Investigation of the Microbiome of Industrial PDO Sfela Cheese and Its Artisanal Variants Using 16S rDNA Amplicon Sequencing and Shotgun Metagenomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cheese Sampling
2.2. Growth Conditions and Strain Isolation
2.3. Identification by MALDI-TOF MS
2.4. DNA Extraction from Sfela Cheese Samples
2.5. Sequencing and Bioinformatics Analysis
3. Results and Discussion
3.1. Identification of Isolates via MALDI-TOF MS Analysis
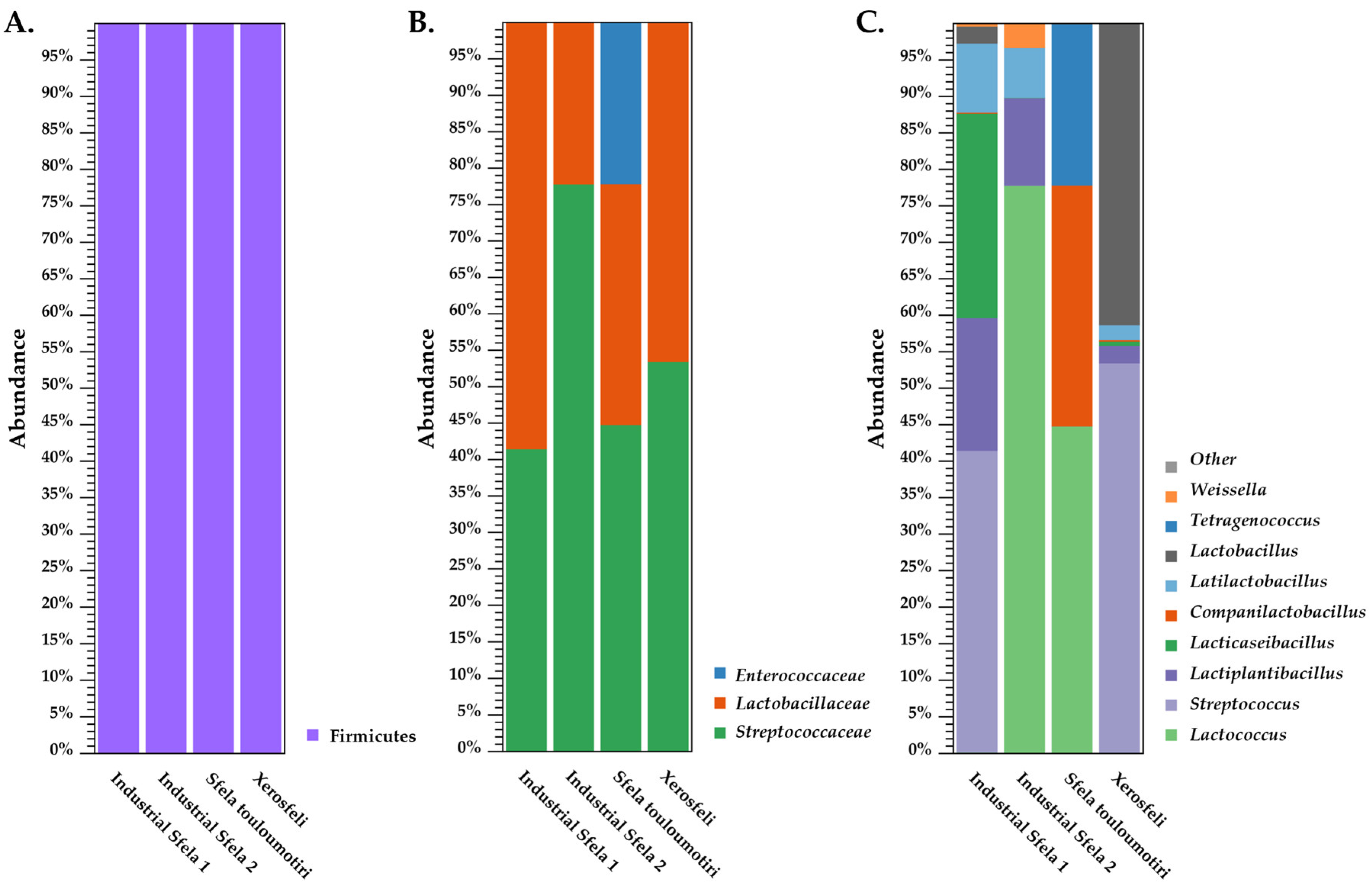
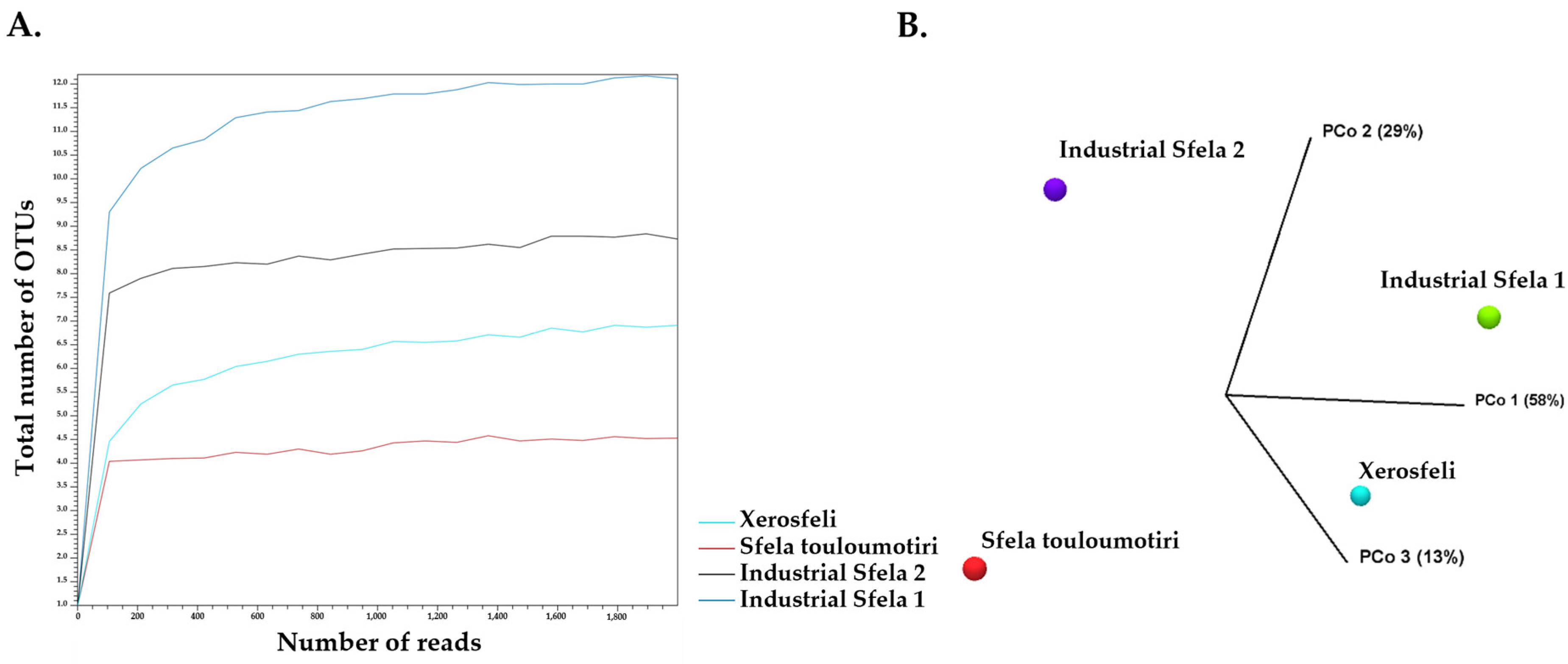
3.2. 16S rDNA Amplicon Sequencing
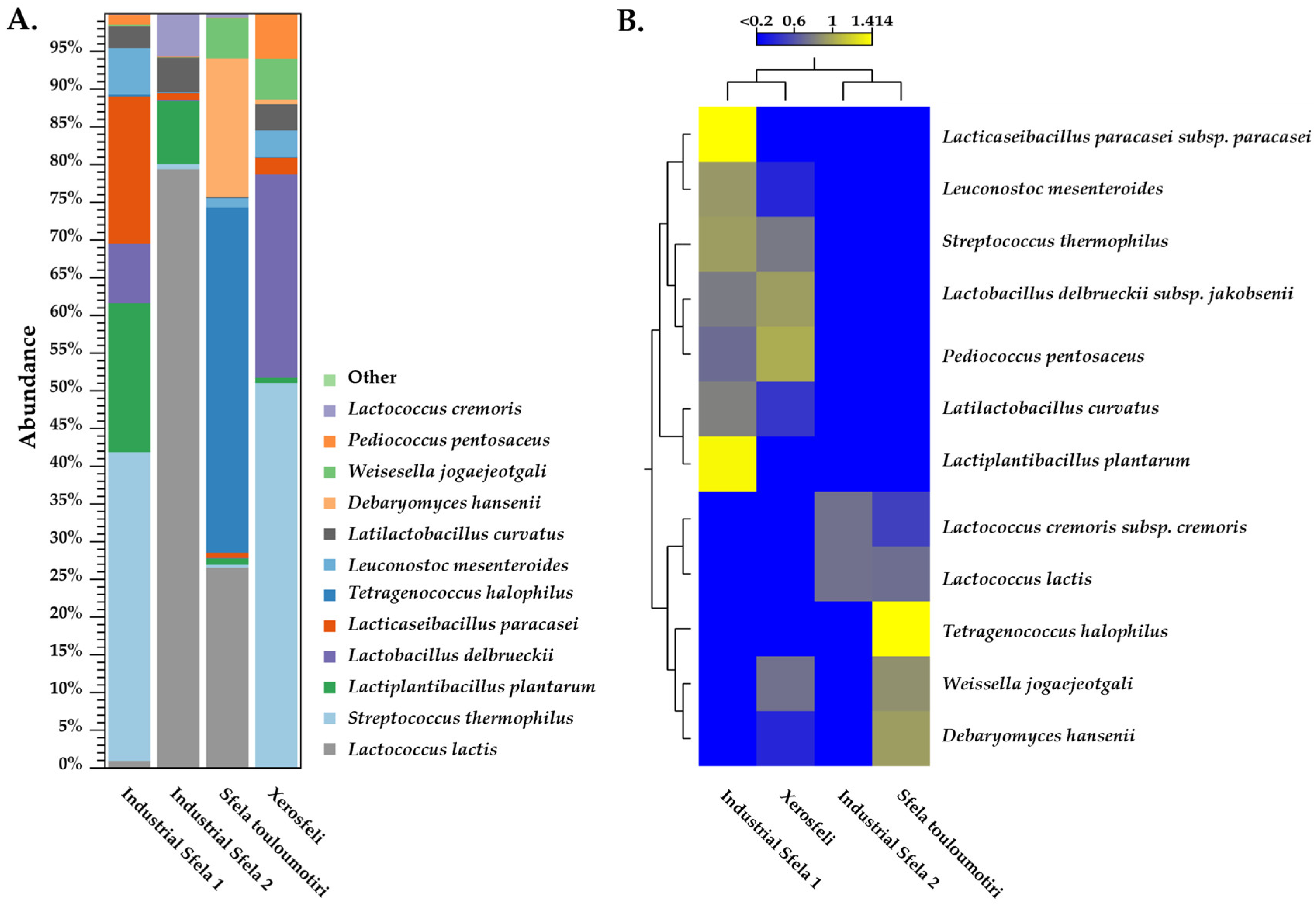
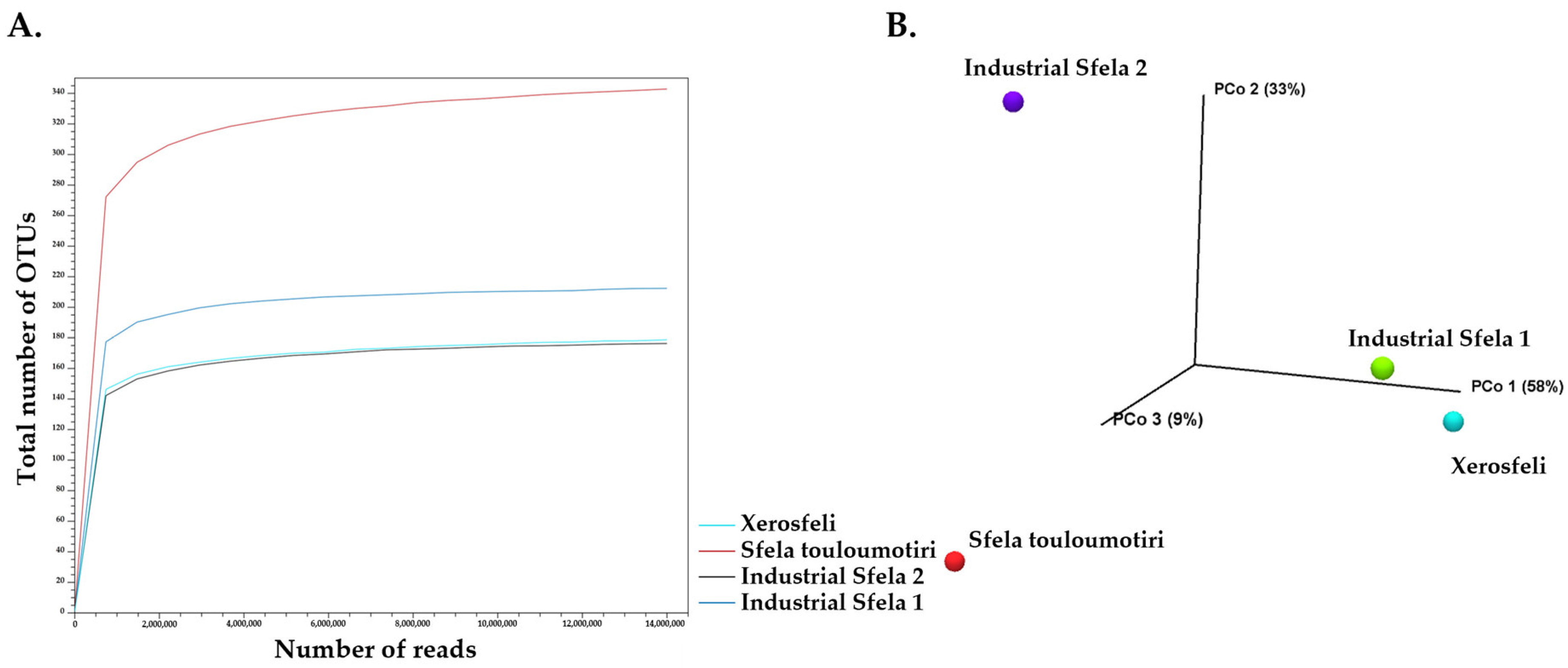
3.3. Identification of Sfela Microbiome at Species Level Using Shotgun Metagenomics Analysis
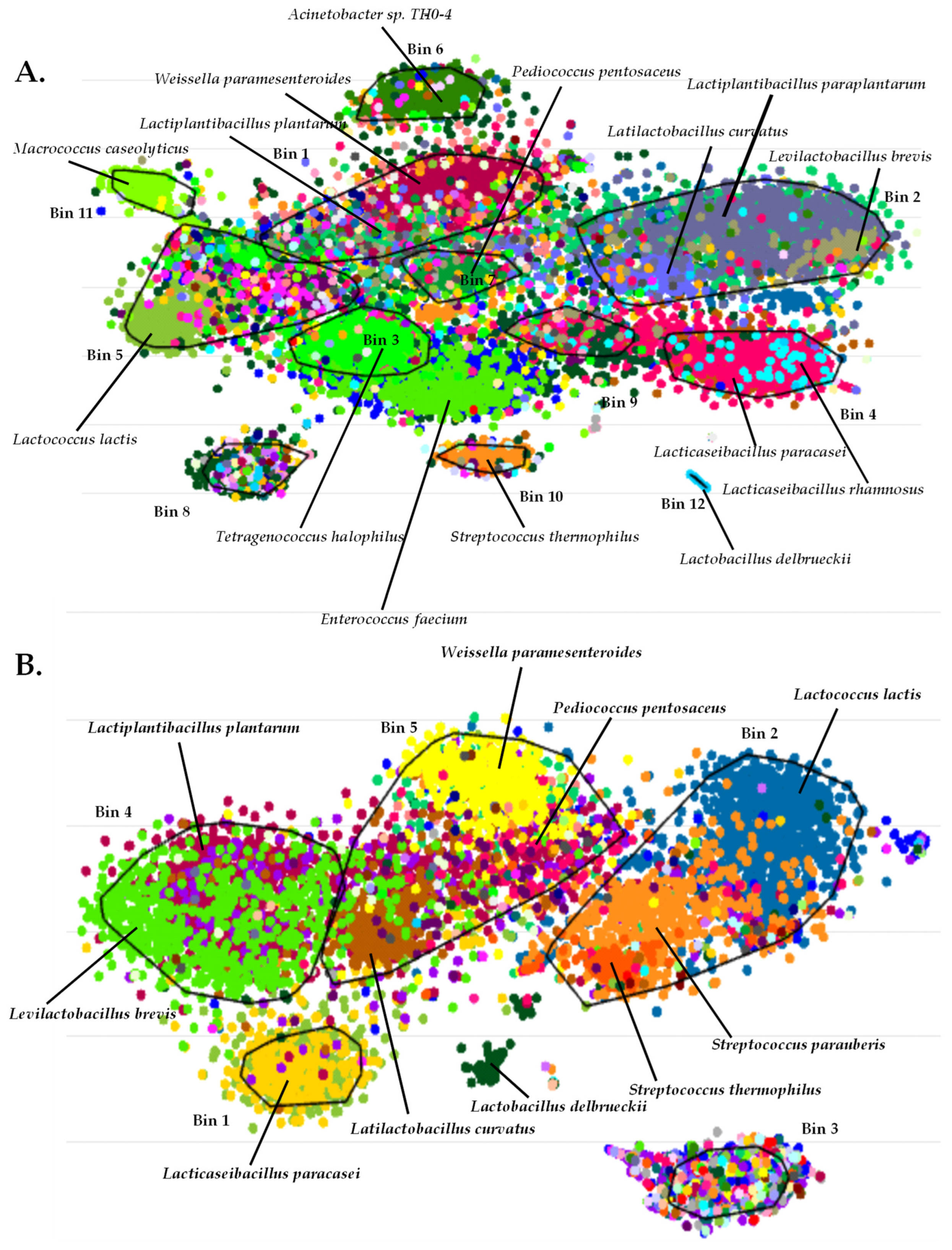
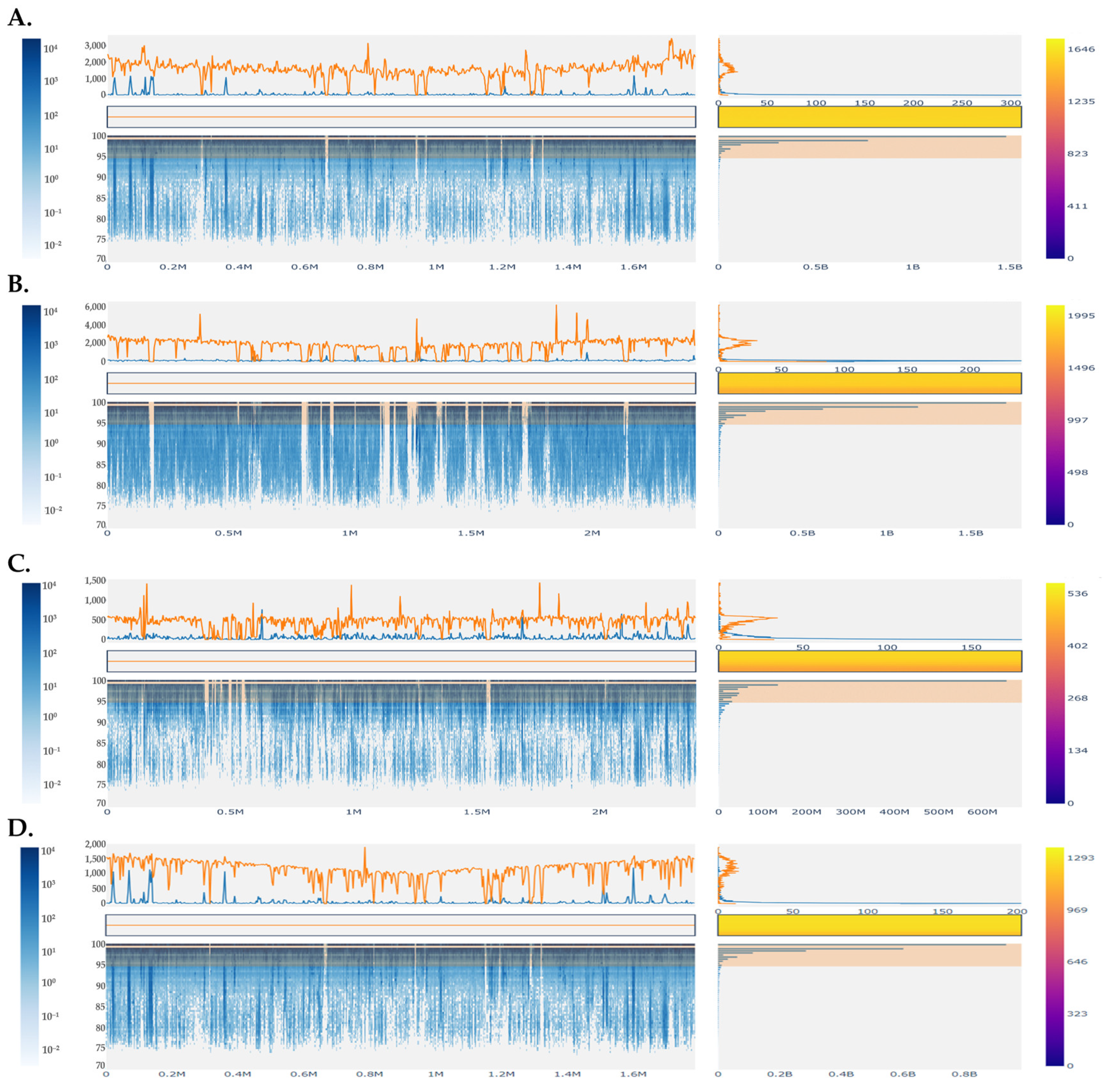
3.4. Binning of Metagenome-Assembled Contigs of Sfela Cheese Samples—Construction of Recruitment Plots
3.5. Functional Analysis of Metagenome Contigs
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McSweeney, P.L.H.; Ottogalli, G.; Fox, P.F. Diversity and classification of cheese varieties: An overview. In Cheese, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: San Diego, CA, USA, 2017; pp. 781–808. [Google Scholar] [CrossRef]
- Hayaloglu, A.A. Cheese varieties ripened under brine. In Cheese, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: San Diego, CA, USA, 2017; pp. 997–1040. [Google Scholar] [CrossRef]
- Moatsou, G.; Govaris, A. White brined cheeses: A diachronic exploitation of small ruminants milk in Greece. Small Rumin. Res. 2011, 101, 113–121. [Google Scholar] [CrossRef]
- Nega, A.; Moatsou, G. Proteolysis and related enzymatic activities in ten Greek cheese varieties. Dairy Sci. Technol. 2011, 92, 57–73. [Google Scholar] [CrossRef]
- Vakrou, A.; Fotopoulos, C.; Mattas, K. Location effects in the production and marketing of traditional Greek cheeses. In Proceedings of the European Association of Agricultural Economists: 52nd Seminar, Parma, Italy, 19–21 June 1997. [Google Scholar] [CrossRef]
- Pappas, C.P.; Kondyli, E.; Voutsinas, L.P.; Mallatou, H. Effects of starter level, draining time and aging on the physicochemical, organoleptic and rheological properties of feta cheese. IJDT 1996, 49, 73–78. [Google Scholar] [CrossRef]
- Litopoulou-Tzanetaki, E.; Tzanetakis, N.; Vafopoulou-Mastrojiannaki, A. Effect of the type of lactic starter on microbiologicalchemical and sensory characteristics of Feta cheese. Food Microbiol. 1993, 10, 31–41. [Google Scholar] [CrossRef]
- Angelopoulou, A.; Alexandraki, V.; Georgalaki, M.; Anastasiou, R.; Manolopoulou, E.; Tsakalidou, E.; Papadimitriou, K. Production of probiotic Feta cheese using Propionibacterium freudenreichii subsp. shermanii as adjunct. Int. Dairy J. 2017, 66, 135–139. [Google Scholar] [CrossRef]
