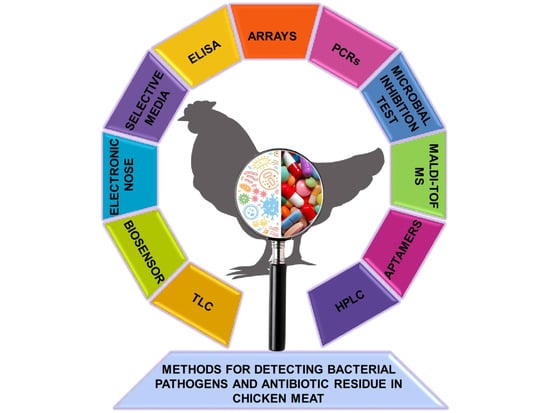
Detection of Bacterial Pathogens and Antibiotic Residues in Chicken Meat: A Review
Abstract
:1. Introduction
2. Source of Bacterial Contamination
3. Conventional Methods of Microbial Detection
3.1. Culture-Based Method
3.2. PCR-Based Method
3.3. Array-Based Method
3.4. ELISA-Based Method
4. Advanced Methods of Detection
4.1. Aptamers Based Method
4.2. Biosensor-Based Method
4.3. Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry-Based Method
4.4. Electronic Nose-Based Method
5. Conventional Methods of Antibiotics Residue Detection
5.1. Microbial Inhibition Test
5.2. ELISA-Based Method
5.3. Thin-Layer Chromatography (TLC)-Based Method
5.4. High-Performance Liquid Chromatography (HPLC)-Based Method
6. Advanced Methods of Detection
Biosensor-Based Method
7. Future Prospect
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mpundu, P.; Mbewe, A.R.; Muma, J.B.; Zgambo, J.; Munyeme, M. Evaluation of Bacterial Contamination in Dressed Chickens in Lusaka Abattoirs. Front. Public Health 2019, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Livestock and Poultry: World Markets and Trade. Available online: https://apps.fas.usda.gov/psdonline/circulars/livestock_poultry.pdf (accessed on 22 April 2020).
- Farrell, D. The Role of Poultry in Human Nutrition. Available online: http://www.fao.org/3/a-al712e.pdf (accessed on 8 December 2019).
- da Silva, M.V. Poultry and Poultry Products—Risks for Human Health. Available online: http://www.fao.org/3/a-al741e.pdf (accessed on 8 December 2019).
- Morton, V.K.; Kearney, A.; Coleman, S.; Viswanathan, M.; Chau, K.; Orr, A.; Hexemer, A. Outbreaks of Salmonella illness associated with frozen raw breaded chicken products in Canada, 2015–2019. Epidemiol. Infect. 2019, 147, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wensley, A.; Padfield, S.; Hughes, G.J. An outbreak of campylobacteriosis at a hotel in England: The ongoing risk due to consumption of chicken liver dishes. Epidemiol. Infect. 2020, 148, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grewal, V.S.; Khera, A. Outbreak of food poisoning in a working men’s hostel: A retrospective cohort study. Med. J. DY Patil Univ. 2017, 10, 517–521. [Google Scholar]
- Gumbo, A.; Bangure, D.; Gombe, N.T.; Mungati, M.; Tshimanga, M.; Hwalima, Z.; Dube, I. Staphylococcus aureus food poisoning among Bulawayo City Council employees, Zimbabwe, 2014. BMC Res. Notes 2015, 8, 485. [Google Scholar] [CrossRef] [Green Version]
- Gieraltowski, L.; Higa, J.; Peralta, V.; Green, A.; Schwensohn, C.; Rosen, H.; Libby, T.; Kissler, B.; Marsden-Haug, N.; Booth, H.; et al. National Outbreak of Multidrug Resistant Salmonella Heidelberg Infections Linked to a Single Poultry Company. PLoS ONE 2016, 11, e0162369. [Google Scholar] [CrossRef] [Green Version]
- Parry, A.; Fearnley, E.; Denehy, E. ‘Surprise’: Outbreak of Campylobacter infection associated with chicken liver pâté at a surprise birthday party, Adelaide, Australia, 2012. West. Pac. Surveill. Response J. 2012, 3, 16–19. [Google Scholar] [CrossRef]
- Tompkins, B.J.; Wirsing, E.; Devlin, V.; Kamhi, L.; Temple, B.; Weening, K.; Cavallo, S.; Allen, L.; Brinig, P.; Goode, B.; et al. Multistate Outbreak of Campylobacter jejuni infections associated with undercooked chicken livers—Northeastern United States, 2012. MMWR Morb. Mortal. Wkly. Rep. 2013, 62, 874–876. [Google Scholar]
