Method of Detecting Microorganisms on the Surface of Mandarin Fish Based on Hyperspectral and Information Fusion
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of Mandarin Fish Samples
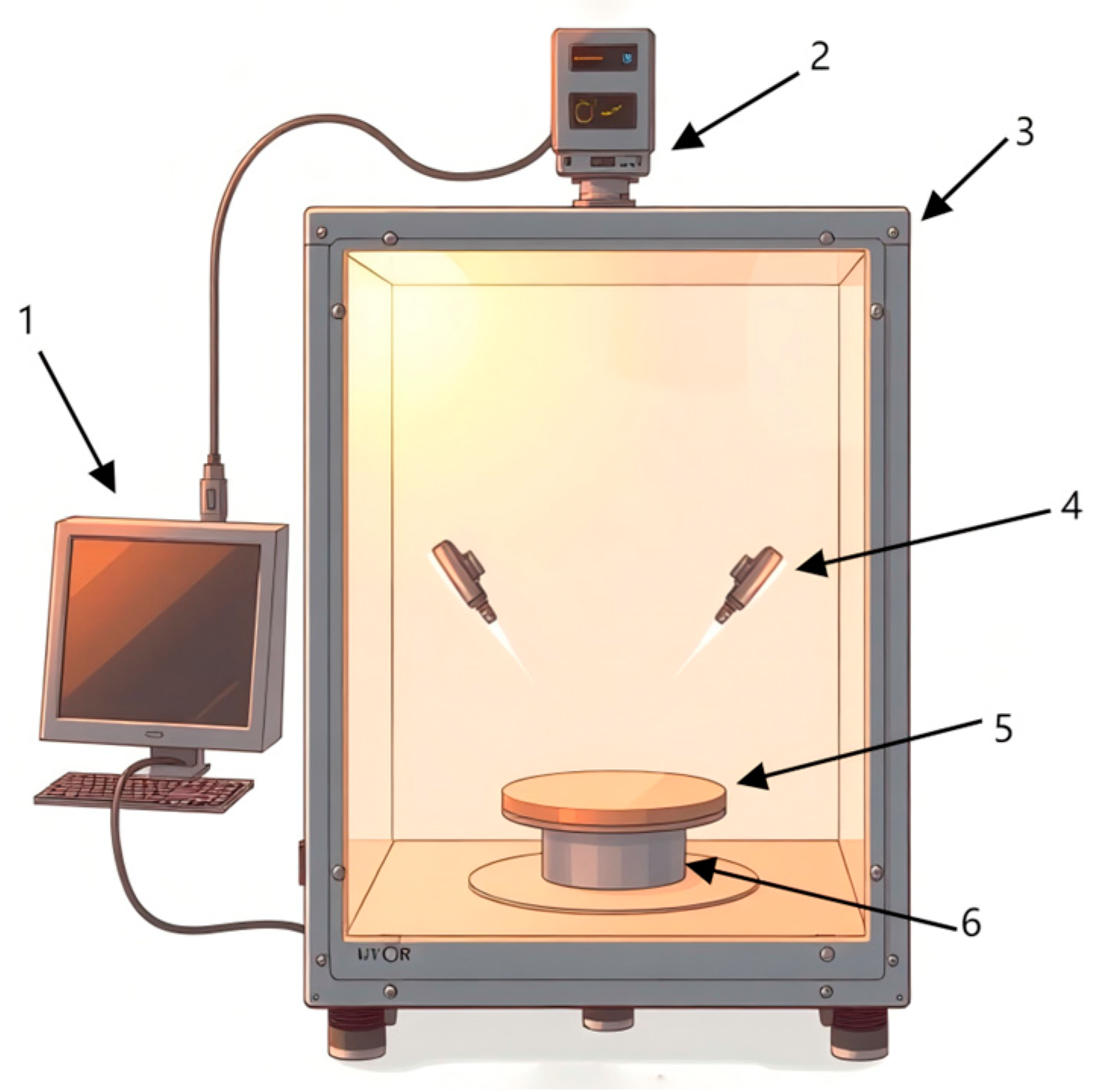
2.2. Collection and Information Extraction of Hyperspectral Images of Mandarin Fish
2.3. Data Processing Methods
2.3.1. Spectral Preprocessing Techniques
2.3.2. Extraction of Texture Feature Parameters
2.3.3. Methodologies for Constructing Quantitative Models
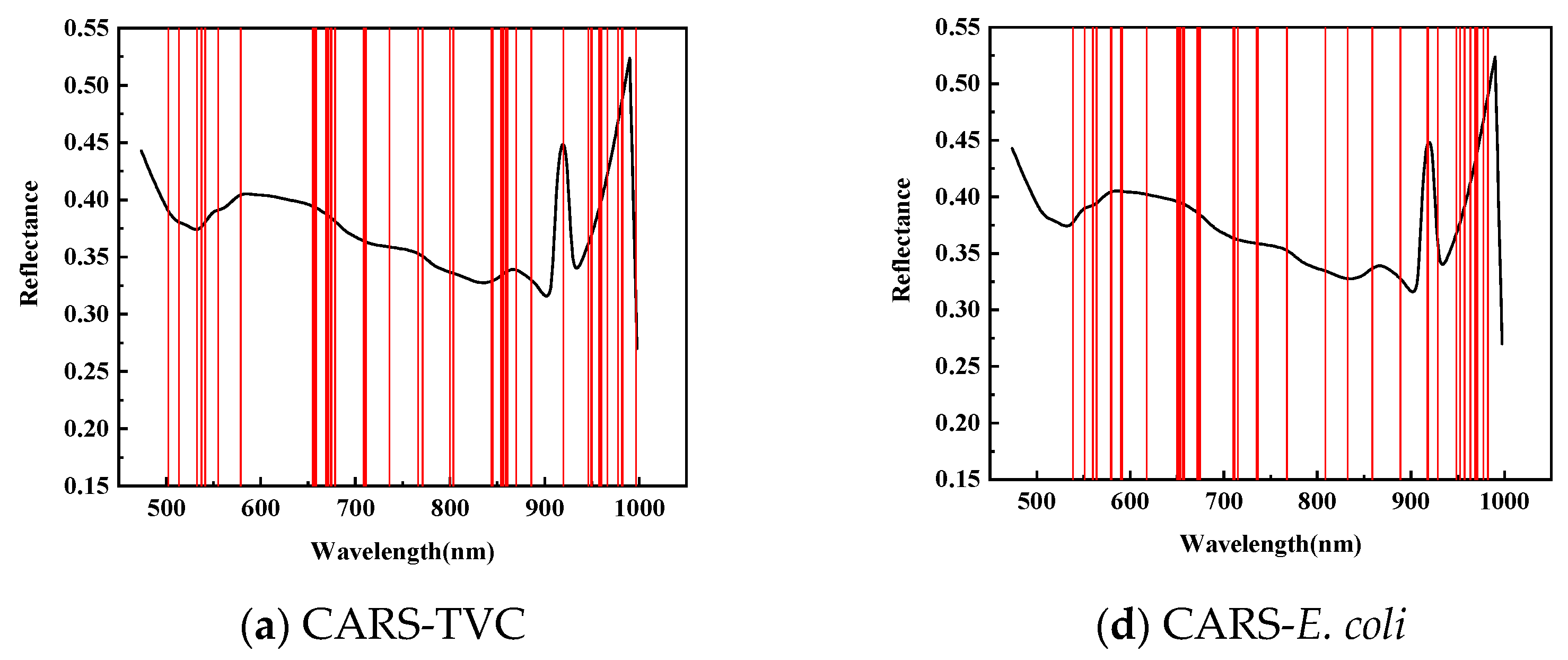
2.3.4. Wavelength Selection Method
2.3.5. Information Fusion
3. Results and Discussion
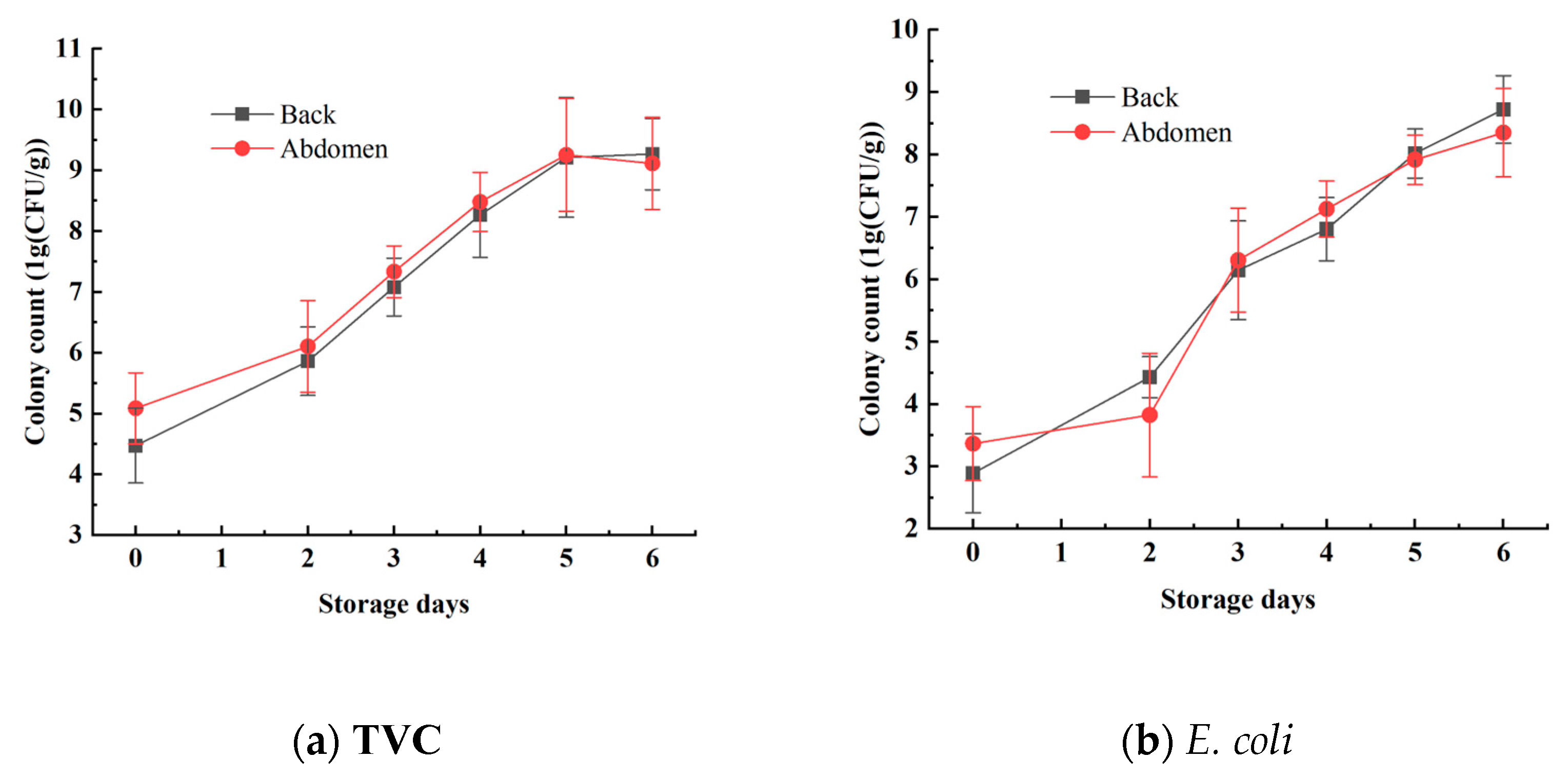
3.1. Analysis of Microbial Spoilage in Mandarin Fish Samples
3.2. Sample Set Division
3.3. Detection of Microorganisms on the Surface of Mandarin Fish Based on Spectral Dimension Analysis
3.3.1. Analysis of a Full-Wavelength Detection Model of Microorganisms on the Surface of Mandarin Fish
3.3.2. Simplified Model Analysis of Surface Microorganism Detection of Mandarin Fish
3.3.3. Comparison and Analysis of Full Wavelength Spectral Data of Different Detection Sites and Simplified Models
3.4. Detection of Surface Microorganisms of Mandarin Fish Based on Texture Information
3.4.1. Mandarin Fish Surface Microbial Detection Model Based on Feature Band Texture Analysis
3.4.2. Regression Model Based on Principal Component Image Texture Analysis
3.5. Surface Microbial Detection of Mandarin Fish Based on Information Fusion
3.5.1. Surface Microbial Detection Model of Mandarin Fish Based on Feature Layer Fusion
3.5.2. Surface Microbial Detection Model of Mandarin Fish Based on Decision-Level Fusion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1st De | 1st Derivative |
| BP | Backpropagation |
| BPA | Basic Probability Assignment |
| BBA | Basic Belief Assignment |
| CARS | Competitive Adaptive Reweighted Sampling |
| E. coli | Escherichia coli |
