Screening, Identification, and Fermentation Characteristics of Lactic Acid Bacteria from Pickled Potherb Mustard and Potential Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material and Treatment
2.2. Isolation of Bacterial Strains
2.3. Molecular Identification
2.4. Fermentation Characteristics of Isolated Bacteria
2.4.1. Growth Curve and Acid Production Capacity
2.4.2. Nitrite Degradation Capacity
2.4.3. Acid and Salt Tolerances
2.5. Assessment of Functional Properties
2.5.1. Antibacterial Capacity
2.5.2. Antioxidant Capacity
2.5.3. Identification of Peptides and Antioxidant Capacity
2.6. Inoculation Fermentation of Potherb Mustard (Brassica juncea var. crispifolia)
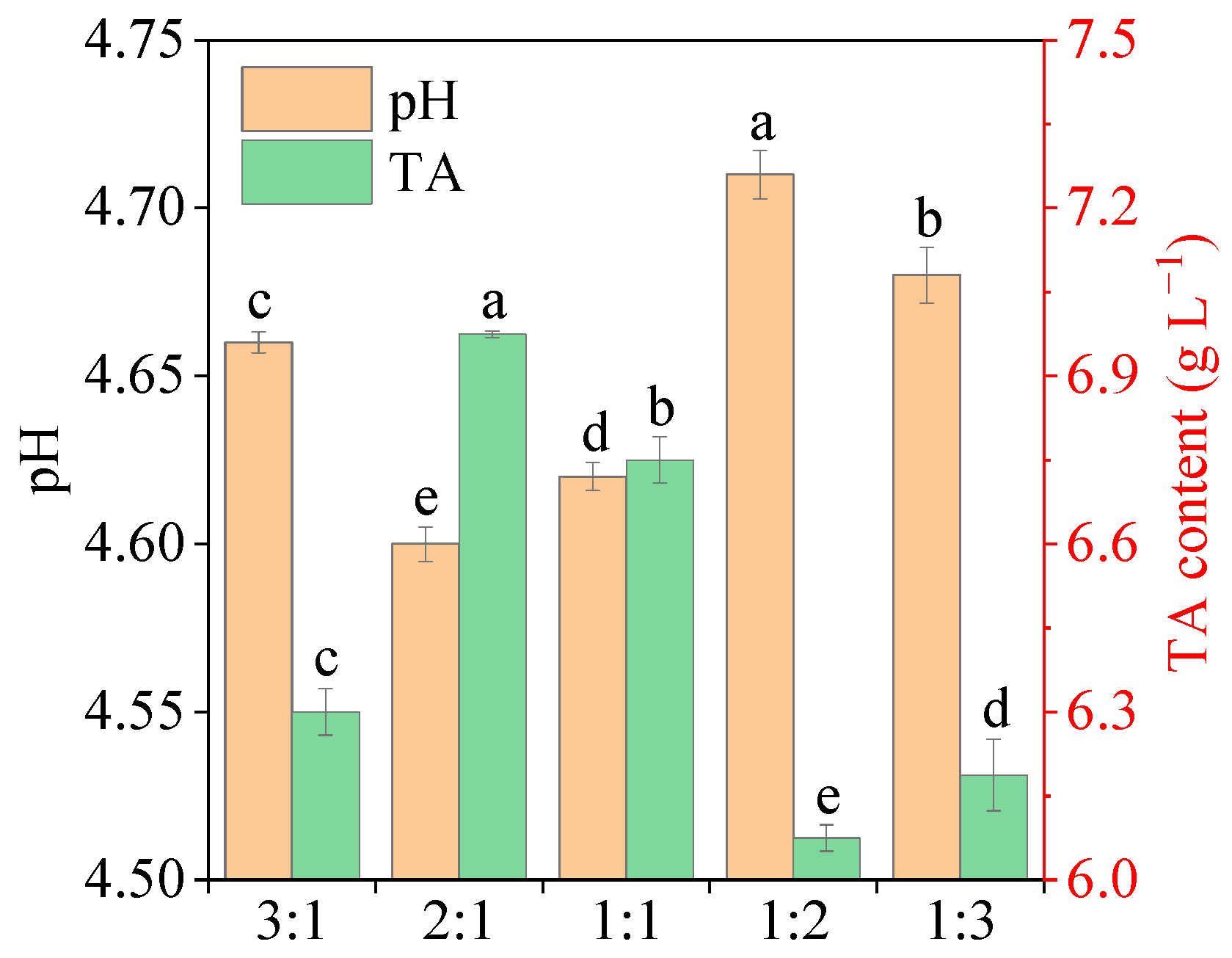
2.7. Chemical Analysis of Fermented Potherb Mustard
2.8. Microbial Diversity Analyses
2.9. Statistical Analyses
3. Results
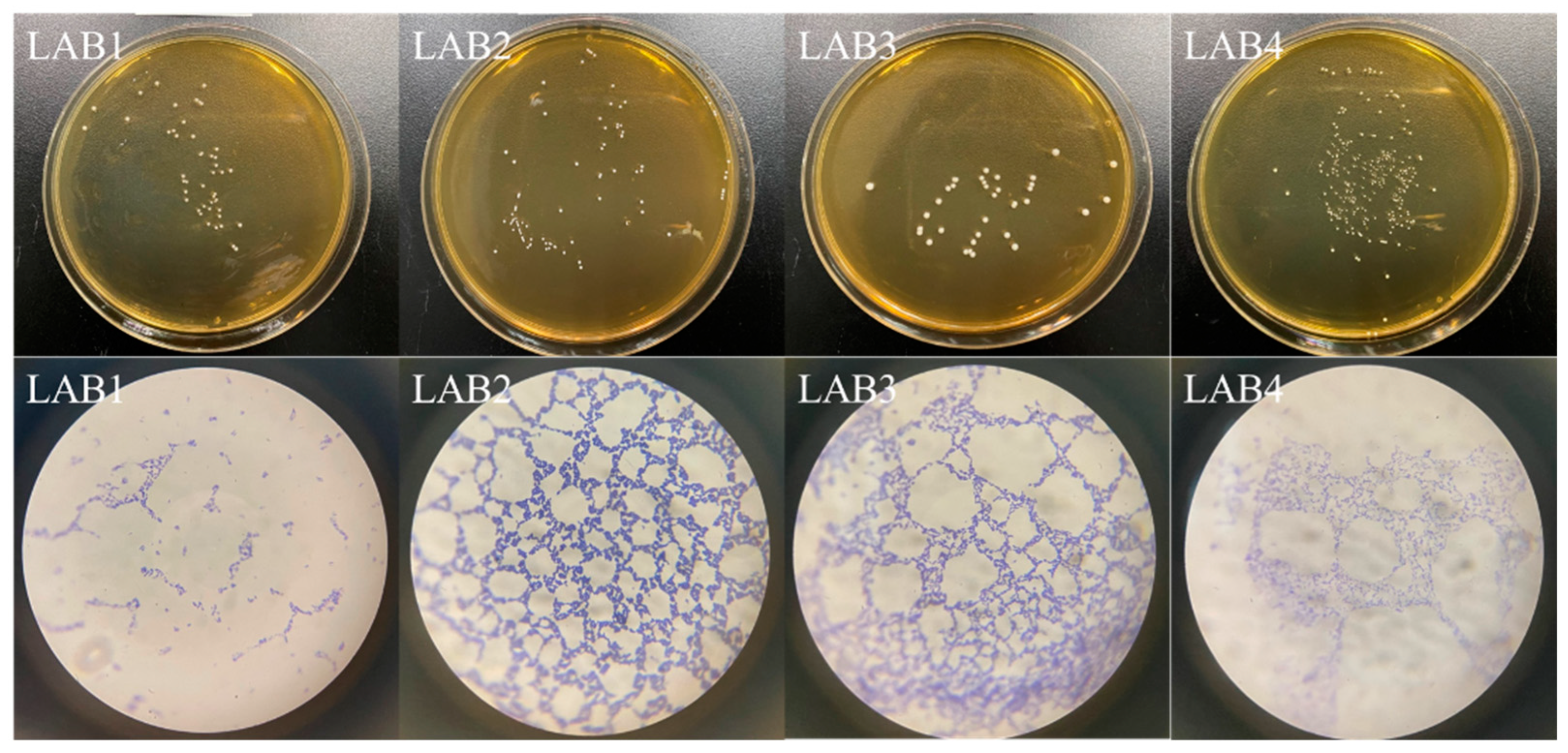
3.1. Isolation of LAB
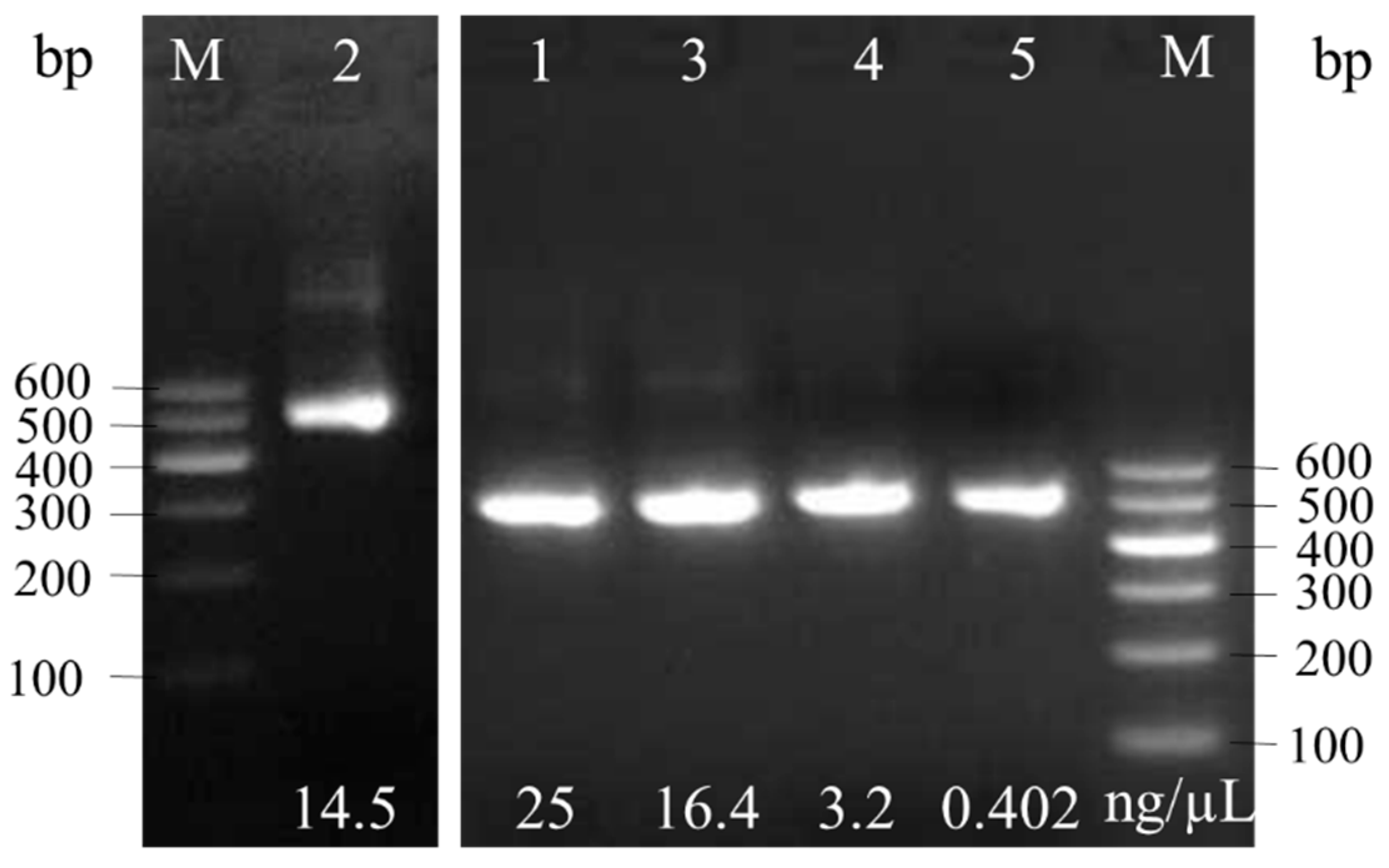
3.2. Molecular Identification
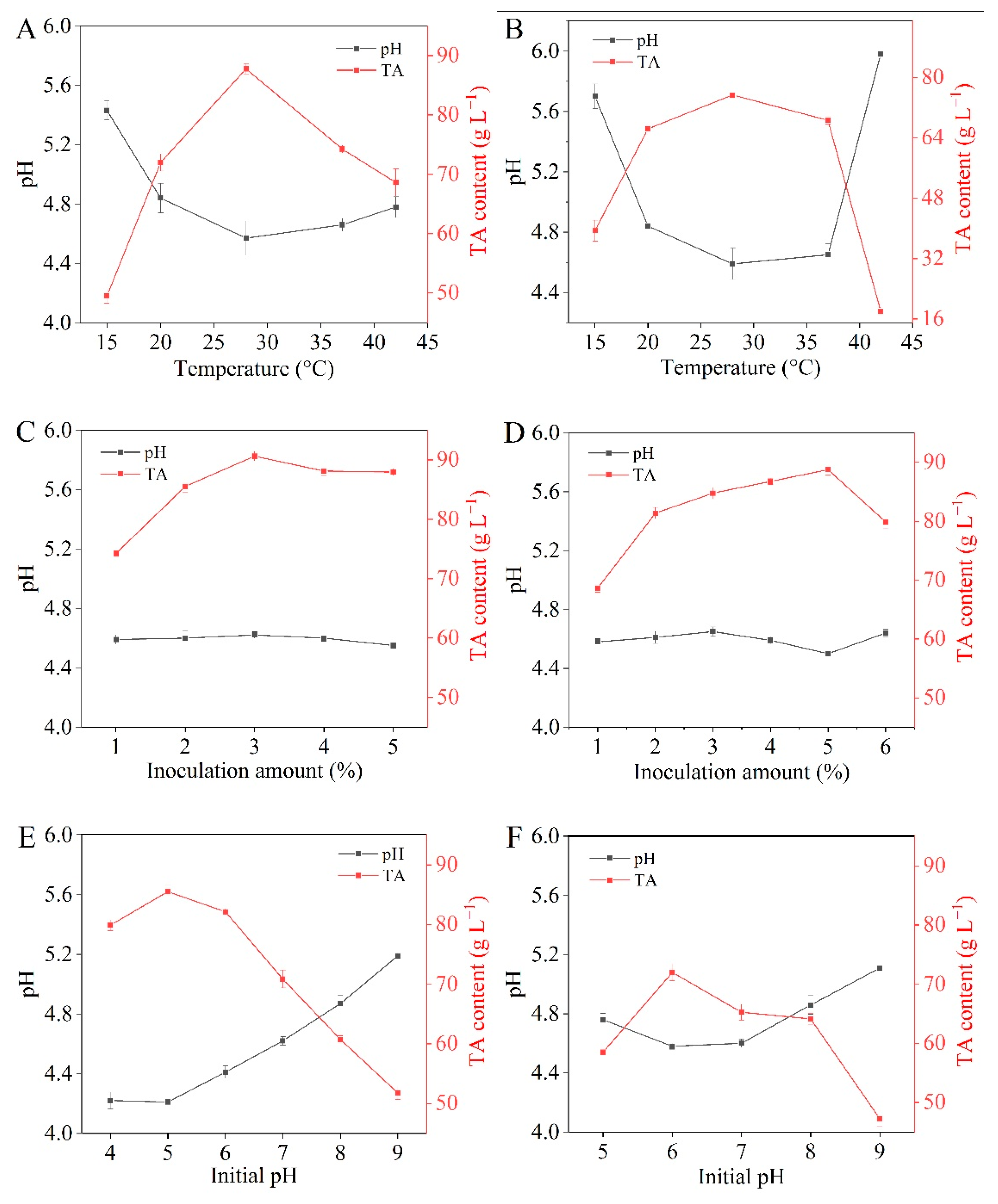
3.3. Fermentation Characteristics of LAB
3.3.1. Growth Curve and Acid Production Capacity
3.3.2. Nitrite Degradation Activity
3.3.3. Acid and Salt Tolerances
3.4. Functional Properties of LAB
3.4.1. Antibacterial Capacity
3.4.2. Antioxidant Capacity
3.4.3. Evaluation of Antioxidant Peptides
3.5. Fermentation of Mustard
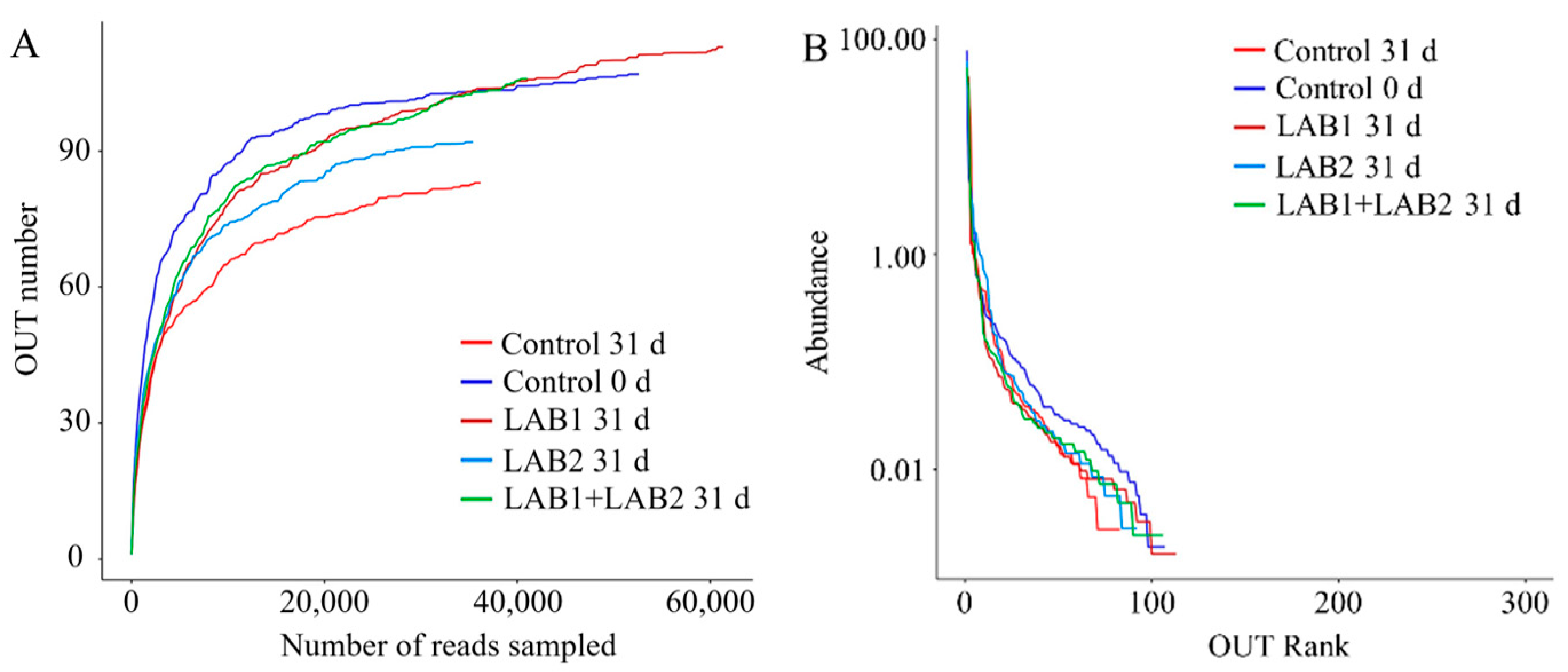
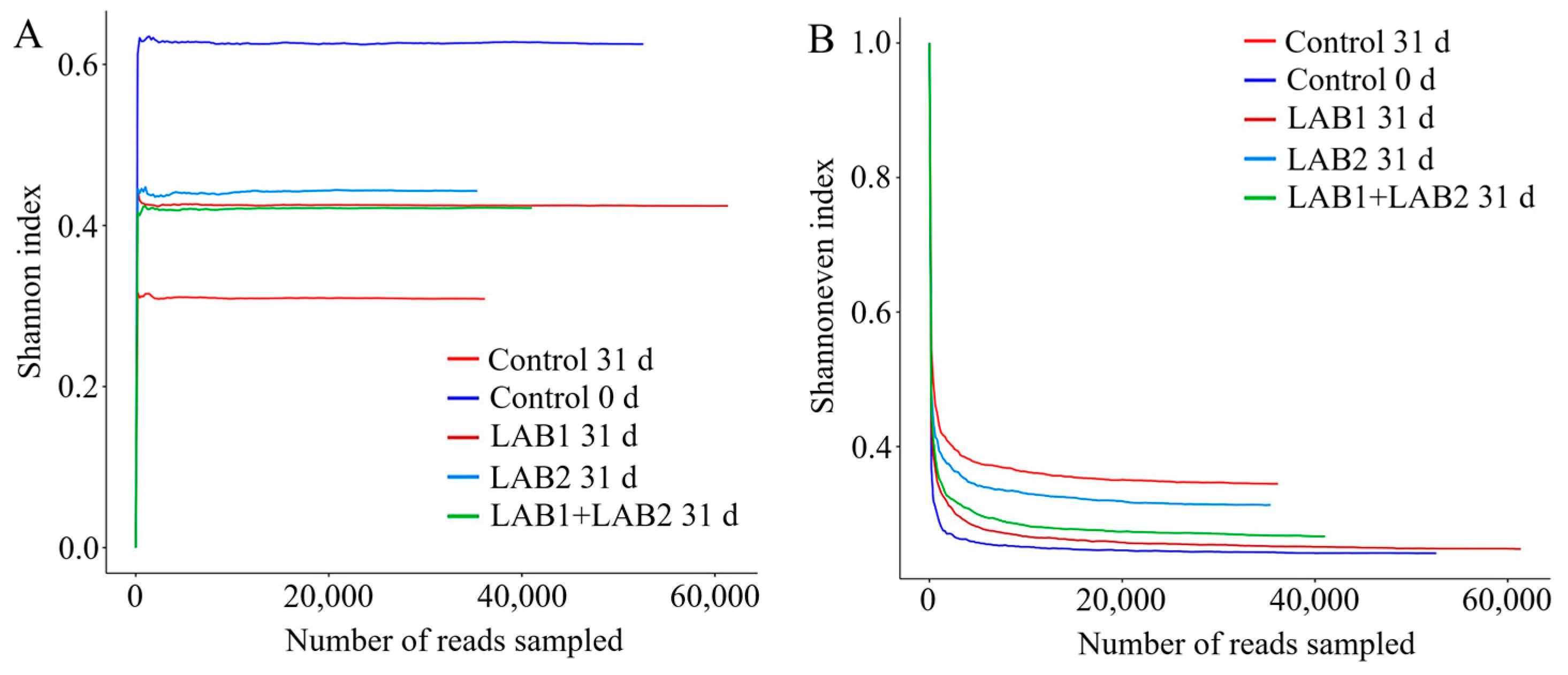
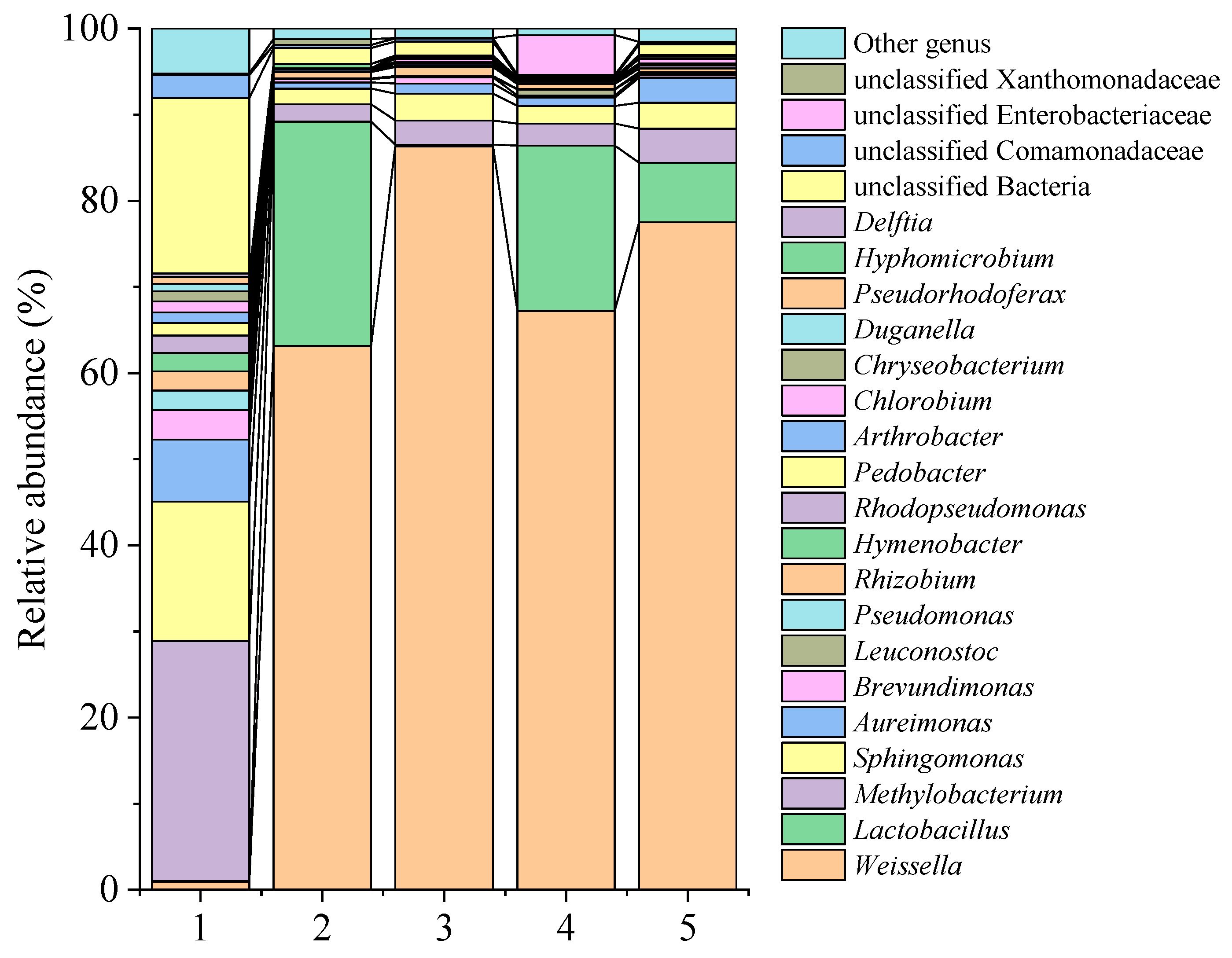
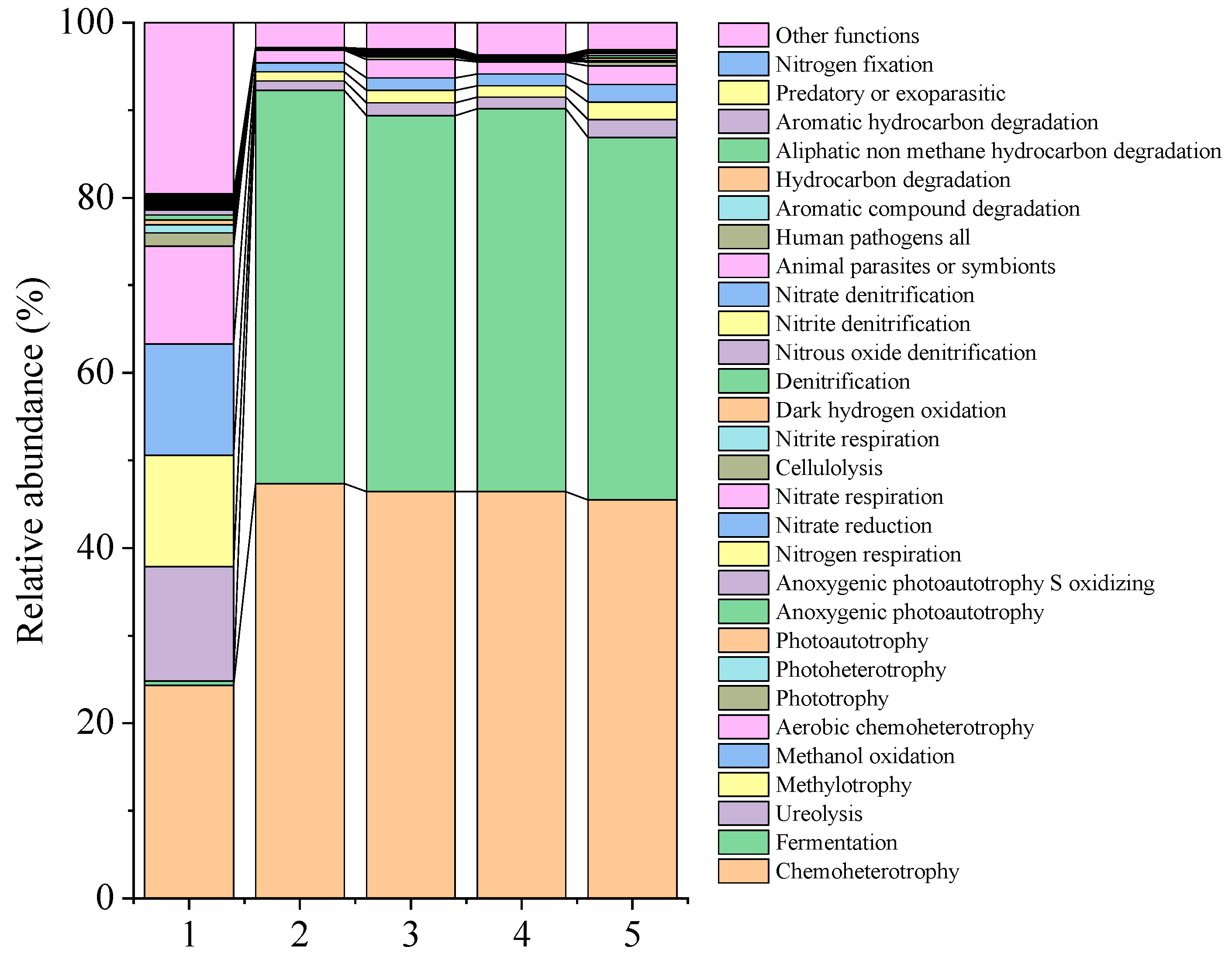
3.6. Microbial Community Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
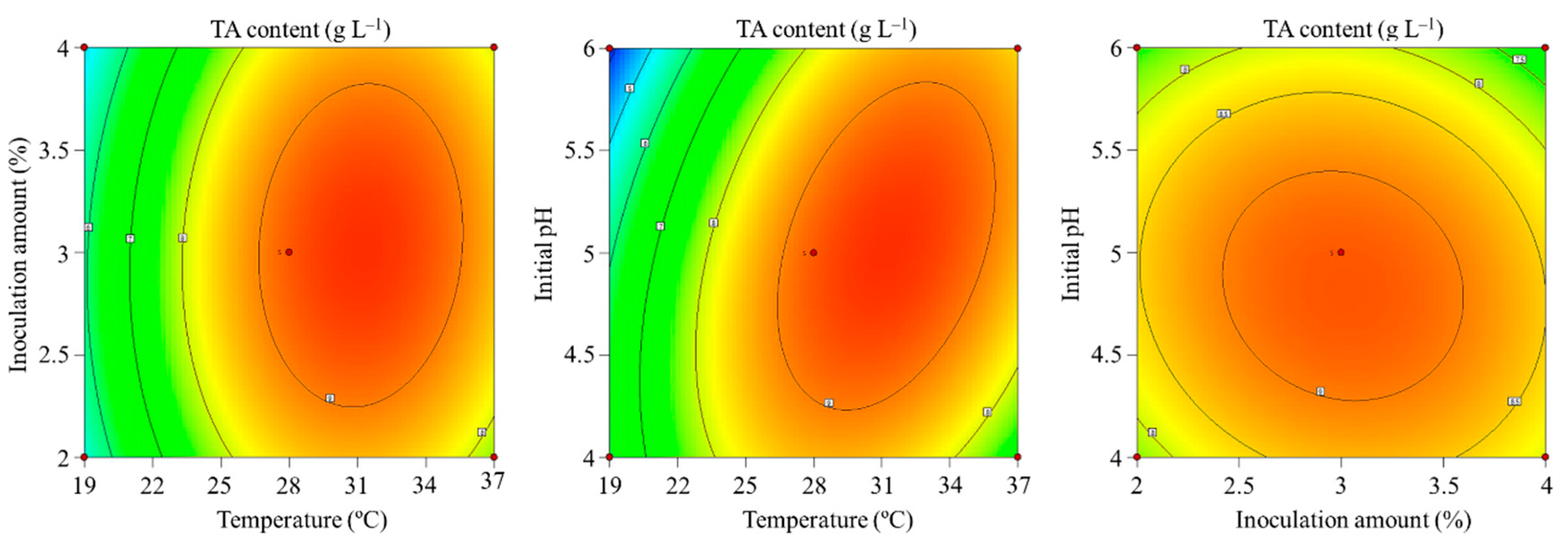
| Independent Variables | Design Factors | Ranges and Levels | ||
|---|---|---|---|---|
| Low Value (−1) | 0 | High Value (+1) | ||
| Temperature (°C) | A | 19.0 | 28.0 | 37.0 |
| Inoculation amount (%) | B | 2.0 | 3.0 | 4.0 |
| Initial pH | C | 4.0 | 5.0 | 6.0 |
| Run | Variables | Responses | ||
|---|---|---|---|---|
| Temperature (A) | Inoculation Amount (B) | Initial pH (C) | Total Acid (g L−1) | |
| 1 | −1 | −1 | 0 | 50.63 |
| 2 | 1 | −1 | 0 | 78.75 |
| 3 | −1 | 1 | 0 | 47.25 |
| 4 | 1 | 1 | 0 | 83.25 |
| 5 | −1 | 0 | −1 | 70.88 |
| 6 | 1 | 0 | −1 | 74.25 |
| 7 | −1 | 0 | 1 | 34.88 |
| 8 | 1 | 0 | 1 | 76.50 |
| 9 | 0 | −1 | −1 | 72.00 |
| 10 | 0 | 1 | −1 | 74.25 |
| 11 | 0 | −1 | 1 | 81.00 |
| 12 | 0 | 1 | 1 | 76.50 |
| 13 | 0 | 0 | 0 | 97.88 |
| 14 | 0 | 0 | 0 | 95.63 |
| 15 | 0 | 0 | 0 | 87.75 |
| 16 | 0 | 0 | 0 | 90.00 |
| 17 | 0 | 0 | 0 | 91.13 |
| Source | Sum of Squares | DF | Mean Square | F Value | p-Value | Significant |
|---|---|---|---|---|---|---|
| Model | 43.47 | 9 | 4.83 | 9.15 | 0.0040 | ** |
| A | 14.89 | 1 | 14.89 | 28.19 | 0.0011 | ** |
| B | 0.0016 | 1 | 0.0016 | 0.0030 | 0.9579 | |
| C | 0.6328 | 1 | 0.6328 | 1.20 | 0.3099 | |
| AB | 0.1550 | 1 | 0.1550 | 0.2936 | 0.6047 | |
| AC | 3.66 | 1 | 3.66 | 6.93 | 0.0338 | * |
| BC | 0.1139 | 1 | 0.1139 | 0.2157 | 0.6564 | |
| A2 | 16.27 | 1 | 16.27 | 30.82 | 0.0009 | ** |
| B2 | 2.59 | 1 | 2.59 | 4.91 | 0.0623 | |
| C2 | 3.18 | 1 | 3.18 | 6.02 | 0.0438 | * |
| Residual | 3.70 | 7 | 0.5280 | |||
| Lack of Fit | 3.00 | 3 | 1.00 | 5.77 | 0.0617 | |
| Pure Error | 0.6936 | 4 | 0.1734 | |||
| Cor Total | 47.17 | 16 |
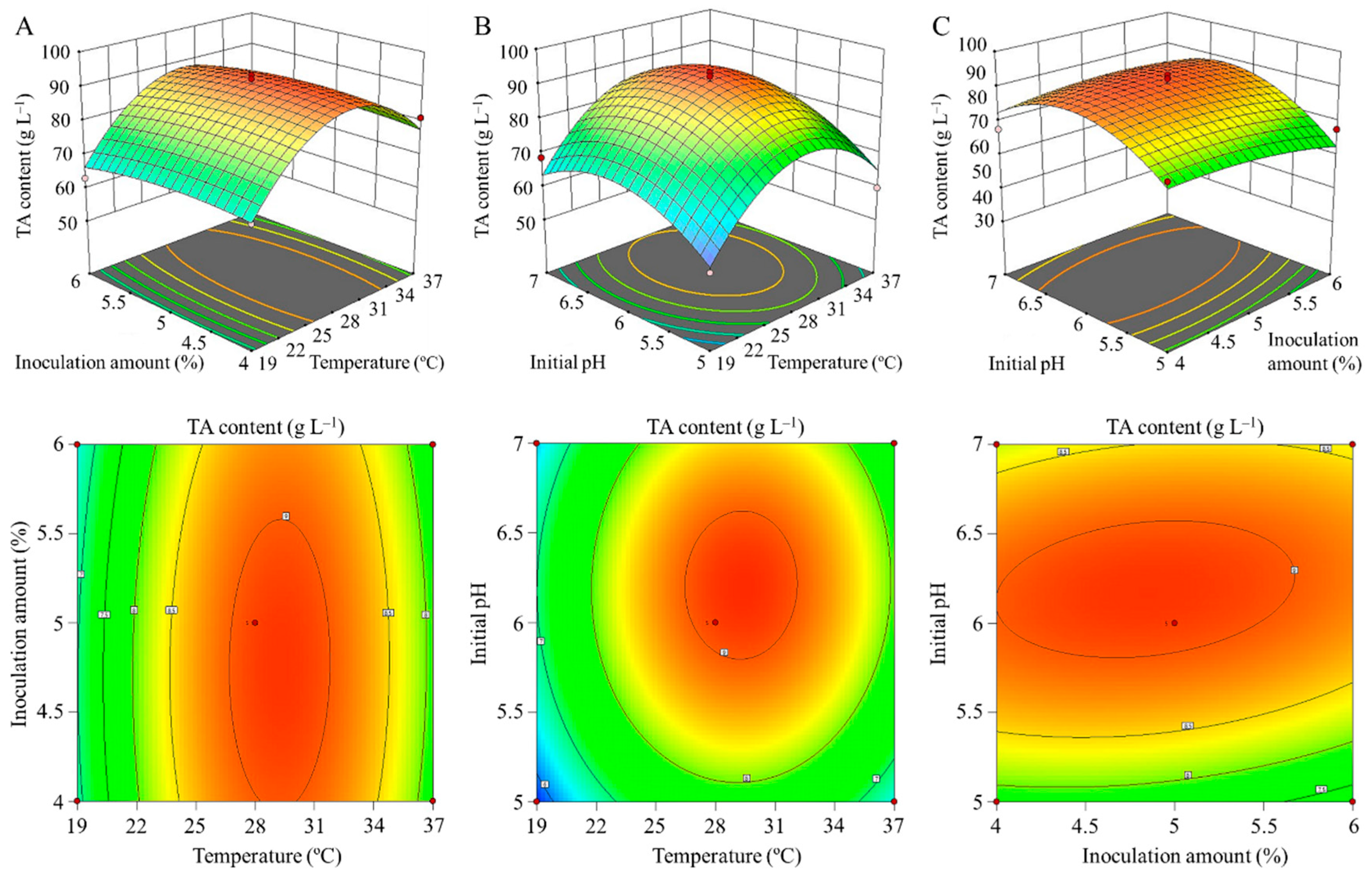
| Independent Variables | Design Factors | Ranges and Levels | ||
|---|---|---|---|---|
| Low Value (−1) | 0 | High Value (+1) | ||
| Temperature (°C) | A | 19.0 | 28.0 | 37.0 |
| Inoculation amount (%) | B | 4.0 | 5.0 | 6.0 |
| Initial pH | C | 5.0 | 6.0 | 7.0 |
| Run | Variables | Responses | ||
|---|---|---|---|---|
| Temperature (A) | Inoculation Amount (B) | Initial pH (C) | Total Acid (g L−1) | |
| 1 | −1 | −1 | 0 | 68.63 |
| 2 | 1 | −1 | 0 | 81.00 |
| 3 | −1 | 1 | 0 | 63.00 |
| 4 | 1 | 1 | 0 | 76.50 |
| 5 | −1 | 0 | −1 | 55.13 |