- Edwards, D.S.; Milne, L.M.; Morrow, K.; Sheridan, P.; Verlander, N.Q.; Mulla, R.; Richardson, J.F.; Pender, A.; Lilley, M.; Reacher, M. Campylobacteriosis outbreak associated with consumption of undercooked chicken liver pâté in the East of England, September 2011: Identification of a dose–response risk. Epidemiol. Infect. 2013, 142, 352–357. [Google Scholar] [CrossRef] [Green Version]
- Farmer, S.; Keenan, A.; Vivancos, R. Food-borne Campylobacter outbreak in Liverpool associated with cross-contamination from chicken liver parfait: Implications for investigation of similar outbreaks. Public Health 2012, 126, 657–659. [Google Scholar] [CrossRef]
- Popović, I.; Heron, B.; Covacin, C. Listeria: An Australian Perspective (2001–2010). Foodborne Pathog. Dis. 2014, 11, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Wensley, A.; Coole, L. Cohort study of a dual-pathogen point source outbreak associated with the consumption of chicken liver pâté, UK, October 2009. J. Public Health 2013, 35, 585–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whittaker, P.J.; Sopwith, W.; Quigley, C.; Gillespie, I.A.; Willshaw, G.A.; Lycett, C.; Surman-Lee, S.; Baxter, D.; Adak, G.K.; Syed, Q. A national outbreak of verotoxin-producing Escherichia coli O157 associated with consumption of lemon-and-coriander chicken wraps from a supermarket chain. Epidemiol. Infect. 2008, 137, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Black, A.P.; Kirk, M.D.; Millard, G. Campylobacter outbreak due to chicken consumption at an Australian Capital Territory restaurant. Commun. Dis. Intell. Q. Rep. 2006, 30, 373–377. [Google Scholar] [PubMed]
- Pearson, A.D.; Greenwood, M.H.; Donaldson, J.; Healing, T.D.; Jones, D.M.; Shahamat, M.; Feltham, R.K.A.; Colwell, R.R. Continuous source outbreak of campylobacteriosis traced to chicken. J. Food Prot. 2000, 63, 309–314. [Google Scholar] [CrossRef] [PubMed]
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific opinion on quantification of the risk posed by broiler meat to human campylobacteriosis in the EU. EFSA J. 2010, 8, 1437. [Google Scholar]
- EFSA Panel on Biological Hazards (BIOHAZ); EFSA Panel on Contaminants in the Food Chain (CONTAM); EFSA Panel on Animal Health and Welfare (AHAW). Scientific opinion on the public health hazards to be covered by inspection of meat (poultry). EFSA J. 2012, 10, 2741. [Google Scholar]
- Mehdi, Y.; Létourneau-Montminy, M.-P.; Gaucher, M.-L.; Chorfi, Y.; Suresh, G.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of antibiotics in broiler production: Global impacts and alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef]
- Cornejo, J.; Yévenes, K.; Avello, C.; Pokrant, E.; Maddaleno, A.; Martín, B.S.; Lapierre, L. Determination of chlortetracycline residues, antimicrobial activity and presence of resistance genes in droppings of experimentally treated broiler chickens. Molecules 2018, 23, 1264. [Google Scholar] [CrossRef] [Green Version]
- Lan, C.; Yin, D.; Yang, Z.; Zhao, W.; Chen, Y.; Zhang, W.; Zhang, S. Determination of Six Macrolide Antibiotics in Chicken Sample by Liquid Chromatography-Tandem Mass Spectrometry Based on Solid Phase Extraction. J. Anal. Methods Chem. 2019, 2019, 1–13. [Google Scholar] [CrossRef]
- Goodnough, M.C.; Johnson, E.A. Control of Salmonella enteritidis infections in poultry by polymyxin B and trimethoprim. Appl. Environ. Microbiol. 1991, 57, 785–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, D.C.; Branion, H.D.; Slinger, S.J.; Anderson, G.W. Influence of environment on the growth response of chicks to Penicillin. Poult. Sci. 1953, 32, 462–466. [Google Scholar] [CrossRef]