| GA | Genetic Algorithm |
| GLCM | Grey level co-occurrence matrix |
| MSC | Multiplicative scattering correction |
| PCA | Principal Component Analysis |
| PCR | Principal Component Regression |
| PLSR | Partial Least Squares Regression |
| R2 | Coefficients of determination |
| RMSE | Root Mean Square Error |
| ROI | Region of interest |
| SNV | Standard normal variate transformation |
| SPXY | Sample set partitioning based on joint x–y distance |
| SVC | Support Vector Classification |
| TVC | Total viable count |
| VN | Vector Normalization |
References
- Yan, A.S.; Xiong, C.X.; Qian, J.W.; Wang, X.D. A study on the rate of flesh content of mandarin fish and nutritional quality of the flesh. J. Huazhong Agric. Univ. 1995, 14, 80–84. (In Chinese) [Google Scholar]
- Zeng, Z.Y.; Ding, Z.L.; Zhou, A.N.; Zhu, C.B.; Yang, S.; Fei, H. Bacterial diseases inSiniperca chuatsi: Status and therapeutic strategies. Vet. Res. Commun. 2024, 48, 3579–3592. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, X.; Tian, Y.; Wang, Q. Effect of a Hypoxia-Controlled Atmosphere Box on Egg Respiration Intensity and Quality. Appl. Sci. 2022, 12, 380. [Google Scholar] [CrossRef]
- Zhou, B.; Fan, X.; Song, J.; Wu, J.; Pan, L.; Tu, K.; Peng, J.; Dong, Q.; Xu, J.; Wu, J. Growth simulation of Pseudomonas fluorescens in pork using hyperspectral imaging. Meat Sci. 2022, 188, 108767. [Google Scholar] [CrossRef]
- Gamilla, H.T.; Yuan, H.; Brad, K. Fresh meat color evaluation a structured light imaging system. Food Res. Int. 2015, 71, 100–107. [Google Scholar]
- Liu, Z.; Huang, Y.; Wang, J.; Yuan, G.; Pang, J. Multiscale Information Fusion Based on Large Model Inspired Bacterial Detection. Big Data Min. Anal. 2025, 8, 1–17. [Google Scholar] [CrossRef]
- GB 4789.2-2022; National Food Safety Standard, Food Microbiological Examination, Aerobic Plate Count. Standardization Administration of the People’s Republic of China: Beijing, China, 2022.
