Evaluation of Nutritional Values of Edible Algal Species Using a Shortwave Infrared Hyperspectral Imaging and Machine Learning Technique
Abstract
:1. Introduction
2. Materials and Methods
2.1. Algae Material
2.2. Proximate Composition
2.3. Hyperspectral Imaging
2.4. Artificial Neural Networks
2.5. Statistical Analysis
3. Results and Discussion
3.1. Exploratory Analysis
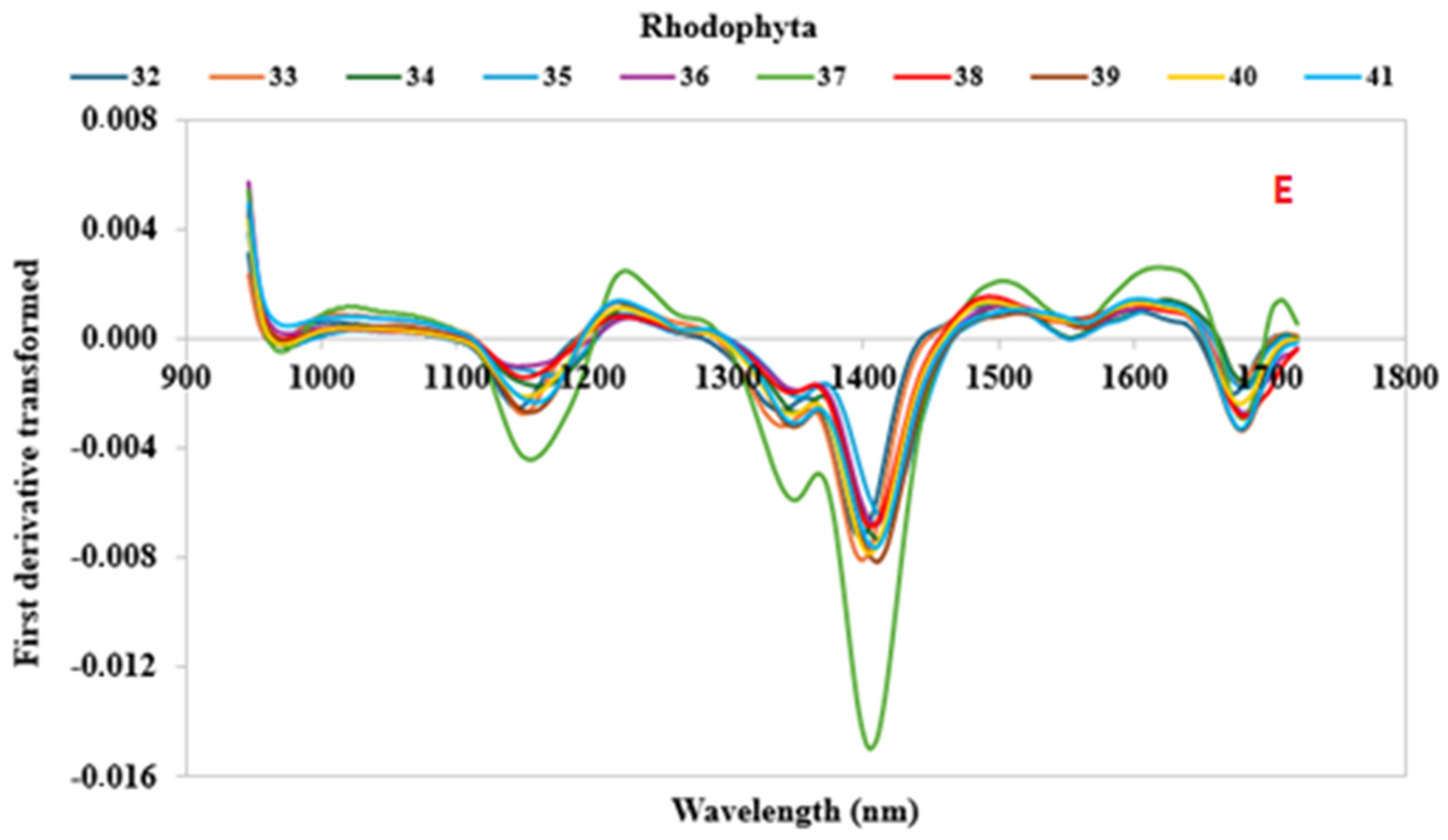
3.2. Spectral Characteristics
3.3. ANN Model Prediction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Salido, M.; Soto, M.; Seoane, S. Seaweed: Nutritional and gastronomic perspective. A review. Algal Res. 2024, 77, 103357. [Google Scholar] [CrossRef]
- Tanniou, A.; Vandanjon, L.; Goncalves, O.; Kervarec, N.; Stiger-Pouvreau, V. Rapid geographical differentiation of the European spread brown macroalga Sargassum muticum using HRMAS NMR and Fourier-Transform Infrared spectroscopy. Talanta 2015, 132, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L. Macroalgae. Encyclopedia 2021, 1, 177–188. [Google Scholar] [CrossRef]
- AlgaeBase. Available online: https://www.algaebase.org (accessed on 3 July 2024).