| 6 | 1 | 0 | −1 | 59.63 |
| 7 | −1 | 0 | 1 | 68.63 |
| 8 | 1 | 0 | 1 | 75.38 |
| 9 | 0 | −1 | −1 | 79.88 |
| 10 | 0 | 1 | −1 | 77.63 |
| 11 | 0 | −1 | 1 | 77.63 |
| 12 | 0 | 1 | 1 | 82.13 |
| 13 | 0 | 0 | 0 | 88.88 |
| 14 | 0 | 0 | 0 | 93.38 |
| 15 | 0 | 0 | 0 | 90.00 |
| 16 | 0 | 0 | 0 | 90.00 |
| 17 | 0 | 0 | 0 | 92.25 |
| Source | Sum of Squares | DF | Mean Square | F Value | p-Value | Significant |
|---|---|---|---|---|---|---|
| Model | 19.93 | 9 | 2.21 | 10.29 | 0.0028 | ** |
| A | 1.72 | 1 | 1.72 | 8.00 | 0.0254 | ** |
| B | 0.0775 | 1 | 0.0775 | 0.3601 | 0.5673 | |
| C | 1.24 | 1 | 1.24 | 5.76 | 0.0474 | |
| AB | 0.0032 | 1 | 0.0032 | 0.0147 | 0.9069 | |
| AC | 0.0127 | 1 | 0.0127 | 0.0588 | 0.8154 | * |
| BC | 0.1139 | 1 | 0.1139 | 0.5292 | 0.4906 | |
| A2 | 11.63 | 1 | 11.63 | 54.05 | 0.0002 | ** |
| B2 | 0.1679 | 1 | 0.1679 | 0.7800 | 0.4064 | |
| C2 | 3.87 | 1 | 3.87 | 17.99 | 0.0038 | * |
| Residual | 1.51 | 7 | 0.2152 | |||
| Lack of Fit | 1.37 | 3 | 0.4567 | 13.36 | 0.0150 | |
| Pure Error | 0.1367 | 4 | 0.0342 | |||
| Cor Total | 21.44 | 16 |
| Strain | Diameter/mm | Appearance | Color | Edge | Adhesion |
|---|---|---|---|---|---|
| LAB1 | 0.76 ± 0.04 | Circular, moist, and glossy | Milk white | Neat and regular | Weak |
| LAB2 | 0.35 ± 0.06 | Circular, smooth, and moist | White | Neat and regular | Weak |
| LAB3 | 1.40 ± 0.08 | Circular, moist, and glossy | Milk white | Neat and regular | Weak |
| LAB4 | 0.17 ± 0.02 | Circular, smooth, and moist | White | Neat and regular | Weak |
| Tests | Characteristics | |||
|---|---|---|---|---|
| LAB1 | LAB2 | LAB3 | LAB4 | |
| Catalase activity | − | − | − | − |
| Growth in 65 g L−1 NaCl | + | + | + | + |
| Bacterial motility test | − | − | − | − |
| Nitrate reduction | − | − | − | − |
| Acid and gas production by fermentation of glucose | + | + | + | + |
| Hydrogen sulfide production | − | − | − | − |
| Gelatin liquefaction | − | − | − | − |
| Indole test | − | − | − | − |
| Bile esculin test | + | + | + | + |
| Culture Duration/h | Nitrite Degradation Rate/% | |||
|---|---|---|---|---|
| LAB1 | LAB2 | LAB3 | LAB4 | |
| 12 | 26.43 ± 4.57 b | 89.63 ± 4.63 a | 8.95 ± 2.58 d | 21.53 ± 3.11 c |
| 24 | 73.55 ± 2.53 b | 90.46 ± 5.31 a | 38.17 ± 3.81 d | 63.06 ± 6.76 c |
| 36 | 87.11 ± 4.34 b | 91.16 ± 5.35 a | 61.94 ± 8.67 d | 77.32 ± 4.17 c |
| 48 | 91.02 ± 2.75 a | 91.72 ± 6.87 a | 79.7 ± 3.14 c | 86.83 ± 6.32 b |
| 60 | 91.58 ± 4.27 a | 91.86 ± 4.36 a | 87.39 ± 6.08 c | 89.9 ± 5.34 b |
| 72 | 91.72 ± 3.65 b | 92 ± 2.72 a | 90.32 ± 6.18 c | 90.32 ± 4.58 c |
| Free Radical Scavenging Rate/% | Strains | ||||
|---|---|---|---|---|---|
| LAB1 | LAB2 | LAB3 | LAB4 | ||
| DPPH | CSs | 33.07 ± 5.91 a | 25.68 ± 1.85 b | 11.67 ± 2.56 d | 15.18 ± 2.20 c |
| CFE | 19.07 ± 3.34 a | 15.95 ± 4.85 b | 4.67 ± 3.73 d | 8.56 ± 1.95 c | |
| OH | CSs | 22.26 ± 3.35 a | 14.46 ± 2.87 c | 16.10 ± 2.03 b | 16.70 ± 2.91 b |
| CFE | 21.61 ± 4.94 a | 13.96 ± 2.84 b | 11.41 ± 3.46 c | 13.29 ± 1.65 b | |
| O2·− | CSs | 52.21 ± 5.53 a | 21.59 ± 7.27 d | 37.52 ± 5.89 b | 27.62 ± 3.00 c |
| CFE | 13.81 ± 4.27 c | 17.00 ± 6.00 a | 9.38 ± 1.69 d | 14.87 ± 2.88 b | |
| Reducing ability | CSs | 54.29 ± 4.84 a | 40.00 ± 5.45 b | 20.00 ± 2.67 d | 22.86 ± 2.26 c |
| CFE | 11.43 ± 2.13 b | 20.00 ± 2.46 a | 2.86 ± 1.94 d | 5.71 ± 2.50 c | |
| Samples | Barcode | Sequence Number | Base Number | Mean Length | Min Length | Max Length | OUTs |
|---|---|---|---|---|---|---|---|
| Control 0 d | AGAACA | 64,812 | 26,373,965 | 406.93 | 356 | 469 | 107.0 |
| Control 31 d | TATCGA | 46,099 | 19,313,956 | 418.97 | 350 | 446 | 83.0 |
| LAB1 31 d | GGTGTG | 72,345 | 30,161,842 | 416.92 | 352 | 446 | 113.0 |
| LAB2 31 d | AGAGAC | 45,003 | 19,164,849 | 425.86 | 352 | 451 | 92.0 |
| LAB1+LAB2 31 d | TATGCA | 47,933 | 19,888,245 | 414.92 | 350 | 453 | 106.0 |