- GB 4789.3-2016; National Food Safety Standard—Food Microbiological Examination—Coliforms Count. Standardization Administration of the People’ s Republic of China: Beijing, China, 2016.
- Liu, S.; Lioe, T.S.; Sun, L.; Adriaenssens, E.M.; McCarthy, A.J.; Sekar, R. Validation of CrAssphage Microbial Source Tracking markers and comparison with Bacteroidales Markers for Detection and Quantification of Faecal Contaminations in Surface Water. Environ. Pollut. 2025, 366, 125403. [Google Scholar] [CrossRef]
- Hansen, P.C.; Jensen, T.K. Smoothing Norm Preconditioning for Regularizing Minimum Inesidual Methods. SIAM J. Matrix Anal. Appl. 2006, 29, 1–14. [Google Scholar] [CrossRef]
- Gillis, N.; Vavasis, S.A. Semidefinite Programming Based Preconditioning for More Robust Near-Separable Nonnegative Matrix Factorization. Siam J. Optimiz. 2015, 25, 677–698. [Google Scholar] [CrossRef]
- Housman, J.A.; Kiris, C.C.; Hafez, M.M. Time-Derivative Preconditioning Methods for Multicomponent Flows—Part II: Two-Dimensional Applications. J. Appl. Mech. 2009, 76, 031013. [Google Scholar] [CrossRef]
- Andriulli, F.P.; Cools, K.; Bagci, H.; Olyslager, F.; Buffa, A.; Christiansen, S. A multiplicative Calderón preconditioner for the electric field integral equation. IEEE Antenna. Propag. 2008, 56, 2398–2412. [Google Scholar] [CrossRef]
- Mizutani, T. Robustness Analysis of Preconditioned Successive Projection Algorithm for General Form of Separable NMF Problem. Linear Algebra Appl. 2016, 497, 1–22. [Google Scholar] [CrossRef]
- Wang, L.; Sun, J.; Li, T. Intelligent sports feature recognition system based on texture feature extraction and SVM parameter selection. J. Intell. Fuzzy Syst. 2020, 39, 4847–4858. [Google Scholar] [CrossRef]
- Ravindran, G.; Titus, T.J.; Ganesh, V.; Devi, V.S.S. Analysis of Image Segmentation Techniques for Texture Feature Extraction. Int. J. Eng. Sci. Res. Technol. 2017, 6, 66–71. [Google Scholar]
- You, J.; Tulpan, D.; Ellis, J.L. Predicting pellet quality using multiple linear regression with Principal Component Analysis (PCA). J. Anim. Sci. 2024, 102, 154–155. [Google Scholar] [CrossRef]
- Nesrstová, V.; Wilms, I.; Palarea-Albaladejo, J.; Filzmoser, P.; Martín-Fernández, J.A.; Friedecký, D.; Hron, K. Principal balances of compositional data for regression and classification using partial least squares. J. Chemom. 2023, 37, e3518. [Google Scholar] [CrossRef]
- Xue, C.; Xu, W.; Bao, Y.; Zhao, L.; Liu, X.; Zheng, R. Prediction of Converter Tapping Weight Based on Principal Component Analysis–Whale Optimization Algorithm–Backpropagation Algorithm Neural Network Model. Steel Res. Int. 2024, 95, 2300410. [Google Scholar] [CrossRef]
- Canova, L.D.S.; Vallese, F.D.; Pistonesi, M.F.; de Araújo Gomes, A. An improved successive projections algorithm version to variable selection in multiple linear regression. Anal. Chim. Acta 2023, 1274, 341560. [Google Scholar] [CrossRef]
- Kabir, E.; Guikema, S.D.; Quiring, S.M. Power outage prediction using data streams: An adaptive ensemble learning approach with a feature- and performance-based weighting mechanism. Risk Anal. 2024, 44, 686–704. [Google Scholar] [CrossRef]
- Jyakhwo, S.; Serov, N.; Dmitrenko, A.; Vinogradov, V.V. Machine Learning Reinforced Genetic Algorithm for Massive Targeted Discovery of Selectively Cytotoxic Inorganic Nanoparticles. Small 2024, 20, e2305375. [Google Scholar] [CrossRef]
