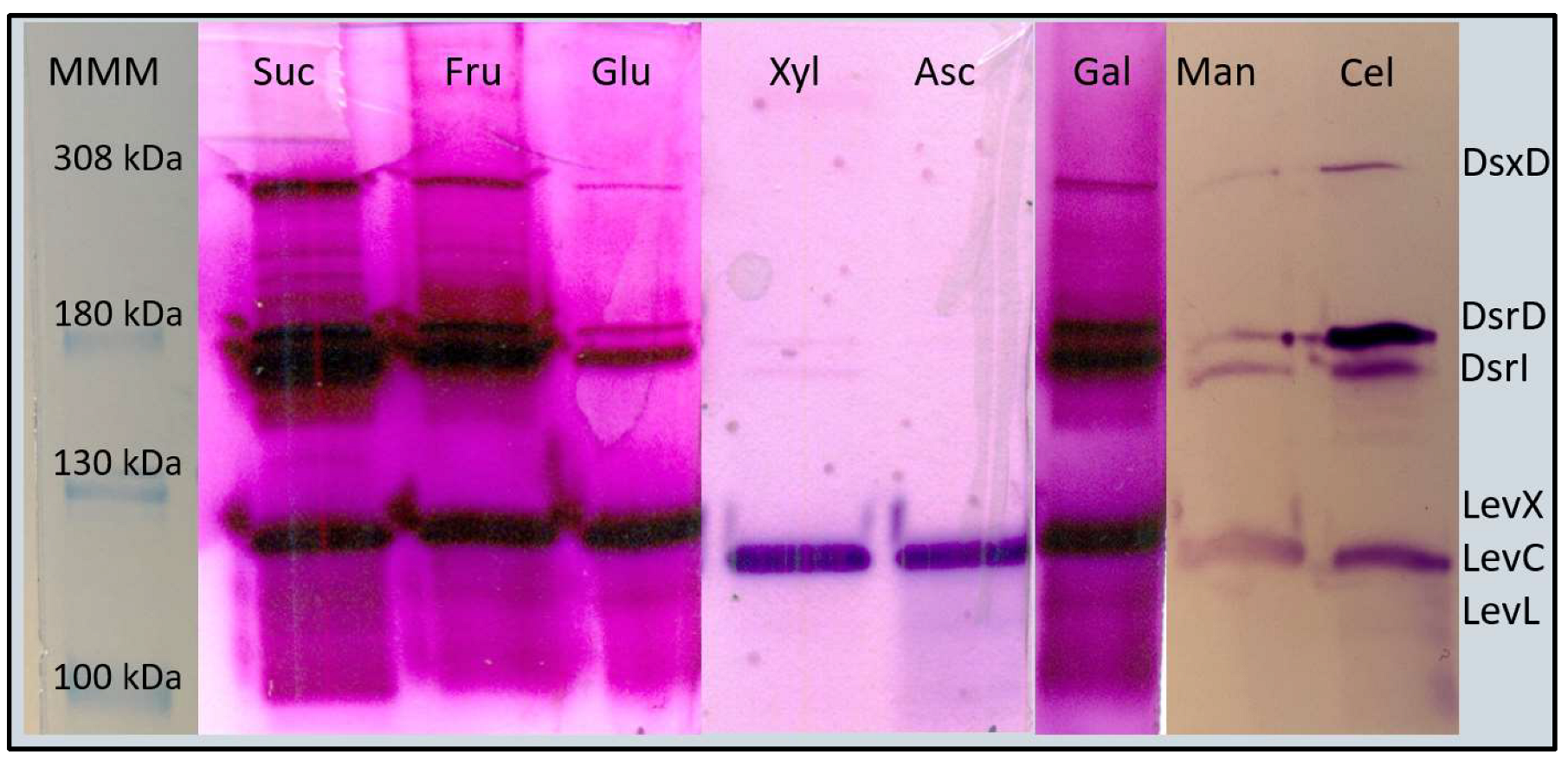
3.1. Specific Total Activity Decreases in CS Different from Sucrose
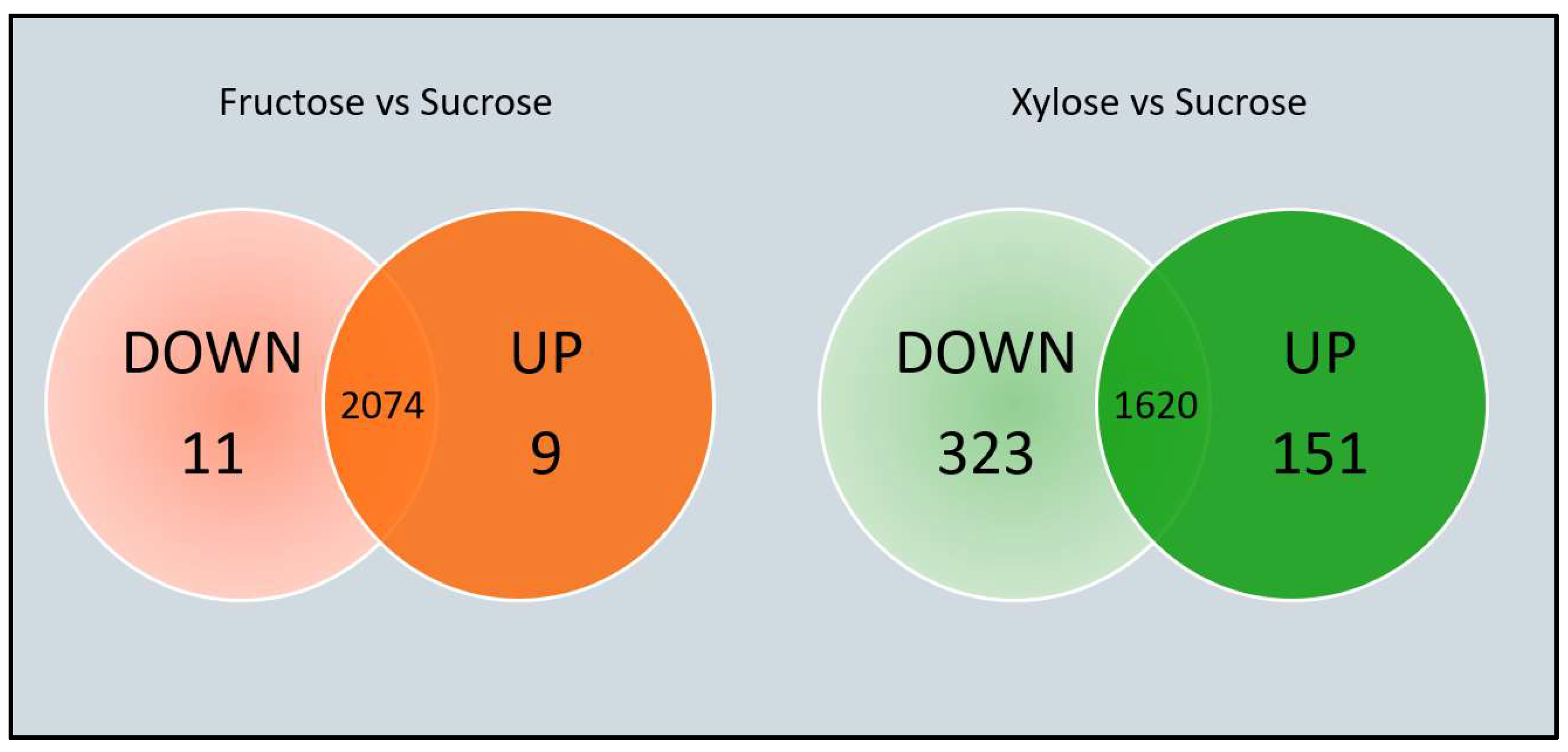
3.2. Differential Expression Analysis (DEA): Using Xylose Produces Major Changes in Gene Expression
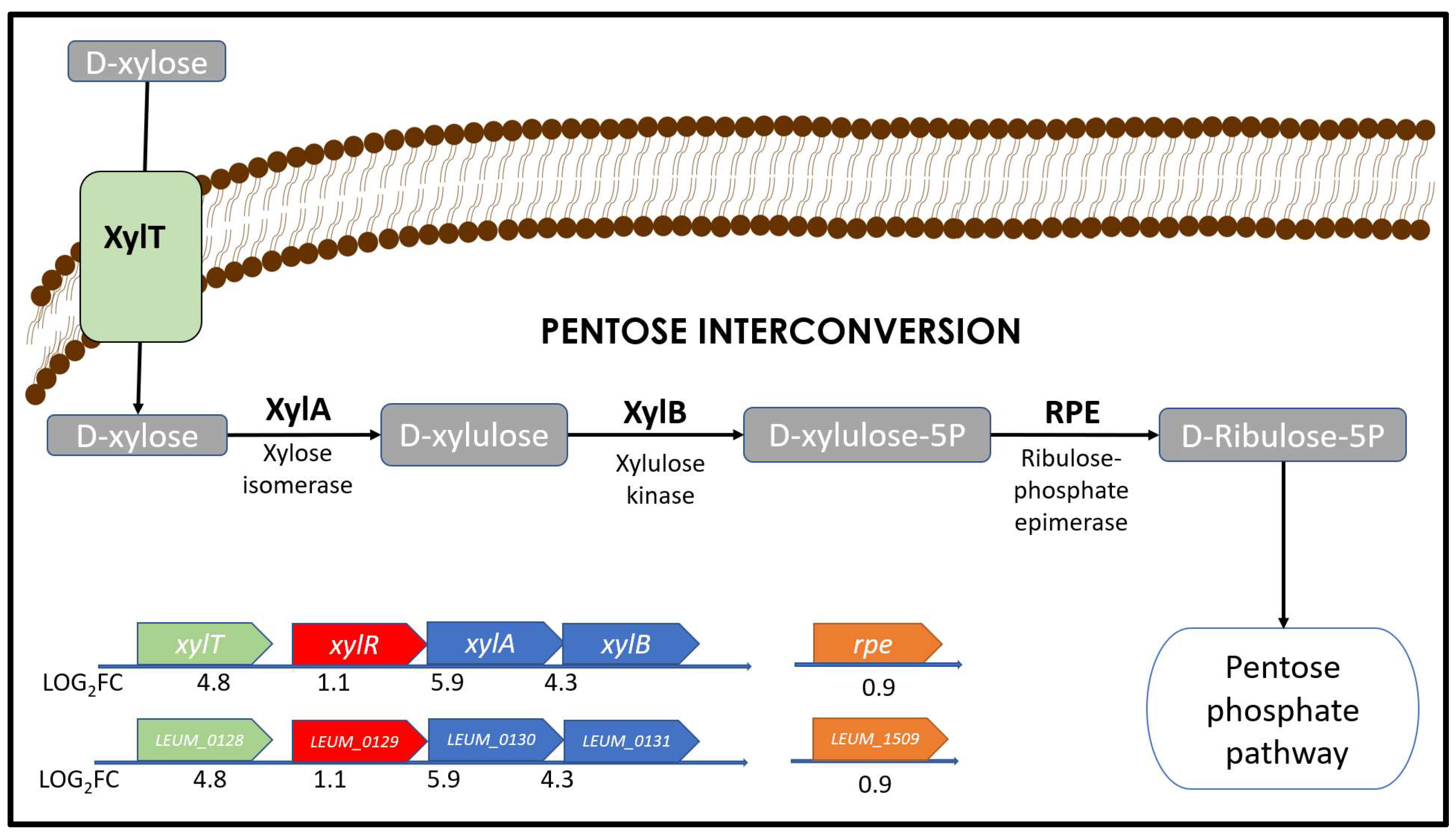
3.3. Exploring Possible Regulation Pathways of Leuconostoc mesenteroides ATCC 8293 GTFs
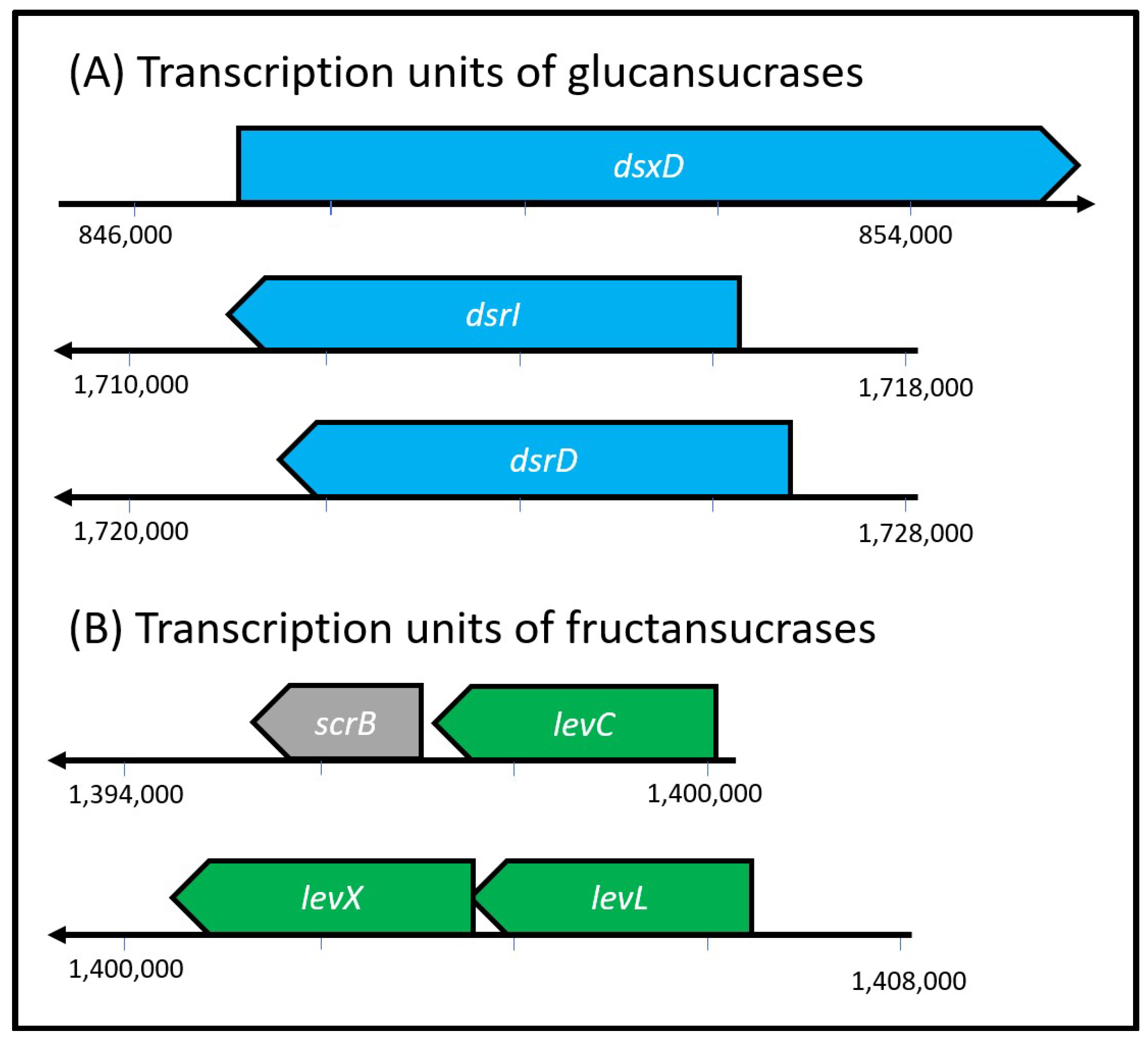
3.4. Genomic Context of GTFs Genes
3.5. The Transcription Factor (TF) CcpA
3.6. The Transcription Factor PerR
3.7. The Transcription Factor LexA
3.8. VicK/VicR Two-Component System
3.9. Transcriptomics Validation by Retro-Transcriptase qPCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shin, S.-Y.; Han, N. Leuconostoc spp. as starters and their beneficial role in fermented foods. In Beneficial Microorganisms in Food and Nutraceuticals; Liong, M.T., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 27, pp. 111–132. [Google Scholar]
- Björkroth, J.; Dicks, L.M.T.; Endo, A.; Holzapfel, H.W. The genus Leuconostoc. In Lactic Acid Bacteria; Holzapfel, W.H., Wood, B.J.B., Eds.; John Wiley & Sons, Ltd: Chichester, UK, 2014; pp. 391–404. [Google Scholar]
- Hemme, D.; Foucaud-Scheunemann, C. Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int. Dairy J. 2004, 14, 467–494. [Google Scholar] [CrossRef]
- van Thieghem, P. Sur la gomme de sucrerie. Ann. Sci. Nat. Bot. 1878, 7, 180–203. [Google Scholar]
- Crow, W.B. The structure and affinities of Leuconostoc mesenteroides (Cienkowsky) van Tieghem. Trans. Br. Mycol. Soc. 1922, 8, 76–84. [Google Scholar] [CrossRef]
- Hehre, E.J.; Sugg, J.Y. Serologically reactive polysaccharides produced through the action of bacterial enzymes: I. Dextran of Leuconostoc mesenteroides from sucrose. J. Exp. Med. 1941, 75, 339–353. [Google Scholar] [CrossRef] [PubMed]
- Grönwall, A.; Ingelman, B. Dextran As a Substitute for Plasma. Nature 1945, 155, 45. [Google Scholar] [CrossRef]