- Feighner, S.D.; Dashkevicz, M.P. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl. Environ. Microbiol. 1987, 53, 331–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baert, K.; De Baere, S.; Croubels, S.; De Backer, P. Pharmacokinetics and Oral Bioavailability of Sulfadiazine and Trimethoprim in Broiler Chickens. Vet. Res. Commun. 2003, 27, 301–309. [Google Scholar] [CrossRef]
- Er, B.; Onurdag, F.K.; Demirhan, B.; Ozgacar, S.O.; Oktem, A.B.; Abbasoglu, U. Screening of quinolone antibiotic residues in chicken meat and beef sold in the markets of Ankara, Turkey. Poult. Sci. 2013, 92, 2212–2215. [Google Scholar] [CrossRef] [PubMed]
- Juan-García, A.; Font, G.; Picó, Y. Determination of quinolone residues in chicken and fish by capillary electrophoresis-mass spectrometry. Electrophoresis 2006, 27, 2240–2249. [Google Scholar] [CrossRef]
- Silfrany, R.O.; Caba, R.E.; Santos, F.S.D.L.; Hanning, I. Detection of quinolones in poultry meat obtained from retail centers in Santiago Province, the Dominican Republic. J. Food Prot. 2013, 76, 352–354. [Google Scholar] [CrossRef]
- PoultryMed. Available online: https://www.poultrymed.com/Aminoglycosides (accessed on 2 January 2020).
- PoultryMed. Available online: https://www.poultrymed.com/Lincosamides (accessed on 2 January 2020).
- Muaz, K.; Riaz, M.; Akhtar, S.; Park, S.; Ismail, A. Antibiotic residues in chicken meat: Global prevalence, threats, and decontamination strategies: A review. J. Food Prot. 2018, 81, 619–627. [Google Scholar] [CrossRef]
- Svobodova, J.; Tůmová, E. Factors affecting microbial contamination of market eggs: A review. Sci. Agric. Bohem. 2015, 45, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Messens, W.; Grijspeerdt, K.; Herman, L. Eggshell penetration by Salmonella: A review. Worlds Poult. Sci. J. 2005, 61, 71–86. [Google Scholar] [CrossRef]
- Vihavainen, E.; Lundström, H.-S.; Susiluoto, T.; Koort, J.; Paulin, L.; Auvinen, P.; Björkroth, K.J. Role of broiler carcasses and processing plant air in contamination of modified-atmosphere-packaged broiler products with psychrotrophic lactic acid bacteria. Appl. Environ. Microbiol. 2006, 73, 1136–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luber, P. Cross-contamination versus undercooking of poultry meat or eggs—Which risks need to be managed first? Int. J. Food Microbiol. 2009, 134, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Warsow, C.R.; Orta-Ramirez, A.; Marks, B.P.; Ryser, E.T.; Booren, A.M. Single Directional Migration of Salmonella into Marinated Whole Muscle Turkey Breast. J. Food Prot. 2008, 71, 153–156. [Google Scholar] [CrossRef]
- Goksoy, E.; Kirkan, S.; Kok, F. Microbiological quality of broiler carcasses during processing in two slaughterhouses in Turkey. Poult. Sci. 2004, 83, 1427–1432. [Google Scholar] [CrossRef]
- Russell, S.M. The effect of an acidic, copper sulfate-based commercial sanitizer on indicator, pathogenic, and spoilage bacteria associated with broiler chicken carcasses when applied at various intervention points during poultry processing. Poult. Sci. 2008, 87, 1435–1440. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, M.; Lagier, J.C.; Raoult, D.; Khelaifia, S. Bacterial culture through selective and non-selective conditions: The evolution of culture media in clinical microbiology. New Microbes New Infect. 2020, 34, 100622. [Google Scholar] [CrossRef]
- Priyanka, B.; Patil, R.K.; Dwarakanath, S. A review on detection methods used for foodborne pathogens. Indian J. Med. Res. 2016, 144, 327–338. [Google Scholar] [CrossRef]