- Tao, L.; Wang, F.; Li, Y.; Wu, J.; Jiang, X.; Xi, Q. A cross-texture haptic model based on tactile feature fusion. Multimed. Syst. 2024, 30, 159. [Google Scholar] [CrossRef]
- Tao, B.; Bai, J.; Wu, H.; Miao, F. Design and research of multi-information fusion RFID sensor for fruit and vegetable quality detection. Microchim. Acta 2024, 191, 568. [Google Scholar] [CrossRef]
- Wang, H.; Huang, Z.; Lou, S.; Li, W.; Liu, G.; Tang, Y. In Silico Prediction of Skin Sensitization for Compounds via Flexible Evidence Combination Based on Machine Learning and Dempster-Shafer Theory. Chem. Res. Toxicol. 2024, 37, 894–909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.; Peng, Y.; Wang, W.; Jiang, F.; Tao, F.; Shan, J. Determination of pork freshness attributes by hyperspectral imaging technique. Trans. Chin. Soc. Agric. Eng. 2012, 28, 254–259. [Google Scholar]
- Han, F.; Huang, X.; Teye, E.; Gu, F.; Gu, H. Nondestructive detection of fish freshness during its preservation by combining electronic nose and electronic tongue techniques in conjunction with chemometric analysis. Anal. Methods 2013, 6, 529–536. [Google Scholar] [CrossRef]
- Huang, X.; Ding, R.; Han, F.K.; Dai, H.; Teye, E.; Xu, F. Rapid and nondestructive evaluation of fish Freshness by near infrared reflectance spectroscopy combined with chemometrics analysis. Anal. Methods 2014, 6, 9675–9683. [Google Scholar]
- Chen, Y.; Zeng, H.B.; Li, Y. Stability analysis of linear delayed systems based on an allowable delay set partitioning approach. Automatica 2024, 163, 111603. [Google Scholar] [CrossRef]
- Baek, I.; Lee, H.; Cho, B.-K.; Mo, C.; Chan, D.E.; Kim, M.S. Shortwave infrared hyperspectral imaging system coupled with multivariable method for TVB-N measurement in pork. Food Control 2020, 124, 107854. [Google Scholar] [CrossRef]
- Ma, J.; Sun, D.-W.; Qu, J.-H.; Pu, H. Prediction of textural changes in grass carp fillets as affected by vacuum freeze drying using hyperspectral imaging based on integrated group wavelengths. Opt. Commun. 2017, 82, 377–385. [Google Scholar] [CrossRef]
- Liu, D.; Pu, H.; Sun, D.-W.; Wang, L.; Zeng, X.-A. Combination of spectra and texture data of hyperspectral imaging for prediction of pH in salted meat. Food Chem. 2014, 160, 330–337. [Google Scholar] [CrossRef]
- Ahmed, F.; Al-Mamun, H.A.; Bari, A.S.M.H.; Hossain, E.; Kwan, P. Classification of crops and weeds from digital images: A support vector machine approach. Crop Prot. 2012, 40, 98–104. [Google Scholar] [CrossRef]

| Location | Microbial Indicator | Minimum Value | The Maximum Value | Average Value | Standard Deviation |
|---|---|---|---|---|---|
| Back | TVC | 3.7475 | 10.6551 | 7.4458 | 1.8965 |
| E. coli | 4.4024 | 10.6590 | 7.6892 | 1.7456 | |
| Abdomen | TVC | 2.0000 | 9.4433 | 6.1308 | 2.2150 |
| E. coli | 2.1761 | 9.4814 | 6.2449 | 2.1027 |
| Location | Microbial Indicator | Training Set | Test Set | ||||
|---|---|---|---|---|---|---|---|
| Variation Range | Mean | Standard Deviation | Variation Range | Mean | Standard Deviation | ||
| Back | TVC | 3.74~10.65 | 7.47 | 1.96 | 4.05~9.98 | 7.38 | 1.77 |
| E. coli | 2.00~9.44 | 6.05 | 2.31 | 2.39~9.17 | 6.29 | 2.17 | |
| Abdomen | TVC | 4.40~10.65 | 7.61 | 1.76 | 4.46~10.49 | 7.85 | 1.69 |