- Tsuchiya, H.M.; Koepsell, H.J.; Corman, J.; Bryant, G.; Bogard, M.O.; Feger, V.H.; Jackson, R.W. The effect of certain cultural factors on production of dextransucrase by Leuconostoc mesenteroides. J. Bacteriol. 1952, 64, 521–526. [Google Scholar] [CrossRef][Green Version]
- Lopez, A.; Monsan, P. Dextran synthesis by immobilized dextran sucrase. Biochimie 1980, 62, 323–329. [Google Scholar] [CrossRef]
- Ajongwen, J.N.; Akitoye, A.; Barker, P.E.; Ganetsos, G.; Shieh, M.T. Large-scale purification of Leuconostoc mesenteriodes NRRL B512F dextransucrase for use in the biosynthesis of dextran by batch and continuous chromatography. Chem. Eng. J. 1993, 51, B43–B50. [Google Scholar] [CrossRef]
- Paul, F.; Auriol, D.; Oriol, E.; Monsan, P. Production and purification of dextransucrase from Leuconostoc mesenteroides NRRL B 512 (F). Ann. N. Y. Acad. Sci. 1984, 434, 267–270. [Google Scholar] [CrossRef]
- Morales-Arrieta, S.; Rodríguez, M.E.; Segovia, L.; López-Munguía, A.; Olvera-Carranza, C. Identification and functional characterization of levS, a gene encoding for a levansucrase from Leuconostoc mesenteroides NRRL B-512 F. Gene 2006, 376, 59–67. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Schoch, C.L.; Sherry, S.T.; Karsch-Mizrachi, I. GenBank. Nucleic Acids Res. 2021, 49, D92–D96. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Mody, K.; Jha, B. Bacterial exopolysaccharides—A perception. J. Basic Microbiol. 2007, 47, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Caggianiello, G.; Kleerebezem, M.; Spano, G. Exopolysaccharides produced by lactic acid bacteria: From health-promoting benefits to stress tolerance mechanisms. Appl. Microbiol. Biotechnol. 2016, 100, 3877–3886. [Google Scholar] [CrossRef] [PubMed]
- Galle, S.; Arendt, E.K. Exopolysaccharides from Sourdough Lactic Acid Bacteria. Crit. Rev. Food Sci. Nutr. 2014, 54, 891–901. [Google Scholar] [CrossRef]
- Zeidan, A.A.; Poulsen, V.K.; Janzen, T.; Buldo, P.; Derkx, P.M.F.; Øregaard, G.; Neves, A.R.N. Polysaccharide production in Lactic Acid Bacteria: From genes to industrial applications. FEMS Microbiol. Rev. 2017, 41, S168–S200. [Google Scholar] [CrossRef][Green Version]