- Esinghal, N.; Ekumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [Green Version]
- Paniel, N.; Noguer, T. Detection of Salmonella in food matrices, from conventional methods to recent aptamer-sensing technologies. Foods 2019, 8, 371. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.K.; Yadav, R. Study of antibiotic resistance in bacteria Isolated from table egg. Int. J. Pharm. Bio Sci. 2017, 8, 668–674. [Google Scholar] [CrossRef]
- Begum, K.; Reza, T.A.; Haque, M.; Hossain, A.; Hassan, F.M.K.; Hasan, S.N.; Akhter, N.; Ahmed, A.; Barua, U. Isolation, identification and antibiotic resistance pattern of Salmonella spp. from chicken eggs, intestines and environmental samples. Bangladesh Pharm. J. 2010, 13, 23–27. [Google Scholar]
- Tessema, K.; Bedu, H.; Ejo, M.; Hiko, A. Prevalence and Antibiotic Resistance of Salmonella Species Isolated from Chicken Eggs by Standard Bacteriological Method. J. Vet. Sci. Technol. 2017, 8, 1–5. [Google Scholar] [CrossRef]
- Akond, M.A.; Hassan, S.M.R.; Alam, S.; Shirin, M. Antibiotic resistance of Escherichia coli isolated from poultry and poultry environment of Bangladesh. Am. J. Environ. Sci. 2009, 5, 47–52. [Google Scholar]
- Akond, M.A.; Shirin, M.; Alam, S.; Hassan, S.; Rahman, M.; Hoq, M. Frequency of drug resistant Salmonella spp. isolated from poultry samples in Bangladesh. Stamford J. Microbiol. 2013, 2, 15–19. [Google Scholar] [CrossRef]
- Ahmed, M.M.; Rahman, M.M.; Mahbub, K.R.; Wahiduzzaman, M. Characterization of antibiotic resistant Salmonella spp isolated from chicken eggs of Dhaka City. J. Sci. Res. 2010, 3, 191. [Google Scholar] [CrossRef] [Green Version]
- Arathy, D.S.; Vanpee, G.; Belot, G.; Mathew, V.; Deallie, C.; Sharma, R. Antimicrobial drug resistance in Escherichia coli isolated from commercial chicken eggs in Grenada, West Indies. West Indian Med. J. 2011, 60, 53–56. [Google Scholar]
- Al-Zenki, S.; Al-Nasser, A.; Al-Safar, A.; Alomirah, H.; Al-Haddad, A.; Hendriksen, R.S.; Aarestrup, F.M. Prevalence and antibiotic resistance of Salmonella isolated from a poultry farm and processing plant environment in the state of Kuwait. Foodborne Pathog. Dis. 2007, 4, 367–373. [Google Scholar] [CrossRef]
- Adesiyun, A.A.; Offiah, N.; Seepersadsingh, N.; Rodrigo, S.; Lashley, V.; Musai, L. Frequency and antimicrobial resistance of enteric bacteria with spoilage potential isolated from table eggs. Food Res. Int. 2006, 39, 212–219. [Google Scholar] [CrossRef]
- Burgos, M.J.G.; Márquez, M.L.F.; Pérez-Pulido, R.; Gálvez, A.; López, R.L. Virulence factors and antimicrobial resistance in Escherichia coli strains isolated from hen egg shells. Int. J. Food Microbiol. 2016, 238, 89–95. [Google Scholar] [CrossRef]
- Akhtar, F.; Hussain, I.; Khan, A.; Rahman, S.U. Prevalence and antibiogram studies of Salmonella enteritidis isolated from human and poultry sources. Pak. Vet. J. 2010, 30, 25–28. [Google Scholar]
- Pyzik, E.; Marek, A. Plasmid profile analysis and evaluation of antibiotic susceptibility of Staphylococcus aureus strains isolated from table chicken eggs. Pol. J. Vet. Sci. 2013, 16, 307–312. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Yadav, A.S.; Singh, S.M.; Bharti, P. Prevalence of Salmonella in chicken eggs collected from poultry farms and marketing channels and their antimicrobial resistance. Food Res. Int. 2010, 43, 2027–2030. [Google Scholar] [CrossRef]