| Samples | Number | Shannon | Chao1 | Ace | Simpson | Shannoneven | Coverage |
|---|---|---|---|---|---|---|---|
| Control 0 d | 52,578.0 | 1.13 | 116.0 | 112.39 | 0.63 | 0.24 | 1.00 |
| Control 31 d | 36,163.0 | 1.52 | 96.0 | 91.77 | 0.31 | 0.34 | 1.00 |
| LAB1 31 d | 61,354.0 | 1.17 | 123.11 | 119.45 | 0.42 | 0.25 | 1.00 |
| LAB2 31 d | 35,390.0 | 1.42 | 95.6 | 96.14 | 0.44 | 0.31 | 1.00 |
| LAB1+LAB2 31 d | 41,042.0 | 1.24 | 121.11 | 115.92 | 0.42 | 0.27 | 1.00 |
| Genus | CK 0 d | CK 31 d | LAB1 31 d | LAB2 31 d | LAB1+LAB2 31 d |
|---|---|---|---|---|---|
| Weissella | 0.94 | 63.14 | 86.31 | 67.22 | 77.52 |
| Lactobacillus | 0.05 | 26.05 | 0.20 | 19.20 | 6.91 |
| Methylobacterium | 27.92 | 2.02 | 2.81 | 2.53 | 3.95 |
| Sphingomonas | 16.16 | 1.82 | 3.09 | 2.06 | 3.02 |
| Aureimonas | 7.22 | 0.72 | 1.21 | 0.98 | 2.92 |
| Brevundimonas | 3.38 | 0.37 | 0.72 | 0.21 | 0.27 |
| Leuconostoc | 0.01 | 0.06 | 0.01 | 0.74 | 0.13 |
| Pseudomonas | 2.30 | 0.03 | 0.15 | 0.04 | 0.21 |
| Rhizobium | 2.21 | 0.75 | 1.04 | 0.61 | 0.41 |
| Hymenobacter | 2.12 | 0.01 | 0.08 | 0.09 | 0.05 |
| Rhodopseudomonas | 2.06 | 0.01 | 0.32 | 0.24 | 0.33 |
| Pedobacter | 1.43 | 0.08 | 0.16 | 0.10 | 0.19 |
| Arthrobacter | 1.27 | 0.27 | 0.03 | 0.05 | 0.04 |
| Chlorobium | 1.27 | 0.00 | 0.35 | 0.06 | 0.56 |
| Chryseobacterium | 1.19 | 0.06 | 0.09 | 0.03 | 0.26 |
| Duganella | 0.86 | 0.03 | 0.13 | 0.04 | 0.07 |
| Pseudorhodoferax | 0.77 | 0.00 | 0.01 | 0.01 | 0.01 |
| Hyphomicrobium | 0.00 | 0.48 | 0.00 | 0.00 | 0.00 |
| Delftia | 0.42 | 0.00 | 0.13 | 0.05 | 0.10 |
| unclassified Bacteria | 20.37 | 1.83 | 1.65 | 0.12 | 1.23 |
| unclassified Comamonadaceae | 2.63 | 0.35 | 0.38 | 0.22 | 0.25 |
| unclassified Enterobacteriaceae | 0.20 | 0.03 | 0.04 | 4.60 | 0.04 |
| unclassified Xanthomonadaceae | 0.04 | 0.66 | 0.01 | 0.00 | 0.01 |
| Other genus | 5.18 | 1.25 | 1.07 | 0.79 | 1.55 |
| Functions | CK 0d | CK 31 d | LAB1 31 d | LAB2 31 d | LAB1+LAB2 31 d |
|---|---|---|---|---|---|
| Chemoheterotrophy | 24.32 | 47.35 | 46.44 | 46.42 | 45.50 |
| Fermentation | 0.51 | 44.92 | 42.94 | 43.77 | 41.39 |
| Ureolysis | 13.04 | 1.05 | 1.47 | 1.30 | 2.04 |
| Methylotrophy | 12.72 | 1.05 | 1.42 | 1.31 | 2.01 |
| Methanol oxidation | 12.72 | 1.05 | 1.42 | 1.31 | 2.01 |
| Aerobic chemoheterotrophy | 11.16 | 1.39 | 2.08 | 1.38 | 2.13 |
| Phototrophy | 1.50 | 0.01 | 0.33 | 0.15 | 0.43 |
| Photoheterotrophy | 0.91 | 0.01 | 0.15 | 0.12 | 0.16 |
| Photoautotrophy | 0.56 | 0.00 | 0.17 | 0.03 | 0.27 |
| Anoxygenic photoautotrophy | 0.56 | 0.00 | 0.17 | 0.03 | 0.27 |
| Anoxygenic photoautotrophy S oxidizing | 0.56 | 0.00 | 0.17 | 0.03 | 0.27 |
| Nitrogen respiration | 0.16 | 0.03 | 0.03 | 0.05 | 0.05 |
| Nitrate reduction | 0.16 | 0.03 | 0.03 | 0.05 | 0.05 |
| Nitrate respiration | 0.16 | 0.03 | 0.03 | 0.05 | 0.05 |
| Cellulolysis | 0.13 | 0.01 | 0.01 | 0.04 | 0.03 |
| Nitrite respiration | 0.13 | 0.01 | 0.01 | 0.04 | 0.03 |
| Dark hydrogen oxidation | 0.13 | 0.01 | 0.01 | 0.04 | 0.03 |
| Denitrification | 0.13 | 0.01 | 0.01 | 0.04 | 0.03 |
| Nitrous oxide denitrification | 0.13 | 0.01 | 0.01 | 0.04 | 0.03 |
| Nitrite denitrification | 0.13 | 0.01 | 0.01 | 0.04 | 0.03 |
| Nitrate denitrification | 0.13 | 0.01 | 0.03 | 0.00 | 0.02 |
| Animal parasites or symbionts | 0.11 | 0.00 | 0.01 | 0.02 | 0.03 |
| Human pathogens (all) | 0.11 | 0.00 | 0.01 | 0.02 | 0.03 |
| Aromatic compound degradation | 0.08 | 0.03 | 0.01 | 0.01 | 0.02 |
| Hydrocarbon degradation | 0.05 | 0.01 | 0.01 | 0.00 | 0.00 |
| Aliphatic non-methane hydrocarbon degradation | 0.05 | 0.01 | 0.01 | 0.00 | 0.00 |
| Aromatic hydrocarbon degradation | 0.05 | 0.01 | 0.01 | 0.00 | 0.00 |
| Predatory or exoparasitic | 0.00 | 0.05 | 0.00 | 0.00 | 0.00 |
| Nitrogen fixation | 0.00 | 0.00 | 0.00 | 0.00 | 0.03 |