- Meng, F.; Lyu, Y.; Chen, X.; Lu, F.; Zhao, H.; Lu, Y.; Zhao, M.; Lu, Z. Maltose-Enhanced Exopolysaccharide Synthesis of Lactiplantibacillus plantarum through CRP-like Protein. J. Agric. Food Chem. 2022, 71, 1113–1121. [Google Scholar] [CrossRef]
- Li, Y.; Burne, R.A. Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofilms in response to pH and carbohydrate. Microbiology 2001, 147, 2841–2848. [Google Scholar] [CrossRef][Green Version]
- Hudson, M.C.; Curtiss, R. Regulation of expression of Streptococcus mutans genes important to virulence. Infect. Immun. 1990, 58, 464–470. [Google Scholar] [CrossRef][Green Version]
- Bivolarski, V.; Vasileva, T.; Dzhambazov, B.; Momchilova, A.; Chobert, J.M.; Ivanova, I.; Iliev, I. Characterization of Glucansucrases and Fructansucrases Produced by Wild Strains Leuconostoc mesenteroides URE13 and Leuconostoc mesenteroides LM17 Grown on Glucose or Fructose Medium as a Sole Carbon Source. Biotechnol. Biotechnol. Equip. 2013, 27, 3811–3820. [Google Scholar] [CrossRef][Green Version]
- Quirasco, M.; López-Munguía, A.; Remaud-Simeon, M.; Monsan, P.; Farrés, A. Induction and transcription studies of the dextransucrase gene in Leuconostoc mesenteroides NRRL B-512F. Appl. Environ. Microbiol. 1999, 65, 5504–5509. [Google Scholar] [CrossRef][Green Version]
- Dols, M.; Chraibi, W.; Remaud-Simeon, M.; Lindley, N.D.; Monsan, P.F. Growth and energetics of Leuconostoc mesenteroides NRRL B-1299 during metabolism of various sugars and their consequences for dextransucrase production. Appl. Environ. Microbiol. 1997, 63, 2159–2165. [Google Scholar] [CrossRef][Green Version]
- Chun, B.H.; Kim, K.H.; Jeon, H.H.; Lee, S.H.; Jeon, C.O. Pan-genomic and transcriptomic analyses of Leuconostoc mesenteroides provide insights into its genomic and metabolic features and roles in kimchi fermentation. Sci. Rep. 2017, 7, 11504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Côté, G.L.; Skory, C.D. Cloning, expression, and characterization of an insoluble glucan-producing glucansucrase from Leuconostoc mesenteroides NRRL B-1118. Appl. Microbiol. Biotechnol. 2012, 93, 2387–2394. [Google Scholar] [CrossRef] [PubMed]