- Vieira, M.V.; Pastrana, L.M.; Fucinos, P. Microalgae Encapsulation Systems for Food, Pharmaceutical and Cosmetics Applications. Mar. Drugs 2020, 18, 644. [Google Scholar] [CrossRef] [PubMed]
- FAO. Fishery and Aquaculture Statistics—Yearbook 2021; FAO Yearbook of Fishery and Aquaculture Statistics: Rome, Italy, 2024. [Google Scholar] [CrossRef]
- Afonso, N.C.; Catarino, M.D.; Silva, A.M.S.; Caroso, S.M. Brown macroalgae as valuable food ingredients. Antioxidants 2019, 8, 365. [Google Scholar] [CrossRef] [PubMed]
- Amoriello, T.; Mellara, F.; Amoriello, M.; Ceccarelli, D.; Ciccoritti, R. Powdered seaweeds as a valuable ingredient for functional breads. Eur. Food Res. Technol. 2021, 247, 2431–2443. [Google Scholar] [CrossRef]
- Ross, A.B.; Jones, J.M.; Kubacki, M.L.; Bridgeman, T. Classification of macroalgae as fuel and its thermochemical behaviour. Bioresour. Technol. 2008, 99, 6494–6504. [Google Scholar] [CrossRef]
- Teas, J.; Braverman, L.E.; Kurzer, M.S.; Pino, S.; Hurley, T.G.; Hebert, J.R. Seaweed and soy: Companion foods in Asian cuisine and their effects on thyroid function in American women. J. Med. Food 2007, 10, 90–100. [Google Scholar] [CrossRef]
- Sarkar, M.S.I.; Kamal, M.; Hasan, M.M.; Hossain, M.I. Present status of naturally occurring seaweed flora and their utilization in Bangladesh. Res. Agric. Livest. Fish. 2016, 3, 203–216. [Google Scholar] [CrossRef]
- Roohinejad, S.; Koubaa, M.; Barba, F.J.; Saljoughian, S.; Amid, M.; Greiner, R. Application of seaweeds to develop new food products with enhanced shelf-life, quality and health-related beneficial properties. Food Res. Int. 2017, 99, 1066–1083. [Google Scholar] [CrossRef]
- Tagliapietra, L.T.; Clerici, M.T.P.S. Brown algae and their multiple applications as functional ingredient in food production. Food Res. Int. 2023, 167, 112655. [Google Scholar] [CrossRef] [PubMed]
- Penalver, R.; Lorenzo, J.M.; Nieto, G. Bioaccessibility, digestibility and nutritional properties of algae and Cyanophyceae as basis of their potential as functional food ingredients. Appl. Food Res. 2024, 4, 100404. [Google Scholar] [CrossRef]
- Stengel, D.B.; Connan, S.; Popper, Z.A. Algal chemodiversity and bioactivity: Sources of natural variability and implications for commercial application. Biotechnol. Adv. 2011, 29, 483–501. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Sheikh, M.A.; Ubaid, M.; Chauhan, P.; Kumar, K.; Choundhary, S. Comprehensive exploration of marine algae diversity, bioactive compounds, health benefits, regulatory issues, and food and drug applications. Meas. Food 2024, 14, 100163. [Google Scholar] [CrossRef]
- Bishop, W.M.; Zubeck, H.M. Evaluation of microalgae for use as nutraceuticals and nutritional supplements. J. Nutr. Food Sci. 2012, 2, 147–152. [Google Scholar] [CrossRef]