| Other functions | 19.54 | 2.86 | 2.97 | 3.70 | 3.05 |





Appendix B
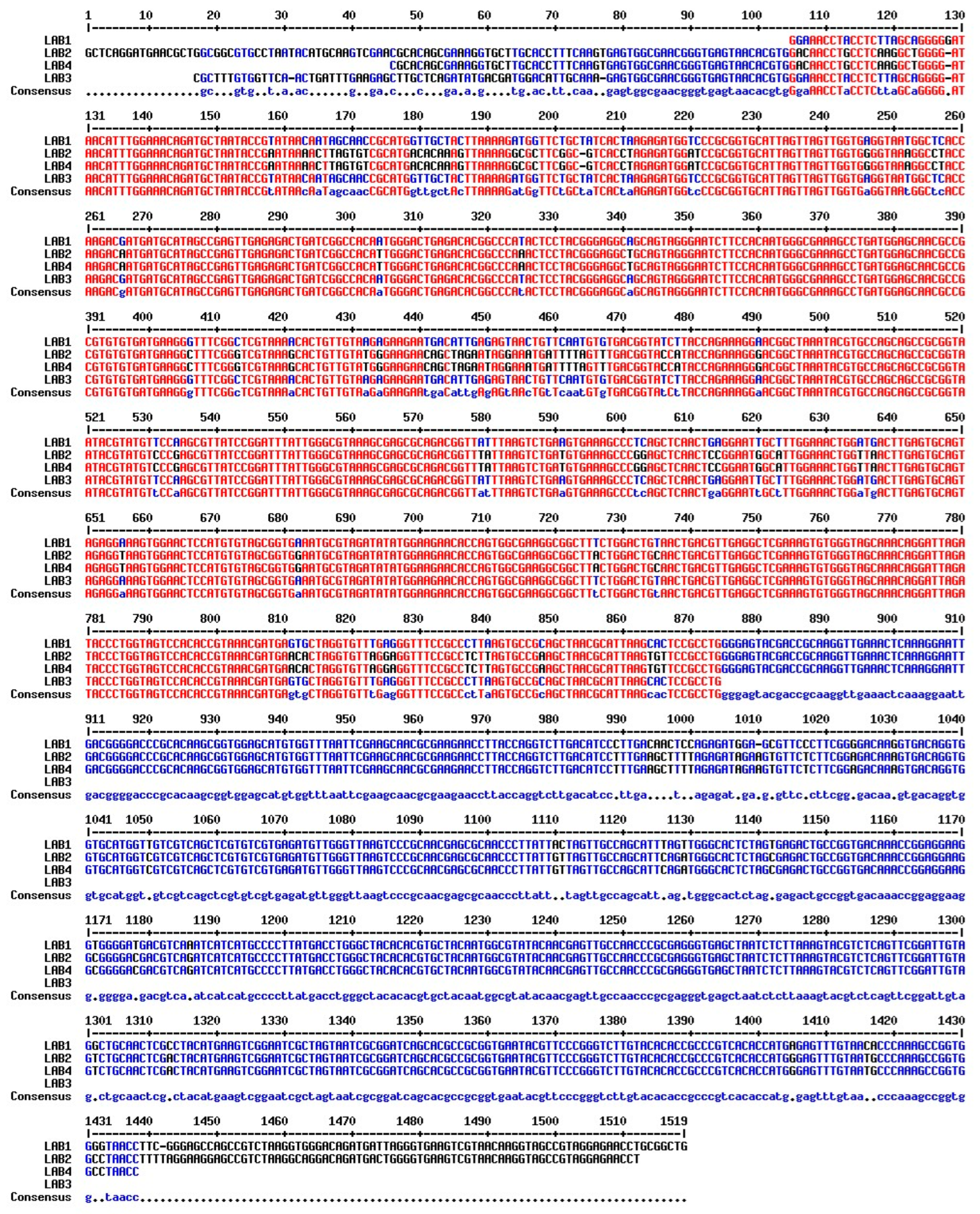
- Deoxyribonucleic acid sequence of LAB1
- GGAAACCTACCTCTTAGCAGGGGGATAACATTTGGAAACAGATGCTAATACCGTATAACAATAGCAACCGCATGGTTGCTACTTAAAAGATGGTTCTGCTATCACTAAGAGATGGTCCCGCGGTGCATTAGTTAGTTGGTGAGGTAATGGCTCACCAAGACGATGATGCATAGCCGAGTTGAGAGACTGATCGGCCACAATGGGACTGAGACACGGCCCATACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCACAATGGGCGAAAGCCTGATGGAGCAACGCCGCGTGTGTGATGAAGGGTTTCGGCTCGTAAAACACTGTTGTAAGAGAAGAATGACATTGAGAGTAACTGTTCAATGTGTGACGGTATCTTACCAGAAAGGAACGGCTAAATACGTGCCAGCAGCCGCGGTAATACGTATGTTCCAAGCGTTATCCGGATTTATTGGGCGTAAAGCGAGCGCAGACGGTTATTTAAGTCTGAAGTGAAAGCCCTCAGCTCAACTGAGGAATTGCTTTGGAAACTGGATGACTTGAGTGCAGTAGAGGAAAGTGGAACTCCATGTGTAGCGGTGAAATGCGTAGATATATGGAAGAACACCAGTGGCGAAGGCGGCTTTCTGGACTGTAACTGACGTTGAGGCTCGAAAGTGTGGGTAGCAAACAGGATTAGATACCCTGGTAGTCCACACCGTAAACGATGAGTGCTAGGTGTTTGAGGGTTTCCGCCCTTAAGTGCCGCAGCTAACGCATTAAGCACTCCGCCTGGGGAGTACGACCGCAAGGTTGAAACTCAAAGGAATTGACGGGGACCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGACATCCCTTGACAACTCCAGAGATGGAGCGTTCCCTTCGGGGACAAGGTGACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTATTACTAGTTGCCAGCATTTAGTTGGGCACTCTAGTGAGACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAAATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGCGTATACAACGAGTTGCCAACCCGCGAGGGTGAGCTAATCTCTTAAAGTACGTCTCAGTTCGGATTGTAGGCTGCAACTCGCCTACATGAAGTCGGAATCGCTAGTAATCGCGGATCAGCACGCCGCGGTGAATACGTTCCCGGGTCTTGTACACACCGCCCGTCACACCATGAGAGTTTGTAACACCCAAAGCCGGTGGGGTAACCTTCGGGAGCCAGCCGTCTAAGGTGGGACAGATGATTAGGGTGAAGTCGTAACAAGGTAGCCGTAGGAGAACCTGCGGCTG
- Deoxyribonucleic acid sequence of LAB2
- GCTCAGGATGAACGCTGGCGGCGTGCCTAATACATGCAAGTCGAACGCACAGCGAAAGGTGCTTGCACCTTTCAAGTGAGTGGCGAACGGGTGAGTAACACGTGGACAACCTGCCTCAAGGCTGGGGATAACATTTGGAAACAGATGCTAATACCGAATAAAACTTAGTGTCGCATGACACAAAGTTAAAAGGCGCTTCGGCGTCACCTAGAGATGGATCCGCGGTGCATTAGTTAGTTGGTGGGGTAAAGGCCTACCAAGACAATGATGCATAGCCGAGTTGAGAGACTGATCGGCCACATTGGGACTGAGACACGGCCCAAACTCCTACGGGAGGCTGCAGTAGGGAATCTTCCACAATGGGCGAAAGCCTGATGGAGCAACGCCGCGTGTGTGATGAAGGCTTTCGGGTCGTAAAGCACTGTTGTATGGGAAGAACAGCTAGAATAGGAAATGATTTTAGTTTGACGGTACCATACCAGAAAGGGACGGCTAAATACGTGCCAGCAGCCGCGGTAATACGTATGTCCCGAGCGTTATCCGGATTTATTGGGCGTAAAGCGAGCGCAGACGGTTTATTAAGTCTGATGTGAAAGCCCGGAGCTCAACTCCGGAATGGCATTGGAAACTGGTTAACTTGAGTGCAGTAGAGGTAAGTGGAACTCCATGTGTAGCGGTGGAATGCGTAGATATATGGAAGAACACCAGTGGCGAAGGCGGCTTACTGGACTGCAACTGACGTTGAGGCTCGAAAGTGTGGGTAGCAAACAGGATTAGATACCCTGGTAGTCCACACCGTAAACGATGAACACTAGGTGTTAGGAGGTTTCCGCCTCTTAGTGCCGAAGCTAACGCATTAAGTGTTCCGCCTGGGGAGTACGACCGCAAGGTTGAAACTCAAAGGAATTGACGGGGACCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGACATCCTTTGAAGCTTTTAGAGATAGAAGTGTTCTCTTCGGAGACAAAGTGACAGGTGGTGCATGGTCGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTATTGTTAGTTGCCAGCATTCAGATGGGCACTCTAGCGAGACTGCCGGTGACAAACCGGAGGAAGGCGGGGACGACGTCAGATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGCGTATACAACGAGTTGCCAACCCGCGAGGGTGAGCTAATCTCTTAAAGTACGTCTCAGTTCGGATTGTAGTCTGCAACTCGACTACATGAAGTCGGAATCGCTAGTAATCGCGGATCAGCACGCCGCGGTGAATACGTTCCCGGGTCTTGTACACACCGCCCGTCACACCATGGGAGTTTGTAATGCCCAAAGCCGGTGGCCTAACCTTTTAGGAAGGAGCCGTCTAAGGCAGGACAGATGACTGGGGTGAAGTCGTAACAAGGTAGCCGTAGGAGAACCT
- Deoxyribonucleic acid sequence of LAB3
- CGCTTTGTGGTTCAACTGATTTGAAGAGCTTGCTCAGATATGACGATGGACATTGCAAAGAGTGGCGAACGGGTGAGTAACACGTGGGAAACCTACCTCTTAGCAGGGGATAACATTTGGAAACAGATGCTAATACCGTATAACAATAGCAACCGCATGGTTGCTACTTAAAAGATGGTTCTGCTATCACTAAGAGATGGTCCCGCGGTGCATTAGTTAGTTGGTGAGGTAATGGCTCACCAAGACGATGATGCATAGCCGAGTTGAGAGACTGATCGGCCACAATGGGACTGAGACACGGCCCATACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCACAATGGGCGAAAGCCTGATGGAGCAACGCCGCGTGTGTGATGAAGGGTTTCGGCTCGTAAAACACTGTTGTAAGAGAAGAATGACATTGAGAGTAACTGTTCAATGTGTGACGGTATCTTACCAGAAAGGAACGGCTAAATACGTGCCAGCAGCCGCGGTAATACGTATGTTCCAAGCGTTATCCGGATTTATTGGGCGTAAAGCGAGCGCAGACGGTTATTTAAGTCTGAAGTGAAAGCCCTCAGCTCAACTGAGGAATTGCTTTGGAAACTGGATGACTTGAGTGCAGTAGAGGAAAGTGGAACTCCATGTGTAGCGGTGAAATGCGTAGATATATGGAAGAACACCAGTGGCGAAGGCGGCTTTCTGGACTGTAACTGACGTTGAGGCTCGAAAGTGTGGGTAGCAAACAGGATTAGATACCCTGGTAGTCCACACCGTAAACGATGAGTGCTAGGTGTTTGAGGGTTTCCGCCCTTAAGTGCCGCAGCTAACGCATTAAGCACTCCGCCTG