- Olvera, C.; Centeno-Leija, S.; López-Munguía, A. Structural and functional features of fructansucrases present in Leuconostoc mesenteroides ATCC 8293. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2006, 92, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.; Slesarev, A.; Wolf, Y.; Sorokin, A.; Mirkin, B.; Koonin, E.; Pavlov, A.; Pavlova, N.; Karamychev, V.; Polouchine, N.; et al. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 2006, 103, 15611–15616. [Google Scholar] [CrossRef][Green Version]
- Escalante, A.; Giles-Gómez, M.; Hernández, G.; Córdova-Aguilar, M.S.; López-Munguía, A.; Gosset, G.; Bolívar, F. Analysis of bacterial community during the fermentation of pulque, a traditional Mexican alcoholic beverage, using a polyphasic approach. Int. J. Food Microbiol. 2008, 124, 126–134. [Google Scholar] [CrossRef]
- Peñas, E.C.; Martinez-Villaluenga, C.; Frias, J. Sauerkraut: Production, Composition, and Health Benefits. In Fermented Foods in Health and Disease Prevention; Peñas, E., Martinez-Villaluenga, C., Frias, J., Eds.; Academic Press: Boston, MA, USA, 2017; pp. 557–576. [Google Scholar] [CrossRef]
- Kyung, K.H.; Medina Pradas, E.; Kim, S.G.; Lee, Y.J.; Kim, K.H.; Choi, J.J.; Cho, J.H.; Chung, C.H.; Barrangou, R.; Breidt, F. Microbial Ecology of Watery Kimchi. J. Food Sci. 2015, 80, M1031–M1038. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vallejo-García, L.C.; Porras-Domínguez, J.R.; López-Munguía, A. Traditional Fermented Foods: Introducing “The Fructan Link”. In The Book of Fructans; Van den Ende, W., Toksoy-Öner, E., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 147–166. [Google Scholar] [CrossRef]
- van Hijum, S.A.; Kralj, S.; Ozimek, L.K.; Dijkhuizen, L.; van Geel-schutten, I.G.H. Structure-Function Relationships of Glucansucrase and Fructansucrase Enzymes from Lactic Acid Bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 157–176. [Google Scholar] [CrossRef][Green Version]