- Dimitrellou, D.; Kandylis, P.; Sidira, M.; Koutinas, A.A.; Kourkoutas, Y. Free and immobilized Lactobacillus casei ATCC 393 on whey protein as starter cultures for probiotic Feta-type cheese production. J. Dairy Sci. 2014, 97, 4675–4685. [Google Scholar] [CrossRef] [PubMed]
- Mantzourani, I.; Terpou, A.; Alexopoulos, A.; Chondrou, P.; Galanis, A.; Bekatorou, A.; Bezirtzoglou, E.; Koutinas, A.A.; Plessas, S. Application of a novel potential probiotic Lactobacillus paracasei strain isolated from kefir grains in the production of Feta-type cheese. Microorganisms 2018, 6, 121. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, O.S.; Argyri, A.A.; Varzakis, E.E.; Tassou, C.C.; Chorianopoulos, N.G. Greek functional Feta cheese: Enhancing quality and safety using a Lactobacillus plantarum strain with probiotic potential. Food Microbiol. 2018, 74, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Tzanetakis, N.; Litopoulou-Tzanetaki, E. Changes in numbers and kinds of lactic acid bacteria in Feta and Teleme, two greek cheeses from ewes’ milk. J. Dairy Sci. 1992, 75, 1389–1393. [Google Scholar] [CrossRef]
- Bintsis, T.; Litopoulou-Tzanetaki, E.; Davies, R.O.N.; Robinson, R. Microbiology of brines used to mature Feta cheese. Int. J. Dairy Technol. 2000, 53, 106–112. [Google Scholar] [CrossRef]
- Anastasiou, R.; Kazou, M.; Georgalaki, M.; Aktypis, A.; Zoumpopoulou, G.; Tsakalidou, E. Omics approaches to assess flavor development in cheese. Foods 2022, 11, 188. [Google Scholar] [CrossRef] [PubMed]
- Hayaloglu, A.A.; Guven, M.; Fox, P.F. Microbiological, biochemical and technological properties of Turkish white cheese ‘Beyaz Peynir’. Int. Dairy J. 2002, 12, 635–648. [Google Scholar] [CrossRef]
- Bhowmik, T.; Marth, E.H. Rote of Micrococcus and Pediococcus Species in cheese ripening: A review. J. Dairy Sci. 1990, 73, 859–866. [Google Scholar] [CrossRef]
- Fadda, M.E.; Cosentino, S.; Deplano, M.; Palmas, F. Yeast populations in Sardinian feta cheese. Int. J. Food Microbiol. 2001, 69, 153–156. [Google Scholar] [CrossRef]
- Psomas, E.; Andrighetto, C.; Litopoulou-Tzanetaki, E.; Lombardi, A.; Tzanetakis, N. Some probiotic properties of yeast isolates from infant faeces and Feta cheese. Int. J. Food Microbiol. 2001, 69, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Urso, R.; Dolci, P.; Comi, G.; Cocolin, L. Microflora of Feta cheese from four Greek manufacturers. Int. J. Food Microbiol. 2008, 126, 36–42. [Google Scholar] [CrossRef]
- El-Sharoud, W.M.; Belloch, C.; Peris, D.; Querol, A. Molecular identification of yeasts associated with traditional Egyptian dairy products. J. Food Sci. 2009, 74, 341–346. [Google Scholar] [CrossRef]
- Jacques, N.; Casaregola, S. Safety assessment of dairy microorganisms: The hemiascomycetous yeasts. Int. J. Food Microbiol. 2008, 126, 321–326. [Google Scholar] [CrossRef]