- Deoxyribonucleic acid sequence of LAB4
- CGCACAGCGAAAGGTGCTTGCACCTTTCAAGTGAGTGGCGAACGGGTGAGTAACACGTGGACAACCTGCCTCAAGGCTGGGGATAACATTTGGAAACAGATGCTAATACCGAATAAAACTTAGTGTCGCATGACACAAAGTTAAAAGGCGCTTCGGCGTCACCTAGAGATGGATCCGCGGTGCATTAGTTAGTTGGTGGGGTAAAGGCCTACCAAGACAATGATGCATAGCCGAGTTGAGAGACTGATCGGCCACATTGGGACTGAGACACGGCCCAAACTCCTACGGGAGGCTGCAGTAGGGAATCTTCCACAATGGGCGAAAGCCTGATGGAGCAACGCCGCGTGTGTGATGAAGGCTTTCGGGTCGTAAAGCACTGTTGTATGGGAAGAACAGCTAGAATAGGAAATGATTTTAGTTTGACGGTACCATACCAGAAAGGGACGGCTAAATACGTGCCAGCAGCCGCGGTAATACGTATGTCCCGAGCGTTATCCGGATTTATTGGGCGTAAAGCGAGCGCAGACGGTTTATTAAGTCTGATGTGAAAGCCCGGAGCTCAACTCCGGAATGGCATTGGAAACTGGTTAACTTGAGTGCAGTAGAGGTAAGTGGAACTCCATGTGTAGCGGTGGAATGCGTAGATATATGGAAGAACACCAGTGGCGAAGGCGGCTTACTGGACTGCAACTGACGTTGAGGCTCGAAAGTGTGGGTAGCAAACAGGATTAGATACCCTGGTAGTCCACACCGTAAACGATGAACACTAGGTGTTAGGAGGTTTCCGCCTCTTAGTGCCGAAGCTAACGCATTAAGTGTTCCGCCTGGGGAGTACGACCGCAAGGTTGAAACTCAAAGGAATTGACGGGGACCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGACATCCTTTGAAGCTTTTAGAGATAGAAGTGTTCTCTTCGGAGACAAAGTGACAGGTGGTGCATGGTCGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTATTGTTAGTTGCCAGCATTCAGATGGGCACTCTAGCGAGACTGCCGGTGACAAACCGGAGGAAGGCGGGGACGACGTCAGATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGCGTATACAACGAGTTGCCAACCCGCGAGGGTGAGCTAATCTCTTAAAGTACGTCTCAGTTCGGATTGTAGTCTGCAACTCGACTACATGAAGTCGGAATCGCTAGTAATCGCGGATCAGCACGCCGCGGTGAATACGTTCCCGGGTCTTGTACACACCGCCCGTCACACCATGGGAGTTTGTAATGCCCAAAGCCGGTGGCCTAACC
References
- Zhong, X.F.; Hu, Y.X.; Liu, D.H.; Chen, J.C.; Ye, X.Q. Changes of phenolic acids and antioxidant activities during potherb mustard (Brassica juncea, Coss.) pickling. Food Chem. 2008, 108, 811–817. [Google Scholar] [CrossRef]
- Zhang, C.C.; Chen, J.B.; Li, X.Q.; Liu, D.Q. Bacterial community and quality characteristics of the fermented potherb mustard (Brassica juncea var. multiceps) under modified atmosphere. Food Res. Int. 2018, 116, 266–275. [Google Scholar] [CrossRef]
- Liu, D.Q.; Zhang, C.C.; Zhang, J.M.; Xin, X.T.; Liao, X.J. Metagenomics reveals the formation mechanism of flavor metabolites during the spontaneous fermentation of potherb mustard (Brassica juncea var. multiceps). Food Res. Int. 2021, 148, 110622. [Google Scholar] [CrossRef]
- Zhou, Q.; Zheng, Z.M.; Wu, Y.P.; Zhang, X.L.; Jia, Z.C.; Zhong, K.; Gao, H. Unraveling the core bacterial community responsible for quality and flavor improvement of the radish paocai during spontaneous fermentation. Food Biosci. 2023, 55, 102956. [Google Scholar] [CrossRef]
- Song, G.; He, Z.; Wang, X.Y.; Zhao, M.W.; Cao, X.Y.; Lin, X.P.; Ji, C.F.; Zhang, S.F.; Liang, H.P. Improving the quality of Suancai by inoculating with Lactobacillus plantarum and Pediococcus pentosaceus. Food Res. Int. 2021, 48, 110581. [Google Scholar] [CrossRef]
- Luis, R.B.J.; Higinio, S.A.; Antonio, L.L.; Amparo, C.D.; Alfredo, M. Microbial community and volatilome changes in brines along the spontaneous fermentation of Spanish-style and natural-style green table olives (Manzanilla cultivar). Food Microbiol. 2023, 113, 104286. [Google Scholar] [CrossRef]
- Ren, Q.L.; Wang, D.Q.; Han, J.; Wang, J.; Liu, Z.M.; Wu, Z.J. Ligilactobacillus cholophilus sp. nov.; isolated from pickled potherb mustard (Brassica juncea Coss.). Int. J. Syst. Evol. Micr. 2023, 73, 006160. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.Y.; Zhang, B.; Li, C.Z.; Xiao, Z.H.; Liu, M.; Fang, F.; Liu, D.M.; Deng, N.; Wang, J.H. Effect of brine fermentation on nitrogen metabolism and microbial community of Allium chinense bulbs and identification of lactic acid bacteria with nitrite degradation function. Food Biosci. 2023, 56, 103107. [Google Scholar] [CrossRef]