- Leemhuis, H.; Pijning, T.; Dobruchowska, J.M.; van Leeuwen, S.S.; Kralj, S.; Dijkstra, B.W.; Dijkhuizen, L. Glucansucrases: Three-dimensional structures, reactions, mechanism, α-glucan analysis and their implications in biotechnology and food applications. J. Biotechnol. 2013, 163, 250–272. [Google Scholar] [CrossRef][Green Version]
- Monsan, P.; Bozonnet, S.; Albenne, C.; Joucla, G.; Willemot, R.-M.; Remaud-Simeón, M. Homopolysaccharides from lactic acid bacteria. Int. Dairy J. 2001, 11, 675–685. [Google Scholar] [CrossRef]
- Zannini, E.; Waters, D.M.; Coffey, A.; Arendt, E.K. Production, properties, and industrial food application of lactic acid bacteria-derived exopolysaccharides. Appl. Microbiol. Biotechnol. 2016, 100, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.M.; Coffey, A.; Arendt, E.K. Exopolysaccharide producing lactic acid bacteria: Their techno-functional role and potential application in gluten-free bread products. Food Res. Int. 2017, 110, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Peshev, D.; Van den Ende, W. Fructans: Prebiotics and immunomodulators. J. Funct. Foods 2014, 8, 348–357. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Hu, Y.; Wu, F.; Jin, Y.; Xu, X.; Gänzle, M.G. Comparison of the Functionality of Exopolysaccharides Produced by Sourdough Lactic Acid Bacteria in Bread and Steamed Bread. J. Agric. Food Chem. 2020, 68, 8907–8914. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Olivares-Illana, V.; Wacher-Odarte, C.; Le Borgne, S.; López-Munguía, A. Characterization of a cell-associated inulosucrase from a novel source: A Leuconostoc citreum strain isolated from Pozol, a fermented corn beverage of Mayan origin. J. Ind. Microbiol. Biotechnol. 2002, 28, 112–117. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef][Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef][Green Version]
- Taboada, B.; Estrada, K.; Ciria, R.; Merino, E. Operon-mapper: A web server for precise operon identification in bacterial and archaeal genomes. Bioinformatics 2018, 34, 4118–4120. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef][Green Version]