| E. coli | 2.17~9.48 | 6.12 | 2.12 | 2.65~9.00 | 6.50 | 2.03 | |
| Microbial Indicator | Model | Pretreatment Method | Rc | RMSEC | Rcv | RMSECV | Rp | RMSEP | |
|---|---|---|---|---|---|---|---|---|---|
| TVC | PLSR | RAW | 0.8403 | 0.9605 | 0.6515 | 1.5041 | 0.8358 | 1.0734 | 0.1129 |
| SG | 0.8398 | 0.9618 | 0.6517 | 1.5037 | 0.8350 | 1.0757 | 0.1139 | ||
| VN | 0.8230 | 1.0060 | 0.6140 | 1.5648 | 0.8067 | 1.1552 | 0.1492 | ||
| 1stDe | 0.8830 | 0.9174 | 0.5750 | 1.6220 | 0.7593 | 1.1530 | 0.2356 | ||
| MSC | 0.8335 | 1.0799 | 0.6549 | 1.4981 | 0.7740 | 1.1219 | 0.0420 | ||
| SNV | 0.8554 | 0.9177 | 0.6124 | 1.5672 | 0.8437 | 1.0492 | 0.1315 | ||
| PCR | RAW | 0.8642 | 0.8914 | 0.6798 | 1.4540 | 0.8402 | 1.0600 | 0.1686 | |
| SG | 0.8643 | 0.8910 | 0.6787 | 1.4558 | 0.8407 | 1.0587 | 0.1677 | ||
| VN | 0.7942 | 1.0767 | 0.6164 | 1.5611 | 0.7675 | 1.2531 | 0.1764 | ||
| 1stDe | 0.7650 | 1.2590 | 0.5156 | 1.6987 | 0.6294 | 1.3769 | 0.1179 | ||
| MSC | 0.8394 | 1.0624 | 0.6867 | 1.4412 | 0.6756 | 1.3062 | 0.2438 | ||
| SNV | 0.8499 | 0.9335 | 0.6515 | 1.5435 | 0.8299 | 1.0900 | 0.1565 | ||
| BP | RAW | 0.7794 | 0.8166 | - | - | 0.5667 | 1.1920 | 0.3754 | |
| SG | 0.7219 | 0.8143 | - | - | 0.5570 | 1.1489 | 0.3346 | ||
| VN | 0.6751 | 0.9616 | - | - | 0.6014 | 1.1559 | 0.1943 | ||
| 1stDe | 0.7784 | 0.7387 | - | - | 0.6762 | 1.0191 | 0.2804 | ||
| MSC | 0.7529 | 0.8578 | - | - | 0.4339 | 1.3035 | 0.4457 | ||
| SNV | 0.6848 | 0.8576 | - | - | 0.8358 | 1.2131 | 0.3555 | ||
| E. coli | PLSR | RAW | 0.8721 | 1.0914 | 0.7455 | 1.5078 | 0.8054 | 1.2882 | 0.1968 |
| SG | 0.8936 | 1.0012 | 0.7648 | 1.4575 | 0.8180 | 1.2501 | 0.2489 | ||
| VN | 0.8716 | 1.0936 | 0.7464 | 1.5057 | 0.7884 | 1.3371 | 0.2435 | ||
| 1stDe | 0.9155 | 0.8972 | 0.6580 | 1.7036 | 0.7465 | 1.4463 | 0.5491 | ||
| MSC | 0.8710 | 1.0960 | 0.7329 | 1.5391 | 0.6218 | 1.7024 | 0.6064 | ||
| SNV | 0.8703 | 1.0986 | 0.6993 | 1.6173 | 0.7756 | 1.3719 | 0.2733 | ||
| PCR | RAW | 0.8871 | 1.0297 | 0.7788 | 1.4189 | 0.8181 | 1.2500 | 0.2203 | |
| SG | 0.8867 | 1.0313 | 0.7775 | 1.4228 | 0.8205 | 1.2423 | 0.2110 | ||
| VN | 0.8720 | 1.0919 | 0.7589 | 1.4733 | 0.7838 | 1.3499 | 0.2580 | ||
| 1stDe | 0.7844 | 1.3836 | 0.5208 | 1.9315 | 0.6472 | 1.6569 | 0.2733 | ||
| MSC | 0.8811 | 1.0549 | 0.7636 | 1.4608 | 0.6715 | 1.6107 | 0.5558 | ||
| SNV | 0.8746 | 1.0814 | 0.7331 | 1.5387 | 0.7824 | 1.3536 | 0.2722 | ||
| BP | RAW | 0.9027 | 0.6400 | - | - | 0.7407 | 1.1924 | 0.5524 | |
| SG | 0.8523 | 0.778 | - | - | 0.8091 | 1.0432 | 0.2652 | ||
| VN | 0.5914 | 1.1994 | - | - | 0.4948 | 1.5425 | 0.3431 | ||
| 1stDe | 0.8452 | 0.7949 | - | - | 0.5394 | 1.4945 | 0.6996 | ||
| MSC | 0.8795 | 0.7077 | - | - | 0.5176 | 1.5187 | 0.8110 | ||
| SNV | 0.7806 | 0.9298 | - | - | 0.8054 | 1.5098 | 0.5800 |
| Microbial Indicator | Model | Pretreatment Method | RMSEC | RMSECV | Rp | RMSEP | |||
|---|---|---|---|---|---|---|---|---|---|
| TVC | PLSR | RAW | 0.9115 | 0.7259 | 0.7995 | 1.0751 | 0.7706 | 1.0797 | 0.3538 |
| SG | 0.9169 | 0.7043 | 0.8043 | 1.0635 | 0.7422 | 1.1352 | 0.4309 | ||
| VN | 0.9008 | 0.7665 | 0.7675 | 1.1474 | 0.7159 | 1.1828 | 0.4163 | ||
| 1stDe | 0.8928 | 0.7949 | 0.7720 | 1.1375 | 0.7950 | 1.0274 | 0.2325 | ||
| MSC | 0.8900 | 0.8047 | 0.6183 | 1.4067 | 0.6938 | 1.2200 | 0.4153 | ||