- Cui, Q.; Li, L.; Huang, H.; Yang, Y.; Chen, S.; Li, C. Novel insight into the formation and improvement mechanism of physical property in fermented tilapia sausage by cooperative fermentation of newly isolated lactic acid bacteria based on microbial contribution. Food Res. Int. 2024, 187, 114456. [Google Scholar] [CrossRef]
- Megur, A.; Daliri, E.B.M.; Balnionytė, T.; Jonita, S.; Lastauskienė, E.; Burokas, A. In vitro screening and characterization of lactic acid bacteria from Lithuanian fermented food with potential probiotic properties. Front. Microbiol. 2023, 14, 1213370. [Google Scholar] [CrossRef]
- Nguyen, N.N.; Nguyen, Q.D. Evolution of kombucha tea from isolated acetic acid bacteria, lactic acid bacteria and yeast in single- and mixed-cultures: Characteristics, bioactivities, fermentation performance and kinetics. Food Biotechnol. 2024, 38, 86–117. [Google Scholar] [CrossRef]
- Li, K.X.; Gu, Q.Y.; Yang, W.H.; Yu, X.B. In vitro screening and probiotic evaluation of anti-obesity and antioxidant lactic acid bacteria. Food Biosci. 2023, 54, 102844. [Google Scholar] [CrossRef]
- Minori, G.; Takashi, K.; Ayane, S.; Zoubia, C.; Ken, Y.; Miki, Y.; Hajime, T.; Bon, K. Induction of superoxide anion radical-scavenging capacity in an argan press cake-suspension by fermentation using Lactobacillus plantarum Argan-L1. LWT-Food Sci. Technol. 2018, 100, 56–61. [Google Scholar] [CrossRef]
- Vaez, N.; Rassoul, M. Exopolysaccharides isolated from fermented milk-associated lactic acid bacteria and applied to produce functional value-added probiotic yogurt. LWT-Food Sci. Technol. 2024, 199, 116116. [Google Scholar] [CrossRef]
- Guo, H.; Hao, Y.; Richel, A.; Everaert, N.; Chen, Y.; Liu, M.; Yang, X.; Ren, G. Antihypertensive effect of quinoa protein under simulated gastrointestinal digestion and peptide characterization. J. Sci. Food Agric. 2020, 100, 5569–5576. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, H.N.; Ma, A.M.; Zhou, J.Z.; Xia, X.D. Synergetic effects of Lactobacillus plantarum and Rhizopus oryzae on physicochemical, nutritional and antioxidant properties of whole-grain oats (Avena sativa L.) during solid-state fermentation. LWT-Food Sci. Technol. 2022, 154, 112687. [Google Scholar] [CrossRef]
- Huang, Y.Y.L.; Ge, L.H.; Lai, H.M.; Wang, Y.L.; Mei, Y.; Zeng, X.Q.; Su, Y.Q.; Shi, Q.; Yuan, H.Y.; Li, H.J.; et al. Seasonal alteration of environmental condition-driven shift in microbiota composition, physicochemical attributes and organic compound profiles in aged Paocai brine during intermittent back-slopping fermentation. Food Biosci. 2022, 50, 102111. [Google Scholar] [CrossRef]
- Zheng, Z.M.; Zhou, Q.; Wu, Y.P.; Zhang, X.H.; Zhang, F.T.; Jia, Z.C.; Zhong, K.; Gao, H. Effects of assorted radishes on the flavor development and bacterial community succession of radish paocai during fermentation. LWT-Food Sci. Technol. 2023, 187, 115377. [Google Scholar] [CrossRef]
- Liang, T.T.; Jiang, T.; Liang, Z.; Zhang, N.; Dong, B.; Wu, Q.P.; Gu, B. Carbohydrate-active enzyme profiles of Lactiplantibacillus plantarum strain 84-3 contribute to flavor formation in fermented dairy and vegetable products. Food Chem. 2023, 20, 101036. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, Y.; Xu, Q.; Yu, Y.; Zheng, G.J.; Wang, Y.; Zhang, Q.P.; Xu, X.Q.; Zhang, N.N.; Chu, J.Y.; et al. Microbial community succession and its correlation with quality characteristics during gray sufu fermentation. Foods 2023, 12, 2767. [Google Scholar] [CrossRef]
- Chen, Q.; Li, Y.; Yan, K.L.; Li, G.H.; Luo, D.H.; Bai, W.D.; Zeng, X.F.; Wu, Q.P.; Deng, J.H.; Dong, H. Variations of volatile flavors and microbial communities in Chinese Chaozhou pickle during natural fermentation revealed by GC-IMS and high-throughput sequencing. LWT-Food Sci. Technol. 2024, 191, 115610. [Google Scholar] [CrossRef]