- Senadheera, M.D.; Guggenheim, B.; Spatafora, G.A.; Huang, Y.C.C.; Choi, J.; Hung, D.C.I.; Treglown, J.S.; Goodman, S.D.; Ellen, R.P.; Cvitkovitch, D.G. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development . J. Bacteriol. 2005, 187, 4064–4076. [Google Scholar] [CrossRef][Green Version]
- Solis-Miranda, J.; Fonseca-García, C.; Nava, N.; Pacheco, R.; Quinto, C. Genome-wide identification of the crrlk1l subfamily and comparative analysis of its role in the legume-rhizobia symbiosis. Genes 2020, 11, 793. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gu, Y.; Ding, Y.; Ren, C.; Sun, Z.; Rodionov, D.A.; Zhang, W.; Yang, S.; Yang, C.; Jiang, W. Reconstruction of xylose utilization pathway and regulons in Firmicutes. BMC Genomics 2010, 11, 255. [Google Scholar] [CrossRef][Green Version]
- Novichkov, P.S.; Kazakov, A.E.; Ravcheev, D.A.; Leyn, S.A.; Kovaleva, G.Y.; Sutormin, R.A.; Kazanov, M.D.; Riehl, W.; Arkin, A.P.; Dubchak, I.; et al. RegPrecise 3.0—A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013, 14, 745. [Google Scholar] [CrossRef]
- Besrour-Aouam, N.; Mohedano, M.L.; Fhoula, I.; Zarour, K.; Najjari, A.; Aznar, R.; Prieto, A.; Ouzari, H.-I.I.; López, P. Different modes of regulation of the expression of dextransucrase in Leuconostoc lactis AV1n and Lactobacillus sakei MN. Front. Microbiol. 2019, 10, 959. [Google Scholar] [CrossRef][Green Version]
- De Bellis, P.; Ferrara, M.; Bavaro, A.R.; Linsalata, V.; Di Biase, M.; Musio, B.; Gallo, V.; Mulè, G.; Valerio, F. Characterization of Dextran Produced by the Food-Related Strain Weissella cibaria C43-11 and of the Relevant Dextransucrase Gene. Foods 2022, 11, 2819. [Google Scholar] [CrossRef] [PubMed]
- Dror, B.; Savidor, A.; Salam, B.; Sela, N.; Lampert, Y.; Teper-Bamnolker, P.; Daus, A. High levels of CO2 induce spoilage by Leuconostoc mesenteroides by upregulating Dextran synthesis genes. Appl. Environ. Microbiol. 2019, 85, 1. [Google Scholar] [CrossRef][Green Version]