- Seiler, H.; Busse, M. The yeasts of cheese brines. Int. J. Food Microbiol. 1990, 11, 289–303. [Google Scholar] [CrossRef]
- Ferrocino, I.; Rantsiou, K.; Cocolin, L. Investigating dairy microbiome: An opportunity to ensure quality, safety and typicity. Curr. Opin. Biotechnol. 2022, 73, 164–170. [Google Scholar] [CrossRef]
- Kahraman-Ilıkkan, Ö.; Bağdat, E. Metataxonomic sequencing to assess microbial safety of Turkish white cheeses. Braz. J. Microbiol. 2022, 53, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Gezginc, Y.; Karabekmez-Erdem, T.; Tatar, H.D.; Dağgeçen, E.C.; Ayman, S.; Akyol, İ. Metagenomics and volatile profile of Turkish artisanal Tulum cheese microbiota. Food Biosci. 2022, 45, 101497. [Google Scholar] [CrossRef]
- Ozturk, H.I.; Demirci, T.; Akin, N.; Ogul, A. Elucidation of the initial bacterial community of Ezine PDO cheese using next-generation sequencing. Arch. Microbiol. 2022, 204, 656. [Google Scholar] [CrossRef] [PubMed]
- Nelli, A.; Venardou, B.; Skoufos, I.; Voidarou, C.C.; Lagkouvardos, I.; Tzora, A. An Insight into goat cheese: The tales of artisanal and industrial Gidotyri microbiota. Microorganisms 2023, 11, 123. [Google Scholar] [CrossRef] [PubMed]
- Kamilari, E.; Tsaltas, D.; Stanton, C.; Ross, R.P. Metataxonomic mapping of the microbial diversity of Irish and Eastern Mediterranean cheeses. Foods 2022, 11, 2483. [Google Scholar] [CrossRef]
- Michailidou, S.; Pavlou, E.; Pasentsis, K.; Rhoades, J.; Likotrafiti, E.; Argiriou, A. Microbial profiles of Greek PDO cheeses assessed with amplicon metabarcoding. Food Microbiol. 2021, 99, 103836. [Google Scholar] [CrossRef]
- Sant’Anna, F.M.; Wetzels, S.U.; Cicco, S.H.S.; Figueiredo, R.C.; Sales, G.A.; Figueiredo, N.C.; Nunes, C.A.; Schmitz-Esser, S.; Mann, E.; Wagner, M.; et al. Microbial shifts in Minas artisanal cheeses from the Serra do Salitre region of Minas Gerais, Brazil throughout ripening time. Food Microbiol. 2019, 82, 349–362. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Kamilaris, A.; Tsaltas, D. Characterizing Halloumi cheese’s bacterial communities through metagenomic analysis. LWT 2020, 126, 109298. [Google Scholar] [CrossRef]
- Papademas, P.; Aspri, M.; Mariou, M.; Dowd, S.E.; Kazou, M.; Tsakalidou, E. Conventional and omics approaches shed light on Halitzia cheese, a long-forgotten white-brined cheese from Cyprus. Int. Dairy J. 2019, 98, 72–83. [Google Scholar] [CrossRef]
- Papadakis, P.; Konteles, S.; Batrinou, A.; Ouzounis, S.; Tsironi, T.; Halvatsiotis, P.; Tsakali, E.; Van Impe, J.F.M.; Vougiouklaki, D.; Strati, I.F.; et al. Characterization of bacterial microbiota of P.D.O. Feta cheese by 16S metagenomic analysis. Microorganisms 2021, 9, 2377. [Google Scholar] [CrossRef]