- Xiang, L.W.; Zhang, N.D.; Lu, Y.; Zhao, Q.H.; Xu, Q.; Rao, Y.; Liu, L.; Zhang, Q. Effect of Weissella cibaria co-inoculation on the quality of Sichuan Pickle fermented by Lactobacillus plantarum. LWT-Food Sci. Technol. 2020, 121, 108975. [Google Scholar] [CrossRef]
- Sharma, S.; Kandasamy, S.; Kavitake, D.; Shetty, P.H. Probiotic characterization and antioxidant properties of Weissella confusa KR780676, isolated from an Indian fermented food. LWT-Food Sci. Technol. 2018, 97, 53–60. [Google Scholar] [CrossRef]
- Ogunremi, O.R.; Freimüller, L.S.; Mischler, S.; Miescher, S.S. Antifungal activity of lactic acid bacteria isolated from kunu-zaki, a cereal-based Nigerian fermented beverage. Food Biosci. 2022, 49, 101648. [Google Scholar] [CrossRef]
- Tenea, G.N.; Hurtado, P.; Ortega, C. A Novel Weissella cibaria strain UTNGt21O isolated from wild Solanum quitoense fruit: Genome sequence and characterization of a peptide with highly inhibitory potential toward gram-negative bacteria. Foods 2020, 9, 1242. [Google Scholar] [CrossRef]
- Debasish, K.D.; Bon, G.K.; Chanchal, S.; Sun, C.K. Characterization of Weissella confusa DDA7 isolated from kimchi. LWT-Food Sci. Technol. 2019, 111, 663–672. [Google Scholar] [CrossRef]
- Souza, L.V.; Rodrigues, S.R.; Falqueto, A.; Fusieger, A.; Martins, E.; Caggia, C.; Randazzo, C.L.; Carvalho, A.F. Evaluation of antifungal activity of lactic acid bacteria against fungi in simulated cheese matrix. LWT-Food Sci. Technol. 2023, 182, 114773. [Google Scholar] [CrossRef]
- Javed, G.A.; Shazadi, K.; Munir, A.; Javed, S.; Ambreen, H.S.; Arshad, N. Weissella confusa MW051433 and Levilactobacillus brevis MW362788 of human milk origin possess antioxidant competency: In vivo evidence. Process Biochem. 2023, 133, 11–19. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.F.; Chen, J.; Zhu, Y.T.; Hu, K.; Ma, Q.; Zuo, Y. A novel propylene glycol alginate gel based colorimetric tube for rapid detection of nitrite in pickled vegetables. Food Chem. 2022, 373, 131678. [Google Scholar] [CrossRef]
- Ding, Z.S.; Suzanne, D.J.; Robert, P.; Rong, R.; Truong, V.D.; Summer, C.P.; Fred, B. Evaluation of nitrate and nitrite contents in pickled fruit and vegetable products. Food Control 2018, 90, 304–311. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Han, J.R.; Zheng, X.G.; Yan, J.Q.; Chen, X.A.; Zhou, Q.Q.; Zhao, X.D.; Gu, Q.; Li, P. Use of Lactiplantibacillus plantarum ZJ316 as a starter culture for nitrite degradation, foodborne pathogens inhibition and microbial community modulation in pickled mustard fermentation. Food Chem. X 2022, 14, 100344. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.W.; Zeng, X.Q.; Zhang, P.; Leng, L.L.; Du, Q.W.; Pan, D.D. Nitrite reductases of lactic acid bacteria: Regulation of enzyme synthesis and activity, and different applications. Food Biosci. 2024, 59, 103833. [Google Scholar] [CrossRef]
- Wang, N.N.; Ren, J.; Wang, L.L.; Wang, Y.H.; Wang, Z.; Guo, D. A preliminary study to explain how Streptomyces pactum (Act12) works on phytoextraction: Soil heavy metal extraction, seed germination, and plant growth. Environ. Monit. Assess. 2023, 195, 757. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Wei, W.L.; Tang, L.; Wang, D.D.; Wang, Y.; Wu, Z.Y.; Zhang, W.X. Characterization of aroma and bacteria profiles of Sichuan industrial paocai by HS-SPME-GC-O-MS and 16S rDNA amplicon sequencing. Food Res. Int. 2021, 148, 110667. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microb. 2021, 77, 2264–2274. [Google Scholar] [CrossRef]
- Kim, S.A.; Lee, D.H.; Ryu, B.H.; Han, N.S. Strain-specific barcode PCR and quantitative PCR assay for identification and enumeration of kimchi starter, Leuconostoc mesenteroides DRC1506. LWT-Food Sci. Technol. 2023, 174, 114460. [Google Scholar] [CrossRef]