- Cardoso, S.M.; Carvalho, L.; Silva, P.J.; Rodrigues, M.S.; Pereira, O.R.; Pereira, L. Bioproducts from seaweeds: A review with special focus on the Iberian Peninsula. Curr. Org. Chem. 2014, 18, 896–917. [Google Scholar] [CrossRef]
- Chen, Y.; You, L.; Sun-Waterhouse, D. Effects of processing on the physicochemical characteristics and health benefits of algae products: Trade-offs among food carbon footprint, nutrient profiles, health properties, and consumer acceptance. Trends Food Sci. Technol. 2024, 147, 104375. [Google Scholar] [CrossRef]
- Garcia-Segovia, P.; Pagan-Moreno, M.J.; Lara, I.F.; Martínez-Monzó, J. Effect of microalgae incorporation on physicochemical and textural properties in wheat bread formulation. Food Sci. Technol. Int. 2017, 23, 437–447. [Google Scholar] [CrossRef]
- Ovando, C.H.; Carvalho, J.C.; de Melo-Pereira, V.G.; Jacques, P.; Soccol, V.T.; Soccol, C.R. Functional properties and health benefits of bioactive peptides derived from Spirulina: A review. Food Rev. Int. 2016, 34, 34–51. [Google Scholar] [CrossRef]
- Willcox, D.C.; Willcox, B.J.; Todoriki, H.; Suzuki, M. The Okinawan diet: Health implications of a low-calorie, nutrient dense, antioxidant-rich dietary pattern low in glycemic load. J. Am. Coll. Nutr. 2009, 28, 500S–516S. [Google Scholar] [CrossRef]
- Ozdogan, G.; Lin, X.; Sun, D.W. Rapid and noninvasive sensory analyses of food products by hyperspectral imaging: Recent application developments. Trends Food Sci. Technol. 2021, 111, 151–165. [Google Scholar] [CrossRef]
- Amoriello, T.; Ciorba, R.; Ruggiero, G.; Amoriello, M.; Ciccoritti, R. A Performance Evaluation of Two Hyperspectral Imaging Systems for the Prediction of Strawberries’ Pomological Traits. Sensors 2024, 24, 174. [Google Scholar] [CrossRef] [PubMed]
- Fatchurrahman, D.; Nosrati, M.; Amodio, M.L.; Chaudhry, M.M.A.; de Chiara, M.L.V.; Mastrandrea, L.; Colelli, G. Comparison Performance of Visible-NIR and Near-Infrared Hyperspectral Imaging for Prediction of Nutritional Quality of Goji Berry (Lycium barbarum L.). Foods 2021, 10, 1676. [Google Scholar] [CrossRef] [PubMed]
- Munera, S.; Rodríguez-Ortega, A.; Aleixos, N.; Cubero, S.; Gómez-Sanchis, J.; Blasco, J. Detection of Invisible Damages in ‘Rojo Brillante’ Persimmon Fruit at Different Stages Using Hyperspectral Imaging and Chemometrics. Foods 2021, 10, 2170. [Google Scholar] [CrossRef] [PubMed]
- Amoriello, T.; Ciccoritti, R.; Ferrante, P. Prediction of Strawberries’ Quality Parameters Using Artificial Neural Networks. Agronomy 2022, 12, 963. [Google Scholar] [CrossRef]
- Naroui Rad, M.R.; Koohkan, S.; Fanaei, H.R.; Pahlavan Rad, M.R. Application of Artificial Neural Networks to predict the final fruit weight and random forest to select important variables in native population of melon (Cucumis melo L.). Sci. Hortic. 2015, 181, 108–112. [Google Scholar] [CrossRef]
- International Association for Cereal Science and Technology; ICC Standard Methods (Methods No. 104/1, 105/2, 110/1, 136); ICC: Vienna, Austria, 2003.