- Spyrelli, E.; Stamatiou, A.; Tassou, C.; Nychas, G.-J.; Doulgeraki, A. Microbiological and metagenomic analysis to assess the effect of container material on the microbiota of Feta cheese during ripening. Fermentation 2020, 6, 12. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Anastasiou, R.; Georgalaki, M.; Bounenni, R.; Paximadaki, A.; Charmpi, C.; Alexandraki, V.; Kazou, M.; Tsakalidou, E. Comparison of the microbiome of artisanal homemade and industrial Feta cheese through amplicon sequencing and shotgun metagenomics. Microorganisms 2022, 10, 1073. [Google Scholar] [CrossRef] [PubMed]
- Schmartz, G.P.; Hirsch, P.; Amand, J.; Dastbaz, J.; Fehlmann, T.; Kern, F.; Muller, R.; Keller, A. BusyBee Web: Towards comprehensive and differential composition-based metagenomic binning. Nucleic Acids Res. 2022, 50, W132–W137. [Google Scholar] [CrossRef] [PubMed]
- Laczny, C.C.; Kiefer, C.; Galata, V.; Fehlmann, T.; Backes, C.; Keller, A. BusyBee Web: Metagenomic data analysis by bootstrapped supervised binning and annotation. Nucleic Acids Res. 2017, 45, 171–179. [Google Scholar] [CrossRef] [PubMed]
- De Roos, J.; Vandamme, P.; De Vuyst, L. Wort substrate consumption and metabolite production during lambic beer fermentation and maturation explain the successive growth of specific bacterial and yeast species. Front. Microbiol. 2018, 9, 2763. [Google Scholar] [CrossRef] [PubMed]
- Verce, M.; De Vuyst, L.; Weckx, S. Shotgun metagenomics of a water kefir fermentation ecosystem reveals a novel oenococcus species. Front. Microbiol. 2019, 10, 479. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, K.; Ruiz-Perez, C.A.; Rodriguez, R.L.; Conrad, R.E.; Konstantinidis, K.T. RecruitPlotEasy: An advanced read recruitment plot tool for assessing metagenomic population abundance and genetic diversity. Front. Bioinform. 2021, 1, 826701. [Google Scholar] [CrossRef] [PubMed]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic. Acids Res. 2015, 43, 566–570. [Google Scholar] [CrossRef] [PubMed]
- Ledina, T.; Golob, M.; Djordjević, J.; Magas, V.; Colovic, S.; Bulajic, S. MALDI-TOF mass spectrometry for the identification of Serbian artisanal cheeses microbiota. JCF 2018, 13, 309–314. [Google Scholar] [CrossRef]
- Sanchez-Juanes, F.; Teixeira-Martin, V.; Gonzalez-Buitrago, J.M.; Velazquez, E.; Flores-Felix, J.D. Identification of species and subspecies of lactic acid bacteria present in Spanish cheeses type “Torta” by MALDI-TOF MS and pheS gene analyses. Microorganisms 2020, 8, 301. [Google Scholar] [CrossRef]
- Gantzias, C.; Lappa, I.K.; Aerts, M.; Georgalaki, M.; Manolopoulou, E.; Papadimitriou, K.; De Brandt, E.; Tsakalidou, E.; Vandamme, P. MALDI-TOF MS profiling of non-starter lactic acid bacteria from artisanal cheeses of the Greek island of Naxos. Int. J. Food Microbiol. 2020, 323, 108586. [Google Scholar] [CrossRef] [PubMed]