- Park, S.E.; Yoo, S.A.; Seo, S.H.; Lee, K.I.; Na, C.S.; Son, H.S. GC–MS based metabolomics approach of Kimchi for the understanding of Lactobacillus plantarum fermentation characteristics. LWT-Food Sci. Technol. 2016, 68, 313–321. [Google Scholar] [CrossRef]
- Yan, P.M.; Jia, J.J.; Zhao, H.W.; Wu, C.D. Changes and driving mechanism of microbial community structure during paocai fermentation. Fermentation 2022, 8, 281. [Google Scholar] [CrossRef]
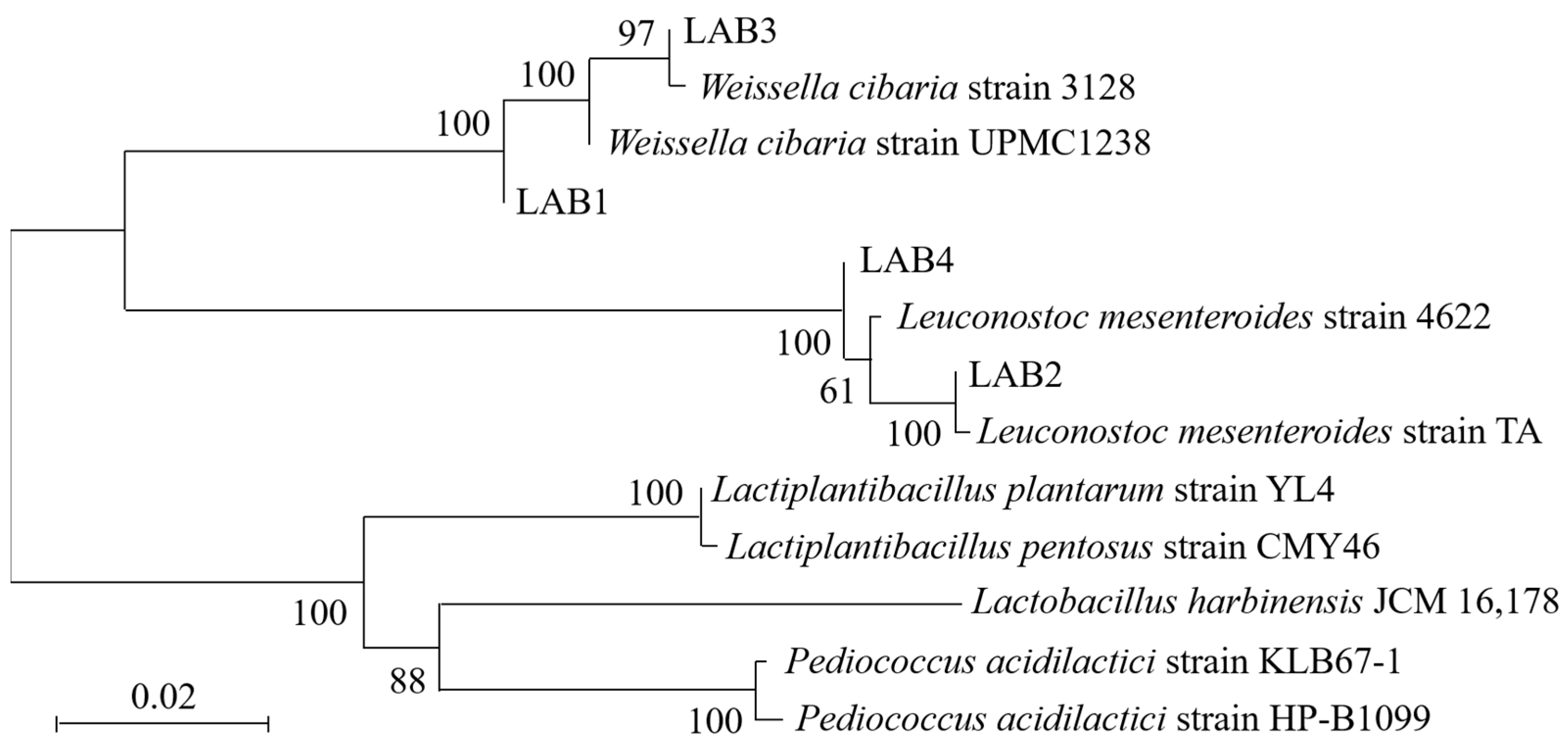
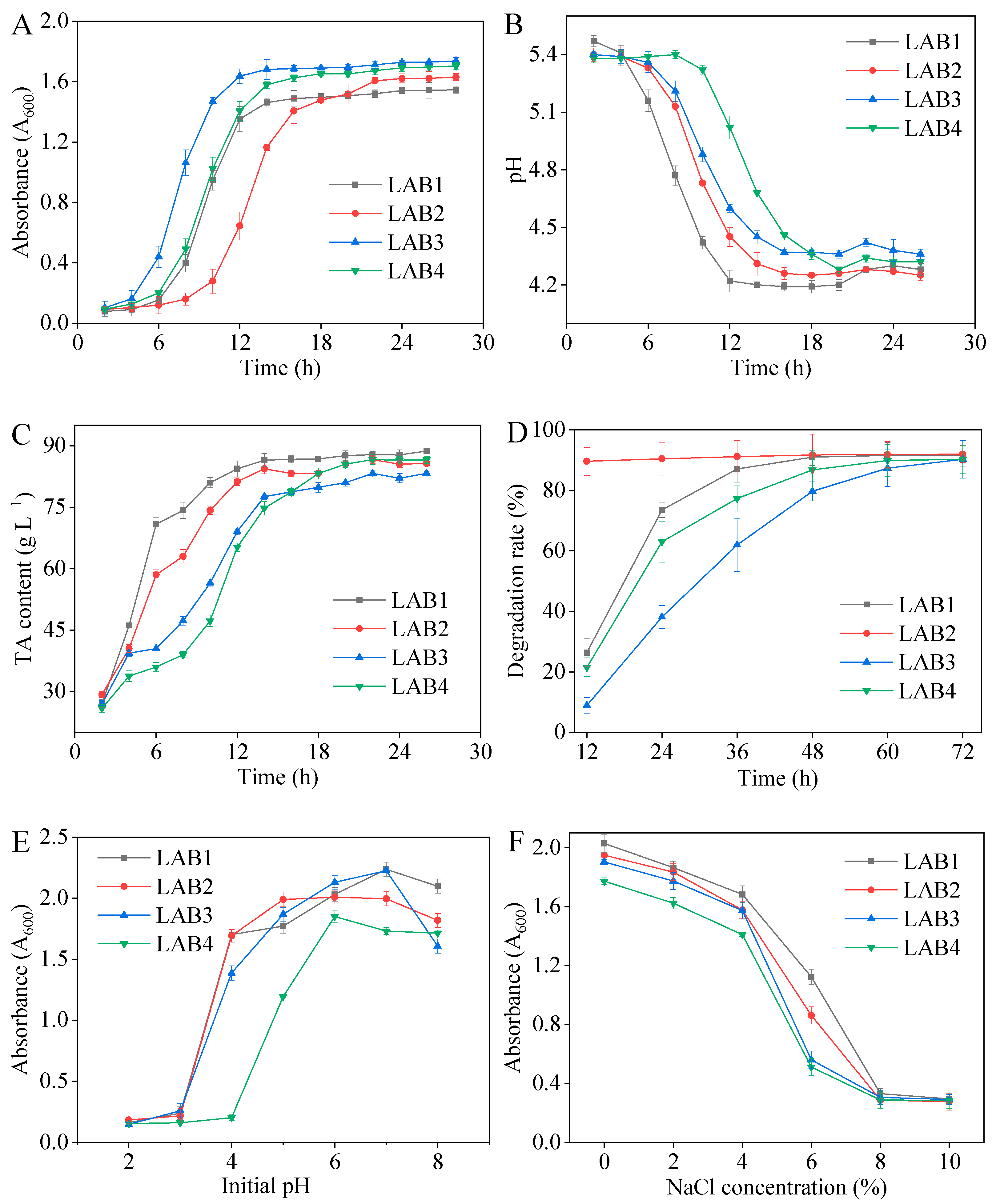
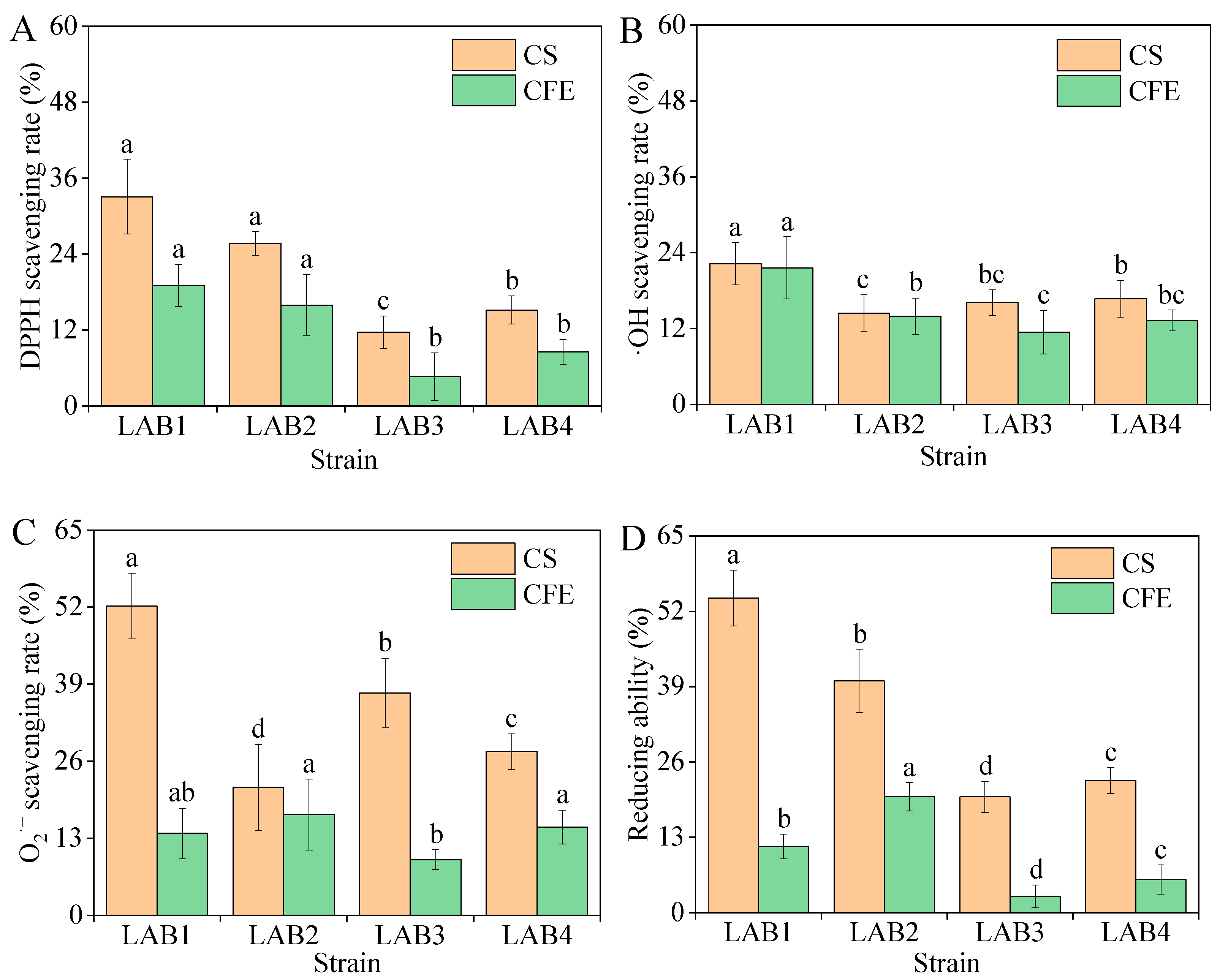
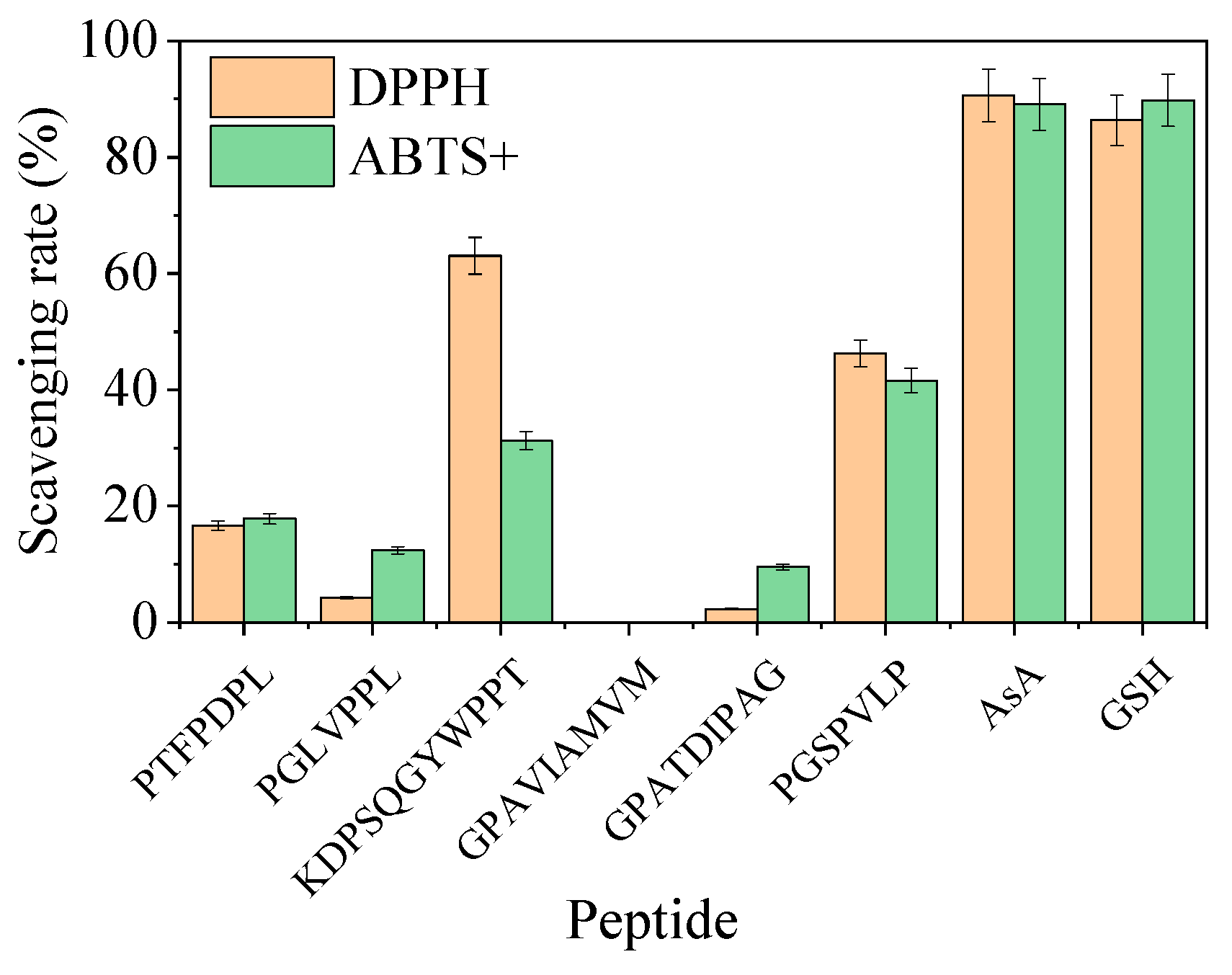

| Strain | Accession | Strain | Sequence Length/bp | Query Cover/% | Percentage of Identification/% |
|---|---|---|---|---|---|
| LAB1 | MN700179.1 | Weissella cibaria strain UPMC1238 | 1413 | 100 | 99.93 |
| LAB2 | KU361186.1 | Leuconostoc mesenteroides strain TA | 1510 | 100 | 100 |
| LAB3 | MT613505.1 | Weissella cibaria strain 3128 | 855 | 100 | 100 |
| LAB4 | MT545113.1 | Leuconostoc mesenteroides strain 4622 | 1391 | 100 | 100 |
| Strain | Antibacterial Circle Diameters/mm | |
|---|---|---|
| E. coli CCTCC AB 2014080 | S. aureus CCTCC AB 2010020 | |
| LAB1 | 14.03 ± 0.58 a | 12.55 ± 0.82 a |
| LAB2 | 12.83 ± 0.74 c | 12.32 ± 0.39 b |
| LAB3 | 13.40 ± 0.81 b | 12.46 ± 0.86 a |
| LAB4 | 11.13 ± 0.63 d | 11.97 ± 0.29 c |
| No. | Protein Accession | Peptides | Peptide Ranker (Bioactive) | Length | Mass | Percentage of HAAs (%) |
|---|---|---|---|---|---|---|
| 1 | tr|A0A1X4JKK3|A0A1X4JKK3_9LACO | S.PTFPDPL.G | 0.865 | 7 | 785.3959 | 71.43 |
| 2 | tr|A0A2S1KSC4|A0A2S1KSC4_9LACO | N.PGLVPPL.S | 0.794 | 7 | 691.4268 | 85.71 |
| 3 | tr|A0A0N9Y818|A0A0N9Y818_9LACO | P.KDPSQGYWPPT.V | 0.739 | 11 | 1274.5931 | 45.45 |
| 4 | tr|A0A0D1LI50|A0A0D1LI50_9LACO | V.GPAVIAMVM.A | 0.701 | 9 | 887.4609 | 88.89 |
| 5 | tr|A0A1X4JKK3|A0A1X4JKK3_9LACO | E.PGSPVLP.H | 0.682 | 7 | 665.3748 | 71.43 |
| 6 | tr|A0A0D1LZ28|A0A0D1LZ28_9LACO | G.EGWTLWNGNPIP.S | 0.674 | 12 | 1382.6619 | 50 |
| 7 | tr|A0A0N9Y5P2|A0A0N9Y5P2_9LACO | L.TFWLDGK.A | 0.604 | 7 | 865.4333 | 42.86 |
| 8 | tr|A0A0D1K432|A0A0D1K432_9LACO | G.KDPVAKPGDGGYWAT.Y | 0.59 | 15 | 1560.7572 | 40 |
| 9 | tr|A0A2S1KTI5|A0A2S1KTI5_9LACO | N.GPATDIPAG.Y | 0.565 | 9 | 797.3919 | 55.56 |
| 10 | tr|A0A2S1KTI5|A0A2S1KTI5_9LACO | N.LNGPYGPK.V | 0.54 | 8 | 844.4443 | 37.5 |
| 11 | tr|A0A1X4JKK3|A0A1X4JKK3_9LACO | Y.TPEGPNGPL.T | 0.512 | 9 | 880.429 | 44.44 |
| 12 | tr|A0A2S1KNI5|A0A2S1KNI5_9LACO | L.VGDVGFG.K | 0.501 | 7 | 649.3071 | 42.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, X.; Zhang, J.; Shen, H.; Shi, N.; Zhou, H.; Li, Y.; Guo, Y.; Luo, H.; Yu, L. Screening, Identification, and Fermentation Characteristics of Lactic Acid Bacteria from Pickled Potherb Mustard and Potential Applications. Foods 2025, 14, 1431. https://doi.org/10.3390/foods14081431
Kong X, Zhang J, Shen H, Shi N, Zhou H, Li Y, Guo Y, Luo H, Yu L. Screening, Identification, and Fermentation Characteristics of Lactic Acid Bacteria from Pickled Potherb Mustard and Potential Applications. Foods. 2025; 14(8):1431. https://doi.org/10.3390/foods14081431
Chicago/Turabian StyleKong, Xiaoxue, Jiaxin Zhang, Hui Shen, Nan Shi, Hui Zhou, Yi Li, Yuxing Guo, Haibo Luo, and Lijuan Yu. 2025. "Screening, Identification, and Fermentation Characteristics of Lactic Acid Bacteria from Pickled Potherb Mustard and Potential Applications" Foods 14, no. 8: 1431. https://doi.org/10.3390/foods14081431
APA StyleKong, X., Zhang, J., Shen, H., Shi, N., Zhou, H., Li, Y., Guo, Y., Luo, H., & Yu, L. (2025). Screening, Identification, and Fermentation Characteristics of Lactic Acid Bacteria from Pickled Potherb Mustard and Potential Applications. Foods, 14(8), 1431. https://doi.org/10.3390/foods14081431