- Shuuluka, D.; Bolton, J.J.; Anderson, R.J. Protein content, amino acid composition and nitrogen-to-protein conversion factors of Ulva rigida and Ulva capensis from natural populations and Ulva lactuca from an aquaculture system, in South Africa. J. Appl. Phycol. 2013, 25, 677–685. [Google Scholar] [CrossRef]
- Angell, A.R.; Mata, L.; de Nys, R.; Paul, N.A. The protein content of seaweeds: A universal nitrogen-to-protein conversion factor of five. J. Appl. Phycol. 2016, 28, 511–524. [Google Scholar] [CrossRef]
- Biancarosa, I.; Espe, M.; Bruckner, C.G.; Heesch, S.; Liland, N.; Waagbø, R.; Torstensen, B.; Lock, E.J. Amino acid composition, protein content, and nitrogen-to-protein conversion factors of 21 seaweed species from Norwegian waters. J. Appl. Phycol. 2017, 29, 1001–1009. [Google Scholar] [CrossRef]
- Niemi, C.; Mortensen, A.M.; Rautenberger, R.; Matsson, S.; Gorzsas, A.; Gentili, F.G. Rapid and accurate determination of protein content in North Atlantic seaweed by NIR and FTIR spectroscopies. Food Chem. 2023, 404, 134700. [Google Scholar] [CrossRef]
- Lee, S.C.; Prosky, L.; DeVries, J.W. Determination of total, soluble, and insoluble, dietary fibre in foods enzymatic-gravimetric method, MES-TRIS buffer: Collaborative study. J. Assoc. Off. Anal. Chem. 1992, 75, 395–416. [Google Scholar]
- Sultana, F.; Wahab, M.A.; Nahiduzzaman, M.; Mohiuddin, M.; Iqbal, M.Z.; Shakil, A.; Mamun, A.A.; Khan, M.S.R.; Wong, L.; Asaduzzaman, M. Seaweed farming for food and nutritional security, climate change mitigation and adaptation, and women empowerment: A review. Aquac. Fish. 2023, 8, 463–480. [Google Scholar] [CrossRef]
- Garcia-Vaquero, M.; Hayes, M. Red and Green Macroalgae for fish, animal feed and human functional food development. Food Rev. Int. 2015, 32, 15–45. [Google Scholar] [CrossRef]
- Wong, K.H.; Cheung, P.C.K. Nutritional evaluation of some subtropical red and green seaweeds Part I—Proximate composition, amino acid profiles and some physico-chemical properties. Food Chem. 2000, 71, 475–482. [Google Scholar] [CrossRef]
- Morales, M.; Aflalo, C.; Bernard, O. Microalgal lipids: A review of lipids potential and quantification for 95 phytoplankton species. Biomass Bioenergy 2021, 150, 106108. [Google Scholar] [CrossRef]
- Breuer, G.; Lamers, P.; Martens, D.; Draaisma, R.; Wijffels, R. Effect of light intensity, pH, and temperature on triacylglycerol (TAG) accumulation induced by nitrogen starvation in Scenedesmus obliquus. Bioresour. Technol. 2013, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Conde, T.; Neves, B.F.; Couto, D.; Melo, T.; Neves, B.; Costa, M.; Silva, J.; Domingues, P.; Domingues, M.R. Microalgae as sustainable bio-factories of healthy lipids: Evaluating fatty acid content and antioxidant activity. Mar. Drugs 2021, 19, 357. [Google Scholar] [CrossRef] [PubMed]