- Tzora, A.; Nelli, A.; Voidarou, C.; Fthenakis, G.; Rozos, G.; Theodorides, G.; Bonos, E.; Skoufos, I. Microbiota “fingerprint” of Greek Feta cheese through ripening. Appl. Sci. 2021, 11, 5631. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic acid bacteria in raw-milk cheeses: From starter cultures to probiotic functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef] [PubMed]
- Laranjo, M.; Potes, M.E. Chapter 6—Traditional Mediterranean cheeses: Lactic acid bacteria populations and functional traits. In Lactic Acid Bacteria in Food Biotechnology; Ray, R.C., Paramithiotis, S., de Carvalho Azevedo, V.A., Montet, D., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 97–124. [Google Scholar] [CrossRef]
- Özer, B. CHEESE|Microflora of white-brined cheeses. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, MS, USA, 2014; pp. 402–408. [Google Scholar] [CrossRef]
- Marino, M.; Innocente, N.; Maifreni, M.; Mounier, J.; Cobo-Diaz, J.F.; Coton, E.; Carraro, L.; Cardazzo, B. Diversity within Italian cheesemaking brine-associated bacterial communities evidenced by massive parallel 16S rRNA gene tag sequencing. Front. Microbiol. 2017, 8, 2119. [Google Scholar] [CrossRef] [PubMed]
- Vermote, L.; Verce, M.; De Vuyst, L.; Weckx, S. Amplicon and shotgun metagenomic sequencing indicates that microbial ecosystems present in cheese brines reflect environmental inoculation during the cheese production process. Int. Dairy J. 2018, 87, 44–53. [Google Scholar] [CrossRef]
- Morales, F.; Morales, J.I.; Hernandez, C.H.; Hernandez-Sanchez, H. Isolation and partial characterization of halotolerant lactic acid bacteria from two Mexican cheeses. Appl. Biochem. Biotechnol. 2011, 164, 889–905. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kim, S.A.; Jo, Y.M.; Seo, H.; Kim, G.Y.; Cheon, S.W.; Yang, S.H.; Jeon, C.O.; Han, N.S. Probiotic potential of Tetragenococcus halophilus EFEL7002 isolated from Korean soy Meju. BMC Microbiol. 2022, 22, 149. [Google Scholar] [CrossRef] [PubMed]
- Fornasari, M.E.; Rossetti, L.; Carminati, D.; Giraffa, G. Cultivability of Streptococcus thermophilus in Grana Padano cheese whey starters. FEMS Microbiol. Lett. 2006, 257, 139–144. [Google Scholar] [CrossRef]
- Sola, L.; Quadu, E.; Bortolazzo, E.; Bertoldi, L.; Randazzo, C.L.; Pizzamiglio, V.; Solieri, L. Insights on the bacterial composition of Parmigiano Reggiano natural whey starter by a culture-dependent and 16S rRNA metabarcoding portrait. Sci. Rep. 2022, 12, 17322. [Google Scholar] [CrossRef]
- Bozoudi, D.; Torriani, S.; Zdragas, A.; Litopoulou-Tzanetaki, E. Assessment of microbial diversity of the dominant microbiota in fresh and mature PDO Feta cheese made at three mountainous areas of Greece. LWT-Food Sci. Technol. 2016, 72, 525–533. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Nisic, A.; Hyden, P.; Cabal, A.; Sucher, J.; Stoger, A.; Allerberger, F.; Martinovic, A. Genetic diversity of Leuconostoc mesenteroides isolates from traditional Montenegrin brine cheese. Microorganisms 2021, 9, 1612. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Ku, H.J.; Ahn, M.J.; Hong, J.S.; Lee, S.H.; Shin, H.; Lee, K.C.; Lee, J.S.; Ryu, S.; Jeon, C.O.; et al. Weissella jogaejeotgali sp. nov., isolated from jogae jeotgal, a traditional Korean fermented seafood. Int. J. Syst. Evol. Microbiol. 2015, 65, 4674–4681. [Google Scholar] [CrossRef] [PubMed]