| SNV | 0.8994 | 0.7705 | 0.7410 | 1.2018 | 0.7420 | 1.1356 | 0.3651 | ||
| PCR | RAW | 0.8996 | 0.7707 | 0.7992 | 1.0756 | 0.7987 | 1.0194 | 0.2487 | |
| SG | 0.9000 | 0.7693 | 0.7994 | 1.0743 | 0.7836 | 1.0523 | 0.2830 | ||
| VN | 0.8841 | 0.8248 | 0.7563 | 1.1709 | 0.7312 | 1.1556 | 0.3308 | ||
| 1stDe | 0.8730 | 0.8606 | 0.7858 | 1.1069 | 0.7974 | 1.0222 | 0.1616 | ||
| MSC | 0.6888 | 1.2795 | 0.5275 | 1.5205 | 0.5818 | 1.3778 | 0.0983 | ||
| SNV | 0.8843 | 0.8239 | 0.7549 | 1.1739 | 0.7406 | 1.1383 | 0.3144 | ||
| BP | RAW | 0.8350 | 0.6474 | - | - | 0.7662 | 0.8889 | 0.2415 | |
| SG | 0.9180 | 0.4666 | - | - | 0.6826 | 1.0109 | 0.5443 | ||
| VN | 0.7625 | 0.7614 | - | - | 0.7243 | 0.9537 | 0.1923 | ||
| 1stDe | 0.9066 | 0.4965 | - | - | 0.6113 | 1.0947 | 0.5982 | ||
| MSC | 0.7842 | 0.7301 | - | - | 0.7706 | 0.9038 | 0.1737 | ||
| SNV | 0.9115 | 0.8868 | - | - | 0.7422 | 1.0955 | 0.2087 | ||
| E. coli | PLSR | RAW | 0.9486 | 0.6724 | 0.7746 | 1.3626 | 0.7802 | 1.2725 | 0.9130 |
| SG | 0.8930 | 0.9559 | 0.7355 | 1.4597 | 0.6266 | 1.5854 | 0.3166 | ||
| VN | 0.8514 | 1.1143 | 0.6947 | 1.5498 | 0.6430 | 1.5581 | 0.4438 | ||
| 1stDe | 0.8927 | 0.9573 | 0.7610 | 1.3978 | 0.7276 | 1.3954 | 0.4381 | ||
| MSC | 0.6640 | 1.5887 | 0.5154 | 1.8466 | 0.5948 | 1.7010 | 0.1123 | ||
| SNV | 0.8012 | 1.2712 | 0.4941 | 1.8733 | 0.4604 | 1.8059 | 0.5347 | ||
| PCR | RAW | 0.8885 | 0.9746 | 0.7560 | 1.4104 | 0.7633 | 1.3323 | 0.3577 | |
| SG | 0.8897 | 0.9699 | 0.7620 | 1.3954 | 0.7549 | 1.3341 | 0.3642 | ||
| VN | 0.8576 | 1.0926 | 0.7075 | 1.5227 | 0.7233 | 1.4048 | 0.3122 | ||
| 1stDe | 0.8786 | 1.0146 | 0.7648 | 1.3882 | 0.6972 | 1.4582 | 0.4436 | ||
| MSC | 0.6037 | 1.6940 | 0.4994 | 1.8668 | 0.5856 | 1.7100 | 0.0160 | ||
| SNV | 0.8500 | 1.1193 | 0.5873 | 1.7440 | 0.6168 | 1.6011 | 0.4818 | ||
| BP | RAW | 0.8216 | 0.8075 | - | - | 0.5780 | 1.3555 | 0.5480 | |
| SG | 0.8281 | 0.7942 | - | - | 0.6767 | 1.2230 | 0.4288 | ||
| VN | 0.7986 | 0.8527 | - | - | 0.6538 | 1.2568 | 0.4041 | ||
| 1stDe | 0.9209 | 0.5522 | - | - | 0.5718 | 1.3628 | 0.8106 | ||
| MSC | 0.7699 | 0.9041 | - | - | 0.7019 | 1.1832 | 0.2791 | ||
| SNV | 0.8101 | 0.8305 | - | - | 0.6003 | 1.3285 | 0.4980 |
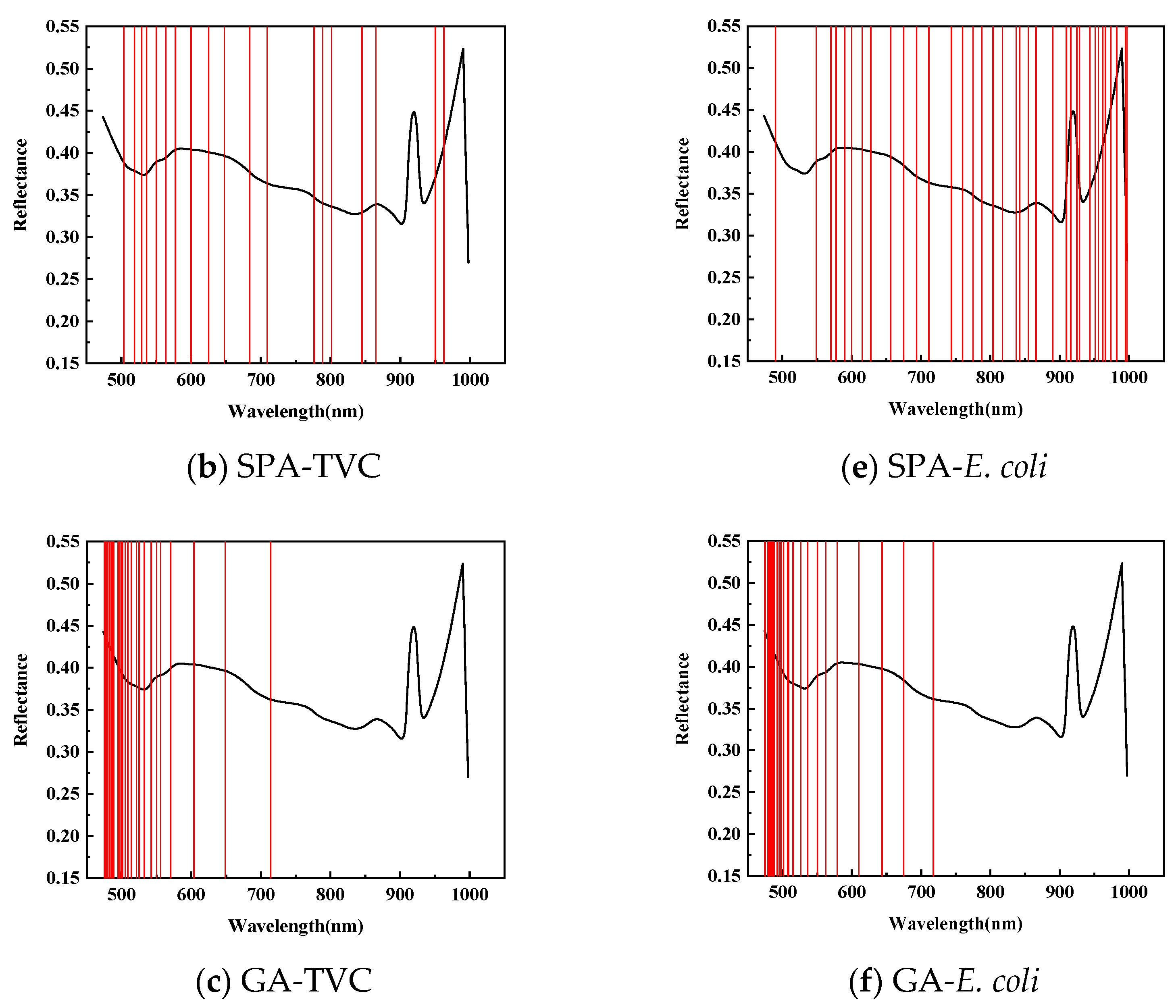
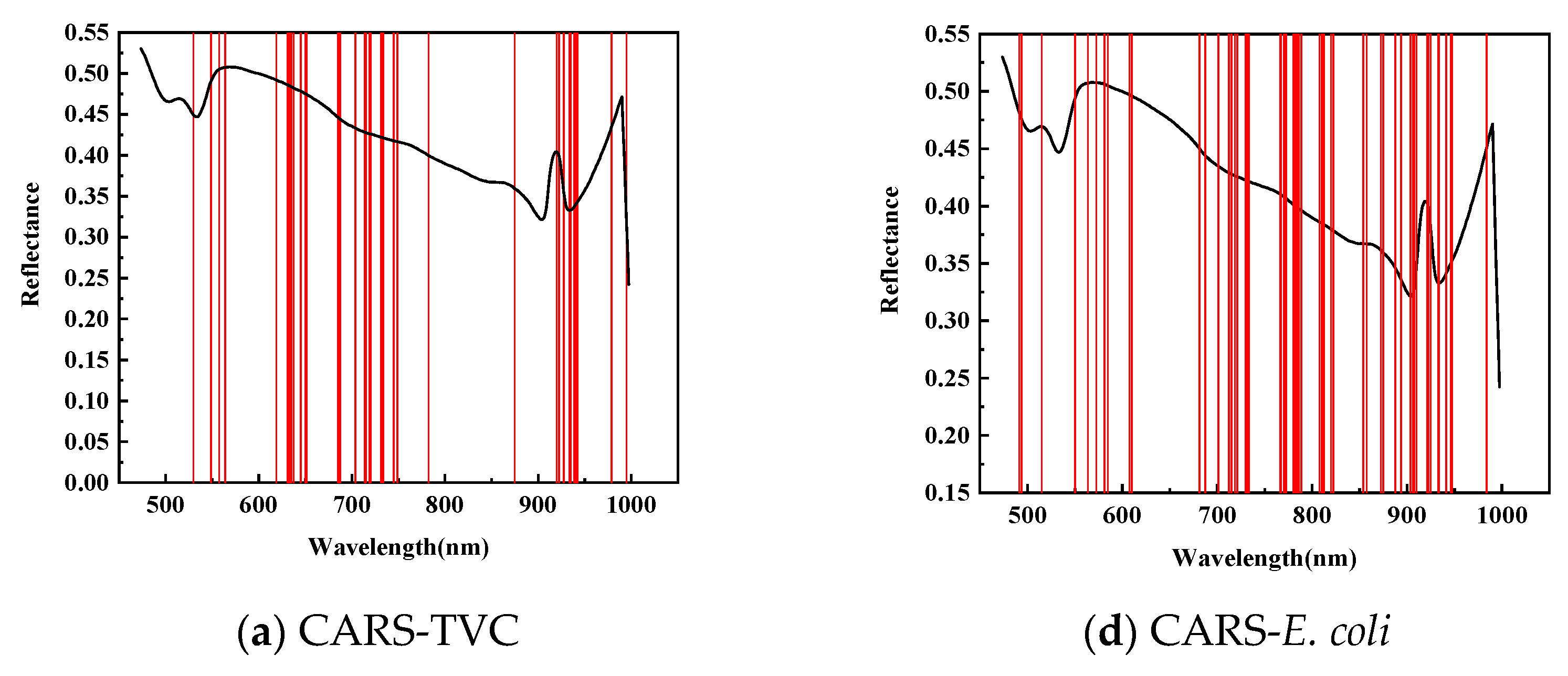
| Index | Wavelength Selection Method | Characteristic Wavelength (nm) | Model | RMSEC | Rcv | RMSECV | Rp | RMSEP | |
|---|---|---|---|---|---|---|---|---|---|
| TVC | CARS | 47 | PLSR | 0.9226 | 0.7541 | 0.8299 | 1.1058 | 0.8437 | 0.9512 |
| PCR | 0.9186 | 0.7723 | 0.8398 | 1.0761 | 0.8344 | 0.9765 | |||
| BP | 0.8688 | 0.6453 | - | - | 0.7476 | 0.9609 | |||
| SPA | 19 | PLSR | 0.8871 | 0.9024 | 0.7758 | 1.2507 | 0.8737 | 0.7721 | |
| PCR | 0.8332 | 1.0809 | 0.6393 | 1.5244 | 0.8737 | 0.7721 | |||
| BP | 0.8570 | 0.6717 | - | - | 0.6567 | 1.0911 | |||
| GA | 26 | PLSR | 0.7981 | 1.1779 | 0.5854 | 1.6072 | 0.6639 | 1.3250 | |
| PCR | 0.8033 | 0.3860 | 1.2464 | 0.3860 | 0.6884 | 1.2851 | |||
| BP | 0.6624 | 0.9764 | - | - | 0.6525 | 1.0963 | |||
| E. coli | CARS | 39 | PLSR | 0.9309 | 0.8147 | 0.8576 | 1.1635 | 0.7632 | 1.4046 |
| PCR | 0.9233 | 0.8569 | 0.8528 | 1.1816 | 0.7663 | 1.3966 | |||
| BP | 0.8213 | 0.8485 | - | - | 0.6129 | 1.4049 | |||
| SPA | 36 | PLSR | 0.8872 | 1.0293 | 0.7617 | 1.4658 | 0.7421 | 1.1355 | |