- Yan, M.; Han, J.; Xu, X.; Liu, L.; Gao, C.; Zheng, H.; Chen, Y.; Tao, Y.; Zhou, H.; Li, Y.; et al. Gsy, a novel glucansucrase from Leuconostoc mesenteroides, mediates the formation of cell aggregates in response to oxidative stress. Sci. Rep. 2016, 6, 38122. [Google Scholar] [CrossRef][Green Version]
- Ramos, A.; Boels, I.C.; De Vos, W.M.; Santos, H. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 2001, 67, 33–41. [Google Scholar] [CrossRef][Green Version]
- Warner, J.B.; Lolkema, J.S. CcpA-Dependent Carbon Catabolite Repression in Bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 475–490. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ravcheev, D.A.; Khoroshkin, M.S.; Laikova, O.N.; Tsoy, O.V.; Sernova, N.V.; Petrova, S.A.; Rakhmaninova, A.B.; Novichkov, P.S.; Gelfand, M.S.; Rodionov, D.A. Comparative genomics and evolution of regulons of the LacI-family transcription factors. Front. Microbiol. 2014, 5, 00294. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Imlay, J.A. Transcription Factors That Defend Bacteria Against Reactive Oxygen Species. Annu. Rev. Microbiol. 2015, 69, 93–108. [Google Scholar] [CrossRef][Green Version]
- Butala, M.; Žgur-Bertok, D.; Busby, S.J.W. The bacterial LexA transcriptional repressor. Cell. Mol. Life Sci. 2009, 66, 82–93. [Google Scholar] [CrossRef]
- Hamada, S.; Slade, H.D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol. Rev. 1980, 44, 331–384. [Google Scholar] [CrossRef]
- Biswas, S. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J. Bacteriol. 2006, 188, 988–998. [Google Scholar] [CrossRef] [PubMed][Green Version]
| ID Locus | Gene | LOG2 FC (Xyl/Suc) | Xylose | LOG2FC (Fru/Suc) | Fructose |
|---|---|---|---|---|---|
| LEUM_1409 | levC | −2.726 | Down-regulated | −1.442 | NCD * |
| LEUM_1410 | levX | −2.135 | Down-regulated | −0.506 | NCD * |
| LEUM_1411 | levL | −2.813 | Down-regulated | −0.500 | NCD * |
| LEUM_0857 | dsxD | −2.059 | Down-regulated | −0.081 | NCD * |
| LEUM_1747 | dsrD | −0.616 | NCD * | −0.193 | NCD * |