- Rustemoglu, M.; Erkan, M.E.; Cengiz, G.; Hajyzadeh, M. Bacterial metagenome profiling of hand-made herby cheese samples utilizing high-throughput sequencing to detect geographical indication and marketing potential. Heliyon 2023, 9, 13334. [Google Scholar] [CrossRef] [PubMed]
- Morea, M.; Baruzzi, F.; Cappa, F.; Cocconcelli, P.S. Molecular characterization of the Lactobacillus community in traditional processing of Mozzarella cheese. Int. J. Food Microbiol. 1998, 43, 53–60. [Google Scholar] [CrossRef] [PubMed]
- El-Salam, M.H.A.; Alichanidis, E.; Zerfiridis, G.K. Domiati and Feta type cheeses. In Cheese: Chemistry, Physics and Microbiology: Volume 2 Major Cheese Groups; Fox, P.F., Ed.; Springer: Boston, MA, USA, 1993; pp. 301–335. [Google Scholar] [CrossRef]
- Papademas, P.; Robinson, R.K. A comparison of the chemical, microbiological and sensory characteristics of bovine and ovine Halloumi cheese. Int. Dairy J. 2000, 10, 761–768. [Google Scholar] [CrossRef]
- Yap, M.; O’Sullivan, O.; O’Toole, P.W.; Cotter, P.D. Development of sequencing-based methodologies to distinguish viable from non-viable cells in a bovine milk matrix: A pilot study. Front. Microbiol. 2022, 13, 1036643. [Google Scholar] [CrossRef]
- Ruggirello, M.; Giordano, M.; Bertolino, M.; Ferrocino, I.; Cocolin, L.; Dolci, P. Study of Lactococcus lactis during advanced ripening stages of model cheeses characterized by GC-MS. Food Microbiol. 2018, 74, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Ammoun, I.; Kothe, C.I.; Mohellibi, N.; Beal, C.; Yaacoub, R.; Renault, P. Lebanese fermented goat milk products: From tradition to meta-omics. Food Res. Int. 2023, 168, 112762. [Google Scholar] [CrossRef]
- Ganesan, B.; Stuart, M.R.; Weimer, B.C. Carbohydrate starvation causes a metabolically active but nonculturable state in Lactococcus lactis. Appl. Environ. Microbiol. 2007, 73, 2498–2512. [Google Scholar] [CrossRef]
- Irlinger, F.; Loux, V.; Bento, P.; Gibrat, J.F.; Straub, C.; Bonnarme, P.; Landaud, S.; Monnet, C. Genome sequence of Staphylococcus equorum subsp. equorum Mu2, isolated from a French smear-ripened cheese. J. Bacteriol. 2012, 194, 5141–5142. [Google Scholar] [CrossRef]
- Place, R.B.; Hiestand, D.; Gallmann, H.R.; Teuber, M. Staphylococcus equorum subsp. linens, subsp. nov., a starter culture component for surface ripened semi-hard cheeses. Syst. Appl. Microbiol. 2003, 26, 30–37. [Google Scholar] [CrossRef]