- Lopes, D.; Melo, T.; Rey, F.; Meneses, J.; Monteiro, F.L.; Helguero, L.A.; Abreu, M.H.; Lillebø, A.I.; Calado, R.; Domingues, M.R. Valuing bioactive lipids from green, red and brown macroalgae from aquaculture, to foster functionality and biotechnological applications. Molecules 2020, 25, 3883. [Google Scholar] [CrossRef]
- Lopes, D.; Rey, F.; Leal, M.C.; Lillebø, A.I.; Calado, R.; Domingues, M.R. Bioactivities of Lipid Extracts and Complex Lipids from Seaweeds: Current Knowledge and Future Prospects. Mar. Drugs 2021, 19, 686. [Google Scholar] [CrossRef]
- Jiménez-Escrig, A.; Sánchez-Muniz, F.J. Dietary fiber from edible seaweeds: Chemical structure, physicochemical properties and effects on cholesterol metabolism. Nutr. Res. 2000, 20, 585–598. [Google Scholar] [CrossRef]
- Dellarosa, N.; Laghi, L.; Martinsdottir, E.; Jonsdottir, R.; Sveinsdottir, K. Enrichment of convenience seafood with omega-3 and seaweed extracts: Effect on lipid oxidation. LWT-Food Sci. Technol. 2015, 62, 746–752. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, L.; He, S.; Cao, X.; Huang, J.; Ji, X.; Wu, M. Use of near-infrared spectroscopy and chemometrics for fast discrimination of Sargassum fusiforme. J. Food Comp. Anal. 2022, 110, 104537. [Google Scholar] [CrossRef]
- Mark, H.; Workman, J. Chemometrics in Spectroscopy; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Ghidini, S.; Varra, M.O.; Dall’Asta, C.; Badiani, A.; Ianieri, A.; Zanardi, E. Rapid authentication of European sea bass (Dicentrarchus labrax L.) according to production method, farming system, and geographical origin by near infrared spectroscopy coupled with chemometrics. Food Chem. 2019, 280, 321–327. [Google Scholar] [CrossRef]
- Shalev, N.T.; Ghermandi, A.; Tchernov, D.; Shemesh, E.; Israel, A.; Brook, A. NIR spectroscopy and artificial neural network for seaweed protein content assessment in-situ. Comput. Electron. Agric. 2022, 201, 107304. [Google Scholar]
- Kim, Y.; Kays, S.E. Near-infrared (NIR) prediction of trans-fatty acids in ground cereal foods. J. Agric. Food Chem. 2009, 57, 8187–8193. [Google Scholar] [CrossRef]
- Pizarroso, J.; Portela, J.; Munoz, A. NeuralSens: Sensitivity Analysis of Neural Networks. J. Stat. Softw. 2022, 10, 1–36. [Google Scholar] [CrossRef]
- Liu, B.; Liu, J.; Chen, T.; Yang, B.; Jiang, Y.; Wei, D.; Chen, F. Rapid characterization of fatty acids in oleaginous microalgae by near-infrared spectroscopy. Int. J. Mol. Sci. 2015, 16, 7045–7056. [Google Scholar] [CrossRef]