| LEUM_1752 | dsrI | −2.220 | Down-regulated | −0.149 | NCD * |
| Condition | Total Activity (U/mg Total Protein) | Biomass (OD600 nm) Reached |
|---|---|---|
| Aerobic | 1.06 ± 0.1 | 6.5 ± 1.2 |
| Anaerobic | 1.04 ± 0.1 | 6.3 ± 0.9 |
| Sequence Name | Strand | Start | End | p-Value | q-Value | Matched Sequence |
|---|---|---|---|---|---|---|
| Upstream levC-scrB | negative | 90 | 106 | 2.81 × 10−5 | 0.118 | tgtttcctctctgttaa |
| Upstream levLX | positive | 408 | 424 | 8.97 × 10−5 | 0.138 | tgtaatactattgaaat |
| RT-qPCR | RNA-Seq | ||
|---|---|---|---|
| ID | Gene | LOG2FC Xyl/Fru | LOG2FC Xyl/Fru |
| LEUM_1409 | levC | −3.73 | −1.67 |
| LEUM_1410 | levX | −4.03 | −2.01 |
| LEUM_1411 | levL | −3.53 | −2.69 |
| LEUM_0857 | dsxD | −6.83 | −2.24 |
| LEUM_1747 | dsrD | −1.61 | −0.93 |
| LEUM_1752 | dsrI | −4.71 | −2.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vallejo-García, L.C.; Sánchez-Olmos, M.d.C.; Gutiérrez-Ríos, R.M.; López Munguía, A. Glycosyltransferases Expression Changes in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 Grown on Different Carbon Sources. Foods 2023, 12, 1893. https://doi.org/10.3390/foods12091893
Vallejo-García LC, Sánchez-Olmos MdC, Gutiérrez-Ríos RM, López Munguía A. Glycosyltransferases Expression Changes in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 Grown on Different Carbon Sources. Foods. 2023; 12(9):1893. https://doi.org/10.3390/foods12091893
Chicago/Turabian StyleVallejo-García, Luz Cristina, María del Carmen Sánchez-Olmos, Rosa María Gutiérrez-Ríos, and Agustín López Munguía. 2023. "Glycosyltransferases Expression Changes in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 Grown on Different Carbon Sources" Foods 12, no. 9: 1893. https://doi.org/10.3390/foods12091893
APA StyleVallejo-García, L. C., Sánchez-Olmos, M. d. C., Gutiérrez-Ríos, R. M., & López Munguía, A. (2023). Glycosyltransferases Expression Changes in Leuconostoc mesenteroides subsp. mesenteroides ATCC 8293 Grown on Different Carbon Sources. Foods, 12(9), 1893. https://doi.org/10.3390/foods12091893