- Yang, X. Moraxellaceae. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Elsevier Ltd.: Amsterdam, The Netherlands, 2014; pp. 826–833. [Google Scholar] [CrossRef]
- Seifert, H.; Strate, A.; Schulze, A.; Pulverer, G. Vascular catheter-related bloodstream infection due to Acinetobacter johnsonii (formerly Acinetobacter calcoaceticus var. lwoffi): Report of 13 cases. Clin. Infect. Dis. 1993, 17, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Bereswill, S.; Fuka, M.M.; Wallisch, S.; Engel, M.; Welzl, G.; Havranek, J.; Schloter, M. Dynamics of bacterial communities during the ripening process of different croatian cheese types derived from raw ewe’s milk cheeses. PLoS ONE 2013, 8, e80734. [Google Scholar] [CrossRef]
- de Garnica, M.L.; Sáez-Nieto, J.A.; González, R.; Santos, J.A.; Gonzalo, C. Diversity of gram-positive catalase-negative cocci in sheep bulk tank milk by comparative 16S rDNA sequence analysis. Int. Dairy J. 2014, 34, 142–145. [Google Scholar] [CrossRef]
- Pitkala, A.; Koort, J.; Bjorkroth, J. Identification and antimicrobial resistance of Streptococcus uberis and Streptococcus parauberis isolated from bovine milk samples. J. Dairy Sci. 2008, 91, 4075–4081. [Google Scholar] [CrossRef] [PubMed]
- Fortina, M.G.; Ricci, G.; Acquati, A.; Zeppa, G.; Gandini, A.; Manachini, P.L. Genetic characterization of some lactic acid bacteria occurring in an artisanal protected denomination origin (PDO) Italian cheese, the Toma piemontese. Food Microbiol. 2003, 20, 397–404. [Google Scholar] [CrossRef]
- Lun, Z.R.; Wang, Q.P.; Chen, X.G.; Li, A.X.; Zhu, X.Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef]
- Danezis, G.P.; Tsiplakou, E.; Pappa, E.C.; Pappas, A.C.; Mavrommatis, A.; Sotirakoglou, K.; Georgiou, C.A.; Zervas, G. Fatty acid profile and physicochemical properties of Greek protected designation of origin cheeses, implications for authentication. Eur. Food Res. Technol. 2020, 246, 1741–1753. [Google Scholar] [CrossRef]
- Pappa, E.C.; Kondyli, E. Descriptive characteristics and cheesemaking technology of Greek cheeses not listed in the EU geographical indications registers. Dairy 2023, 4, 43–67. [Google Scholar] [CrossRef]


| Identified Species via MALDI-TOF MS | Total Number |
|---|---|
| Enterococcus faecium | 62/140 (44.3%) |
| Lactiplantibacillus plantarum | 33/140 (23.6%) |
| Levilactobacillus brevis | 14/140 (10%) |
| Pediococcus pentosaceus | 12/140 (8.6%) |
| Streptococcus thermophilus | 7/140 (5%) |
| Leuconostoc mesenteroides | 5/140 (3.6%) |
| Lacticaseibacillus paracasei | 2/140 (1.4%) |
| Enterococcus gallinarum | 1/140 (0.7%) |
| Latilactobacillus curvatus | 1/140 (0.7%) |
| Lacticaseibacillus rhamnosus | 1/140 (0.7%) |
| Streptococcus gallolyticus | 1/140 (0.7%) |
| Staphylococcus epidermidis | 1/140 (0.7%) |
| Sample | Bin | Completeness * | Contamination * | Strain Heterogeneity * |