- Chu, B.; Li, C.; Wang, S.; Jin, W.; Li, X.; He, G.; Xiao, G. Nondestructive determination and visualization of protein and carbohydrate concentration of Chlorella pyrenoidosa in situ using hyperspectral imaging technique. Comput. Electron. Agric. 2023, 206, 107684. [Google Scholar] [CrossRef]
- Zhou, C.; Jiang, W.; Via, B.K.; Fasina, O.; Han, G. Prediction of mixed hardwood lignin and carbohydrate content using ATR-FTIR and FT-NIR. Carbohydr. Polym. 2015, 121, 336–341. [Google Scholar] [CrossRef]
- Gholipoor, M.; Nadali, F. Fruit yield prediction of pepper using artificial neural network. Sci. Hortic. 2019, 250, 249–253. [Google Scholar] [CrossRef]
- Huang, X.; Wang, H.; Luo, W.; Xue, S.; Hayat, F.; Gao, Z. Prediction of loquat soluble solids and titratable acid content using fruit mineral elements by artificial neural network and multiple linear regression. Sci. Hortic. 2021, 278, 109873. [Google Scholar] [CrossRef]
- Ribeiro, R.; Casanova, D.; Teixeira, M.; Wirth, A.; Gomes, H.M.; Borges, A.P.; Enembreck, F. Generating action plans for poultry management using artificial neural networks. Comput. Electron. Agric. 2019, 161, 131–140. [Google Scholar] [CrossRef]
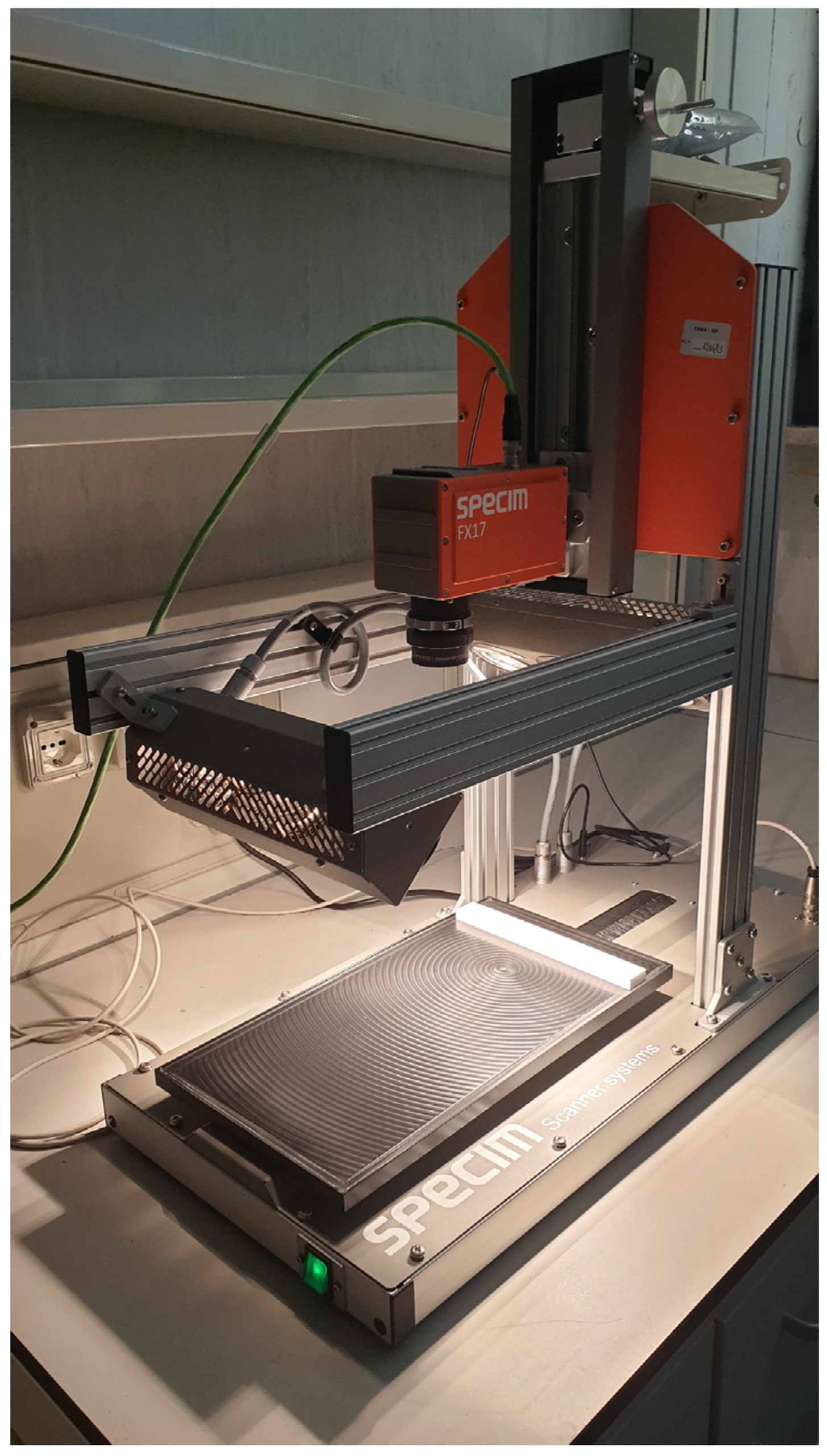





| Sample Number | Phylum | Species | Origin |
|---|---|---|---|
| 1 | Chlorophyta (green algae) | Auxenochlorella pyrenidosa | Northwest France |
| 2 | Chlorella vulgaris | China | |
| 3 | Ulva lactuca | Northwest Spain | |
| 4 | Ulva lactuca | Northwest France | |
| 5 | Ulva lactuca | Italy | |
| 6 | Ulva lactuca var. spiralis | Ireland | |
| 7 | Ulva pertusa | Japan | |
| 8 | Cyanophyta (blue-green algae) | Limnospira platensis | Italy |
| 9 | Limnospira platensis | China | |
| 10 | Limnospira platensis | Italy | |
| 11 | Phaeophyceae (brown algae) | Eisenia bicyclis | Japan |
| 12 | Eisenia bicyclis | Japan | |
| 13 | Eisenia bicyclis | Japan | |
| 14 | Himanthalia elongata | Northwest Spain | |
| 15 | Himanthalia elongata | Northwest France | |
| 16 | Himanthalia elongata | Northwest France | |
| 17 | Laminaria digitata | Northwest France | |
| 18 | Laminaria digitata | North Atlantic | |
| 19 | Laminaria digitata | Japan | |
| 20 | Laminaria longissima | Japan | |
| 21 | Laminaria longissima | Japan | |
| 22 | Laminaria ochroleuca | Atlantic | |
| 23 | Laminaria ochroleuca | Atlantic | |
| 24 | Sargassum fusiforme | Atlantic | |
| 25 | Undaria pinnatifida | Japan | |
| 26 | Undaria pinnatifida | Northwest Spain | |
| 27 | Undaria pinnatifida | Atlantic | |
| 28 | Undaria pinnatifida | Korea | |
| 29 | Undaria pinnatifida | Northwest Spain | |