| PCR | 0.8759 | 1.0764 | 0.7677 | 1.4498 | 0.7421 | 1.1355 | |||
| BP | 0.8608 | 0.741 | - | - | 0.6765 | 1.3072 | |||
| GA | 26 | PLSR | 0.7592 | 1.4520 | 0.6130 | 1.7874 | 0.7324 | 1.4882 | |
| PCR | 0.7929 | 1.3595 | 0.6284 | 1.7598 | 0.7982 | 1.3094 | |||
| BP | 0.7922 | 0.9077 | - | - | 0.7318 | 1.2097 |
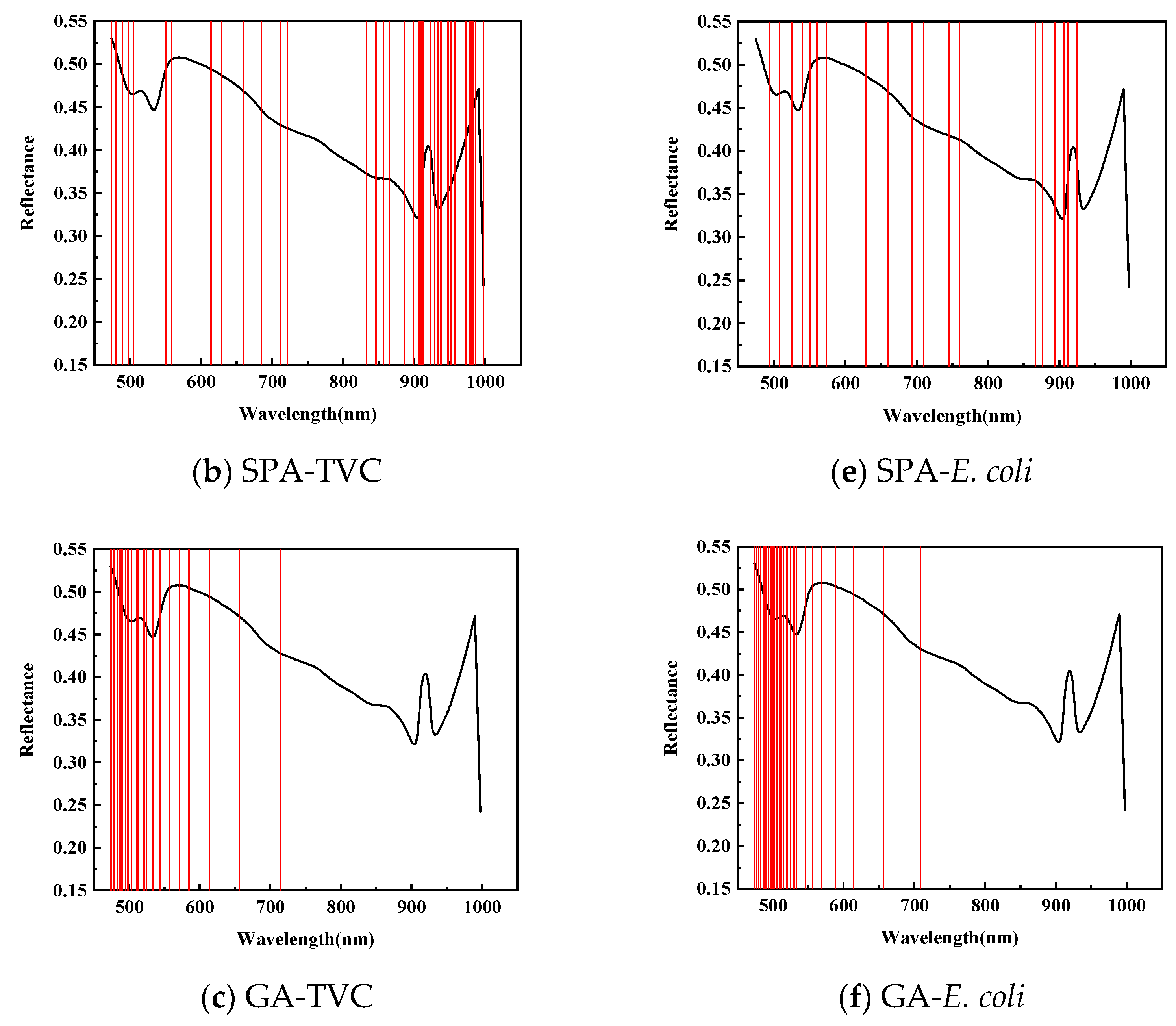
| Index | Wavelength Selection Method | Characteristic Wavelength (nm) | Model | RMSEC | Rcv | RMSECV | RMSEP | ||
|---|---|---|---|---|---|---|---|---|---|
| TVC | CARS | 38 | PLSR | 0.9437 | 0.5836 | 0.8935 | 0.8037 | 0.8464 | 0.9474 |
| PCR | 0.9376 | 0.6136 | 0.8882 | 0.8222 | 0.7517 | 1.1172 | |||
| BP | 0.8744 | 0.5710 | - | - | 0.7015 | 0.9818 | |||
| SPA | 36 | PLSR | 0.8571 | 0.9090 | 0.7367 | 1.2102 | 0.7635 | 1.4038 | |
| PCR | 0.8642 | 0.8880 | 0.7477 | 1.1885 | 0.7635 | 1.4038 | |||
| BP | 0.8700 | 0.5802 | - | - | 0.6829 | 1.0106 | |||
| GA | 24 | PLSR | 0.8218 | 1.005 | 0.7199 | 1.2423 | 0.8168 | 0.9774 | |
| PCR | 0.8162 | 1.0197 | 0.7206 | 1.2410 | 0.8052 | 1.0046 | |||
| BP | 0.8398 | 0.6388 | 0.8935 | - | 0.7519 | 0.912 | |||
| E. coli | CARS | 58 | PLSR | 0.9516 | 0.6528 | 0.8868 | 0.9956 | 0.7239 | 1.4035 |
| PCR | 0.8745 | 1.0306 | 0.7429 | 1.4423 | 0.7721 | 1.2927 | |||
| BP | 0.8209 | 0.8091 | - | - | 0.5847 | 1.3475 | |||
| SPA | 19 | PLSR | 0.8888 | 0.9735 | 0.7800 | 1.3484 | 0.7587 | 1.3251 | |
| PCR | 0.8969 | 0.9395 | 0.7887 | 1.3246 | 0.7744 | 1.2870 | |||
| BP | 0.8191 | 0.8126 | - | - | 0.7415 | 1.1146 | |||
| GA | 28 | PLSR | 0.8212 | 1.2125 | 0.6891 | 1.5615 | 0.8075 | 1.200 | |
| PCR | 0.8016 | 1.2703 | 0.6829 | 1.5741 | 0.7471 | 1.3522 | |||
| BP | 0.8423 | 0.7635 | - | - | 0.7624 | 1.0749 |
| Microbial Indicator | Location | Model | Number of Wavelengths | RMSEC | RMSEP | ||
|---|---|---|---|---|---|---|---|
| TVC | Back | RAW-PCR | 420 | 0.8642 | 0.8914 | 0.8402 | 1.0600 |
| RAW-CARS-PLSR | 47 | 0.9226 | 0.7541 | 0.8437 | 0.9512 | ||
| Abdomen | RAW-PCR | 420 | 0.8871 | 1.0297 | 0.8181 | 1.2500 | |
| RAW-CARS-PLSR | 38 | 0.9437 | 0.5836 | 0.8464 | 0.9474 | ||
| E. coli | Back | RAW-PCR | 420 | 0.9115 | 0.7259 | 0.7706 | 1.0797 |
| RAW-CARS-PLSR | 39 | 0.9309 | 0.8147 | 0.7632 | 1.4046 | ||
| Abdomen | RAW-PCR | 420 | 0.9486 | 0.6724 | 0.7802 | 1.2725 | |
| RAW-CARS-PLSR | 58 | 0.9516 | 0.6528 | 0.7239 | 1.4035 |
| Microbial Indicator | Location | Model | RMSEC | RMSECV | RMSEP | ||||
|---|---|---|---|---|---|---|---|---|---|
| TVC | Back | PLSR | 0.7288 | 1.1428 | 0.6458 | 0.5047 | 0.7104 | 1.3763 | 0.2335 |
| PCR | 0.7324 | 1.1411 | 0.6471 | 0.5138 | 0.7168 | 1.3571 | 0.2160 | ||
| BP | 0.6884 | 1.2851 | 0.6224 | 0.4564 | 0.6756 | 1.3387 | 0.0536 | ||
| Abdomen | PLSR | 0.7448 | 1.3964 | 0.6672 | 0.5294 | 0.7276 | 1.4212 | 0.0248 | |
| PCR | 0.7535 | 1.4471 | 0.6728 | 0.5047 | 0.7104 | 1.2748 | 0.1277 | ||
| BP | 0.7224 | 1.161 | 0.6268 | 0.4648 | 0.6818 | 1.3145 | 0.1535 | ||
| E. coli | Back | PLSR | 0.7015 | 1.4764 | 0.6037 | 1.6684 | 0.6943 | 1.5941 | 0.1177 |
| PCR | 0.7221 | 1.4241 | 0.6418 | 1.3987 | 0.7097 | 1.4635 | 0.0394 | ||
| BP | 0.6639 | 1.3250 | 0.6113 | 1.6329 | 0.6168 | 1.6011 | 0.2761 | ||
| Abdomen | PLSR | 0.7193 | 1.4360 | 0.6151 | 1.6475 | 0.7024 | 1.5782 | 0.1422 | |
| PCR | 0.7549 | 1.3341 | 0.6646 | 1.3352 | 0.7401 | 1.3789 | 0.0448 | ||