|---|---|---|---|---|
| Industrial Sfela 1 | 1 | 93.69 | 127.03 | 7.69 |
| 2 | 95.50 | 225.23 | 9.83 | |
| 3 | 80.18 | 36.94 | 8.77 | |
| 4 | 82.88 | 9.91 | 60.00 | |
| 5 | 87.39 | 95.50 | 2.67 | |
| 6 | 81.08 | 17.12 | 13.04 | |
| 7 | 72.97 | 9.91 | 7.69 | |
| 8 | 29.73 | 0.90 | 0.00 | |
| 9 | 8.11 | 0.00 | 0.00 | |
| 10 | 45.95 | 0.00 | 0.00 | |
| 11 | 91.89 | 63.06 | 7.80 | |
| 12 | 91.89 | 0.00 | 0.00 | |
| Industrial Sfela 2 | 1 | 20.72 | 1.80 | 50.00 |
| 2 | 79.28 | 122.52 | 14.80 | |
| 3 | 11.71 | 3.60 | 100.00 | |
| 4 | 93.69 | 80.18 | 12.73 | |
| 5 | 94.59 | 161.26 | 10.29 | |
| Sfela touloumotiri | 1 | 48.65 | 11.71 | 40.00 |
| 2 | 94.59 | 207.21 | 6.35 | |
| 3 | 57.66 | 14.41 | 18.75 | |
| 4 | 82.88 | 8.11 | 33.33 | |
| 5 | 67.57 | 25.23 | 28.57 | |
| 6 | 91.89 | 81.08 | 62.04 | |
| 7 | 86.49 | 12.61 | 50.00 | |
| 8 | 93.69 | 74.77 | 6.52 | |
| 9 | 72.07 | 18.02 | 40.00 | |
| 10 | 54.95 | 5.41 | 44.44 | |
| 11 | 91.89 | 46.85 | 8.33 | |
| 12 | 95.50 | 36.94 | 12.24 | |
| 13 | 72.97 | 0.90 | 0.00 | |
| Xerosfeli | 1 | 54.95 | 29.73 | 26.32 |
| 2 | 94.59 | 87.39 | 63.79 | |
| 3 | 95.50 | 85.59 | 10.17 | |
| 4 | 90.09 | 0.00 | 0.00 | |
| 5 | 95.50 | 138.74 | 3.70 | |
| 6 | 93.69 | 9.01 | 25.00 | |
| 7 | 68.47 | 2.70 | 0.00 | |
| 8 | 95.50 | 3.60 | 0.00 | |
| 9 | 61.26 | 18.02 | 39.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsouggou, N.; Slavko, A.; Tsipidou, O.; Georgoulis, A.; Dimov, S.G.; Yin, J.; Vorgias, C.E.; Kapolos, J.; Papadelli, M.; Papadimitriou, K. Investigation of the Microbiome of Industrial PDO Sfela Cheese and Its Artisanal Variants Using 16S rDNA Amplicon Sequencing and Shotgun Metagenomics. Foods 2024, 13, 1023. https://doi.org/10.3390/foods13071023
Tsouggou N, Slavko A, Tsipidou O, Georgoulis A, Dimov SG, Yin J, Vorgias CE, Kapolos J, Papadelli M, Papadimitriou K. Investigation of the Microbiome of Industrial PDO Sfela Cheese and Its Artisanal Variants Using 16S rDNA Amplicon Sequencing and Shotgun Metagenomics. Foods. 2024; 13(7):1023. https://doi.org/10.3390/foods13071023
Chicago/Turabian StyleTsouggou, Natalia, Aleksandra Slavko, Olympia Tsipidou, Anastasios Georgoulis, Svetoslav G. Dimov, Jia Yin, Constantinos E. Vorgias, John Kapolos, Marina Papadelli, and Konstantinos Papadimitriou. 2024. "Investigation of the Microbiome of Industrial PDO Sfela Cheese and Its Artisanal Variants Using 16S rDNA Amplicon Sequencing and Shotgun Metagenomics" Foods 13, no. 7: 1023. https://doi.org/10.3390/foods13071023
APA StyleTsouggou, N., Slavko, A., Tsipidou, O., Georgoulis, A., Dimov, S. G., Yin, J., Vorgias, C. E., Kapolos, J., Papadelli, M., & Papadimitriou, K. (2024). Investigation of the Microbiome of Industrial PDO Sfela Cheese and Its Artisanal Variants Using 16S rDNA Amplicon Sequencing and Shotgun Metagenomics. Foods, 13(7), 1023. https://doi.org/10.3390/foods13071023