| 30 | Undaria pinnatifida | Atlantic | |
| 31 | Undaria pinnatifida | Northwest Pacific | |
| 32 | Rhodophyta (red algae) | Chondrus crispus | Ireland |
| 33 | Chondrus crispus | Ireland | |
| 34 | Palmaria palmata | Atlantic | |
| 35 | Palmaria palmata | Atlantic | |
| 36 | Palmaria palmata | Ireland | |
| 37 | Palmaria palmata | Atlantic | |
| 38 | Porphyra umbilicalis | Southwest Atlantic | |
| 39 | Porphyra umbilicalis | Argentina | |
| 40 | Porphyra umbilicalis | Atlantic | |
| 41 | Pyropia yezoensis | Northwest France |
| Sample Number | Species | Protein (g 100 g−1 dw) | Lipid (g 100 g−1 dw) | Fiber (g 100 g−1 dw) |
|---|---|---|---|---|
| 1 | Auxenochlorella pyrenidosa | 59.0 ± 0.2 c | 10.1 ± 0.1 a | 9.0 ± 0.2 r |
| 2 | Chlorella vulgaris | 57.9 ± 0.2 d | 6.7 ± 0.1 c | 12.2 ± 0.2 q |
| 3 | Ulva lactuca | 16.9 ± 0.2 l | 0.6 ± 0.1 m | 34.6 ± 0.6 i |
| 4 | Ulva lactuca | 16.5 ± 0.1 l | 1.2 ± 0.7 il | 34.6 ± 0.6 i |
| 5 | Ulva lactuca | 16.9 ± 0.3 l | 0.7 ± 0.1 m | 35.0 ± 0.3 i |
| 6 | Ulva lactuca var. spiralis | 12.8 ± 0.2 n | 0.9 ± 0.1 l | 40.7 ± 0.7 f |
| 7 | Ulva pertusa | 27.4 ± 0.3 f | 1.5 ± 0.2 i | 43.1 ± 0.3 e |
| 8 | Limnospira platensis | 63.3 ± 0.4 b | 8.1 ± 0.2 b | 6.9 ± 0.3 s |
| 9 | Limnospira platensis | 65.1 ± 0.3 b | 5.9 ± 0.2 d | 2.5 ± 0.3 t |
| 10 | Limnospira platensis | 58.0 ± 0.4 d | 8.3 ± 0.2 b | 7.1 ± 0.3 s |
| 11 | Eisenia bicyclis | 9.1 ± 0.3 op | 1.6 ± 0.1 i | 63.2 ± 0.2 b |
| 12 | Eisenia bicyclis | 7.0 ± 0.2 r | 1.2 ± 0.1 l | 66.9 ± 0.3 a |
| 13 | Eisenia bicyclis | 7.0 ± 0.3 r | 1.1 ± 0.2 l | 66.6 ± 0.3 a |
| 14 | Himanthalia elongata | 7.8 ± 0.3 q | 1.0 ± 0.1 l | 35.9 ± 0.3 h |
| 15 | Himanthalia elongata | 10.2 ± 0.1 o | 2.6 ± 0.1 g | 31.2 ± 0.3 n |
| 16 | Himanthalia elongata | 10.0 ± 0.2 o | 2.6 ± 0.2 g | 31.1 ± 0.2 n |
| 17 | Laminaria digitata | 8.8 ± 0.2 p | 1.8 ± 0.1 i | 35.9 ± 0.3 h |
| 18 | Laminaria digitata | 9.2 ± 0.3 o | 1.1 ± 0.1 l | 33.4 ± 0.3 m |
| 19 | Laminaria digitata | 8.8 ± 0.2 p | 1.8 ± 0.1 i | 36.1 ± 0.1 h |
| 20 | Laminaria longissima | 7.9 ± 0.3 q | 3.1 ± 0.1 f | 32.6 ± 0.3 m |
| 21 | Laminaria longissima | 8.8 ± 0.1 p | 1.7 ± 0.2 i | 36.0 ± 0.1 h |
| 22 | Laminaria ochroleuca | 7.2 ± 0.1 r | 2.1 ± 0.1 h | 35.1 ± 0.3 i |
| 23 | Laminaria ochroleuca | 2.2 ± 0.2 s | 0.0 ± 0.0 n | 9.4 ± 0.1 r |
| 24 | Sargassum fusiforme | 6.8 ± 0.2 r | 1.2 ± 0.1 l | 61.4 ± 0.3 c |
| 25 | Undaria pinnatifida | 17.5 ± 0.2 i | 3.2 ± 0.2 f | 31.0 ± 0.1 n |
| 26 | Undaria pinnatifida | 16.9 ± 0.1 l | 2.2 ± 0.1 h | 33.0 ± 0.1 m |
| 27 | Undaria pinnatifida | 16.9 ± 0.2 l | 2.7 ± 0.2 g | 31.1 ± 0.2 n |
| 28 | Undaria pinnatifida | 14.2 ± 0.2 m | 1.7 ± 0.1 i | 35.1 ± 0.1 i |