| BP | 0.7011 | 1.2464 | 0.6288 | 1.4485 | 0.6877 | 1.2957 | 0.0493 |
| Microbial Indicator | Location | Model | RMSEC | RMSECV | RMSEP | ||||
|---|---|---|---|---|---|---|---|---|---|
| TVC | Back | PLSR | 0.7244 | 1.3122 | 0.6423 | 1.5941 | 0.7024 | 1.3946 | 0.0824 |
| PCR | 0.7318 | 1.3998 | 0.6654 | 1.517 | 0.7074 | 1.4122 | 0.0124 | ||
| BP | 0.7469 | 1.1126 | 0.7055 | 1.6047 | 0.7242 | 1.1491 | 0.0365 | ||
| Abdomen | PLSR | 0.7620 | 1.3954 | 0.6972 | 1.4582 | 0.7347 | 1.4047 | 0.0093 | |
| PCR | 0.7633 | 1.3323 | 0.6943 | 1.4378 | 0.7394 | 1.3875 | 0.0552 | ||
| BP | 0.7705 | 1.0273 | 0.7054 | 1.2243 | 0.6681 | 1.0615 | 0.0342 | ||
| E. coli | Back | PLSR | 0.6901 | 1.2216 | 0.6349 | 1.4244 | 0.6801 | 1.3345 | 0.1129 |
| PCR | 0.7079 | 1.2064 | 0.6383 | 1.4211 | 0.6831 | 1.3273 | 0.1209 | ||
| BP | 0.7047 | 1.2408 | 0.6026 | 1.6422 | 0.6163 | 1.6153 | 0.3745 | ||
| Abdomen | PLSR | 0.7472 | 1.1578 | 0.6502 | 1.5714 | 0.7243 | 1.2199 | 0.0621 | |
| PCR | 0.7635 | 1.1138 | 0.6896 | 1.4699 | 0.7459 | 1.2511 | 0.1373 | ||
| BP | 0.7312 | 1.1386 | 0.6765 | 1.3105 | 0.6966 | 1.2799 | 0.1413 |
| Microbial Indicator | Location | Rc | RMSEC | Rcv | RMSECV | Rp | RMSEP |
|---|---|---|---|---|---|---|---|
| TVC | Back | 0.6966 | 0.9836 | 0.6170 | 1.2958 | 0.6819 | 1.0277 |
| Abdomen | 0.7057 | 0.9302 | 0.6299 | 1.2714 | 0.6886 | 0.9887 | |
| E. coli | Back | 0.7033 | 0.9349 | 0.6287 | 1.2748 | 0.6823 | 1.0269 |
| Abdomen | 0.7078 | 0.9265 | 0.6335 | 1.2622 | 0.6954 | 0.9495 |
| Microbial Indicator | Method | Location | Rc | RMSEC | RMSECV | Rp | RMSEP | |
|---|---|---|---|---|---|---|---|---|
| TVC | Direct consolidation | Back | 0.8915 | 0.9675 | 0.8864 | 0.9721 | 0.8291 | 1.1061 |
| Abdomen | 0.9036 | 0.9634 | 0.8932 | 0.9664 | 0.8218 | 1.2084 | ||
| The D-S theory | Back | 0.9315 | 0.7469 | 0.8822 | 0.9337 | 0.8389 | 1.1651 | |
| Abdomen | 0.9401 | 0.7387 | 0.8903 | 0.9026 | 0.8443 | 1.1464 | ||
| E. coli | Direct consolidation | Back | 0.8885 | 0.9746 | 0.8576 | 1.0926 | 0.8116 | 1.2882 |
| Abdomen | 0.8897 | 0.9699 | 0.8786 | 1.0146 | 0.8180 | 1.2501 | ||
| The D-S theory | Back | 0.9315 | 0.7559 | 0.8871 | 0.9024 | 0.8398 | 1.1638 | |
| Abdomen | 0.9469 | 0.7443 | 0.8969 | 0.8995 | 0.8512 | 1.1224 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, T.; Ma, Y.; Guo, Z.; Wang, Y.; Kong, L.; Feng, Y.; Liu, H.; Meng, L. Method of Detecting Microorganisms on the Surface of Mandarin Fish Based on Hyperspectral and Information Fusion. Foods 2025, 14, 1468. https://doi.org/10.3390/foods14091468
Yuan T, Ma Y, Guo Z, Wang Y, Kong L, Feng Y, Liu H, Meng L. Method of Detecting Microorganisms on the Surface of Mandarin Fish Based on Hyperspectral and Information Fusion. Foods. 2025; 14(9):1468. https://doi.org/10.3390/foods14091468
Chicago/Turabian StyleYuan, Tao, Yixiao Ma, Zuyu Guo, Yijian Wang, Liqin Kong, Yaoze Feng, Haopeng Liu, and Liang Meng. 2025. "Method of Detecting Microorganisms on the Surface of Mandarin Fish Based on Hyperspectral and Information Fusion" Foods 14, no. 9: 1468. https://doi.org/10.3390/foods14091468
APA StyleYuan, T., Ma, Y., Guo, Z., Wang, Y., Kong, L., Feng, Y., Liu, H., & Meng, L. (2025). Method of Detecting Microorganisms on the Surface of Mandarin Fish Based on Hyperspectral and Information Fusion. Foods, 14(9), 1468. https://doi.org/10.3390/foods14091468