| 29 | Undaria pinnatifida | 17.1 ± 0.3 l | 2.0 ± 0.3 h | 34.0 ± 0.2 l |
| 30 | Undaria pinnatifida | 16.8 ± 0.2 l | 2.6 ± 0.1 g | 30.8 ± 0.3 n |
| 31 | Undaria pinnatifida | 17.9 ± 0.2 h | 4.0 ± 0.1 e | 29.2 ± 0.2 o |
| 32 | Chondrus crispus | 16.7 ± 0.1 l | 2.3 ± 0.1 h | 30.6 ± 0.1 n |
| 33 | Chondrus crispus | 14.1 ± 0.1 m | 0.0 ± 0.0 n | 46.0 ± 0.2 d |
| 34 | Palmaria palmata | 18.2 ± 0.1 h | 0.5 ± 0.1 m | 31.1 ± 0.3 n |
| 35 | Palmaria palmata | 27.0 ± 0.1 f | 1.2 ± 0.1 l | 43.0 ± 0.2 e |
| 36 | Palmaria palmata | 17.9 ± 0.1 h | 0.6 ± 0.1 m | 31.0 ± 0.2 n |
| 37 | Palmaria palmata | 27.2 ± 0.2 f | 1.2 ± 0.2 l | 42.9 ± 0.1 e |
| 38 | Porphyra umbilicalis | 22.1 ± 0.1 g | 0.0 ± 0.0 n | 36.6 ± 0.1 h |
| 39 | Porphyra umbilicalis | 29.0 ± 0.1 e | 2.7 ± 0.2 g | 26.2 ± 0.1 p |
| 40 | Porphyra umbilicalis | 28.8 ± 0.2 e | 2.6 ± 0.2 g | 26.0 ± 0.2 p |
| 41 | Pyropia yezoensis | 22.0 ± 0.2 g | 0.6 ± 0.1 m | 39.1 ± 0.2 g |
| Protein (g 100 g−1 dw) | Lipid (g 100 g−1 dw) | Fiber (g 100 g−1 dw) | ||
|---|---|---|---|---|
| Activation Function | Hidden Neurons | Tahn | Exp | Exp |
| Output Neurons | Tahn | Logistic | Identity | |
| Training Set | r | 0.9994 | 0.9972 | 0.9988 |
| R2 | 0.9988 | 0.9944 | 0.9976 | |
| MAE | 0.0396 | 0.0027 | 0.0473 | |
| RMSE | 0.5593 | 0.1606 | 0.6350 | |
| RSE | 2.7248 | 7.0000 | 1.9405 | |
| Test Set | r | 0.9976 | 0.9883 | 0.9914 |
| R2 | 0.9952 | 0.9767 | 0.9828 | |
| MAE | 0.2590 | 0.0834 | 0.7968 | |
| RMSE | 1.2891 | 0.4096 | 2.3032 | |
| RSE | 5.8128 | 15.6521 | 7.5954 | |
| Validation Set | r | 0.9978 | 0.9691 | 0.9833 |
| R2 | 0.9956 | 0.9392 | 0.9670 | |
| MAE | 0.0250 | 0.0146 | 0.2701 | |
| RMSE | 0.9340 | 0.4534 | 2.7609 | |
| RSE | 5.0772 | 24.9689 | 7.7873 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amoriello, T.; Mellara, F.; Amoriello, M.; Ciccoritti, R. Evaluation of Nutritional Values of Edible Algal Species Using a Shortwave Infrared Hyperspectral Imaging and Machine Learning Technique. Foods 2024, 13, 2277. https://doi.org/10.3390/foods13142277
Amoriello T, Mellara F, Amoriello M, Ciccoritti R. Evaluation of Nutritional Values of Edible Algal Species Using a Shortwave Infrared Hyperspectral Imaging and Machine Learning Technique. Foods. 2024; 13(14):2277. https://doi.org/10.3390/foods13142277
Chicago/Turabian StyleAmoriello, Tiziana, Francesco Mellara, Monica Amoriello, and Roberto Ciccoritti. 2024. "Evaluation of Nutritional Values of Edible Algal Species Using a Shortwave Infrared Hyperspectral Imaging and Machine Learning Technique" Foods 13, no. 14: 2277. https://doi.org/10.3390/foods13142277