- Singh, R.; Yadav, A.; Tripathi, V.; Singh, R.P. Antimicrobial resistance profile of Salmonella present in poultry and poultry environment in north India. Food Control 2013, 33, 545–548. [Google Scholar] [CrossRef]
- Geidam, Y.A.; Zakaria, Z.; Aziz, S.A.; Bejo, S.K.; Abu, J.; Omar, S. High prevalence of multi-drug resistant bacteria in selected poultry farms in Selangor, Malaysia. Asian J. Anim. Vet. Adv. 2012, 7, 891–897. [Google Scholar] [CrossRef]
- Khatun, M.N.; Mahbub-E-Elahi, A.T.M.; Ahmed, S.; Parvej, M.S.; Akhter, S.; Ansari, W.K.; Ali, M.S. Frequency of drug resistant Escherichia coli isolated from commercial broiler chicken in Bangladesh. Int. J. Nat. Soc. Sci. 2015, 2, 1–5. [Google Scholar]
- Abdi, R.D.; Mengstie, F.; Beyi, A.F.; Beyene, T.; Waktole, H.; Mammo, B.; Ayana, D.; Abunna, F. Determination of the sources and antimicrobial resistance patterns of Salmonella isolated from the poultry industry in Southern Ethiopia. BMC Infect. Dis. 2017, 17, 352. [Google Scholar] [CrossRef]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, K.H.M.N.H.; Noreddin, A.; Rahman, T.; El Zowalaty, M.E.; Rahman, M. Molecular detection of multidrug resistant Salmonella species isolated from broiler farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.; Mannan, S.; Ali, Y.; Bayzid, M.; Ahad, A.; Bupasha, Z.B. Antibiotic resistance of Escherichia coli isolated from broilers sold at live bird markets in Chattogram, Bangladesh. J. Adv. Vet. Anim. Res. 2019, 6, 272–277. [Google Scholar] [CrossRef]
- Abd-ElGhany, S.M.; Sallam, K.I.; Abd-Elkhalek, A.; Tamura, T. Occurrence, genetic characterization and antimicrobial resistance of Salmonella isolated from chicken meat and giblets. Epidemiol. Infect. 2014, 143, 997–1003. [Google Scholar] [CrossRef] [Green Version]
- Hidano, A.; Yamamoto, T.; Hayama, Y.; Muroga, N.; Kobayashi, S.; Nishida, T.; Tsutsui, T. Unraveling antimicrobial resistance genes and phenotype patterns among Enterococcus faecalis isolated from retail chicken products in Japan. PLoS ONE 2015, 10, e0121189. [Google Scholar] [CrossRef]
- Hayes, J.R.; English, L.L.; Carter, P.J.; Proescholdt, T.; Lee, K.Y.; Wagner, D.D.; White, D.G. prevalence and antimicrobial resistance of Enterococcus species isolated from retail meats. Appl. Environ. Microbiol. 2003, 69, 7153–7160. [Google Scholar] [CrossRef] [Green Version]
- Momtaz, H.; Jamshidi, A. Shiga toxin-producing Escherichia coli isolated from chicken meat in Iran: Serogroups, virulence factors, and antimicrobial resistance properties. Poult. Sci. 2013, 92, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Nazer, A.H.K.; Dadras, H.; Eskandari, S. Aerobic bacteria isolated from eggs and day-old chicks and their antibacterial resistance in Shiraz, Iran. Iran J. Vet. Res. 2006, 7, 20–30. [Google Scholar]
- Luber, P.; Wagner, J.; Hahn, H.; Bartelt, E. Antimicrobial resistance in Campylobacter jejuni and Campylobacter coli strains isolated in 1991 and 2001–2002 from poultry and humans in Berlin, Germany. Antimicrob. Agents Chemother. 2003, 47, 3825–3830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waters, A.E.; Contente-Cuomo, T.; Buchhagen, J.; Liu, C.M.; Watson, L.; Pearce, K.; Foster, J.T.; Bowers, J.; Driebe, E.M.; Engelthaler, D.M.; et al. Multidrug-resistant Staphylococcus aureus in US meat and poultry. Clin. Infect. Dis. 2011, 52, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- White, A.K.; Heyries, K.A.; Doolin, C.; VanInsberghe, M.; Hansen, C.L. High-throughput microfluidic single-cell digital polymerase chain reaction. Anal. Chem. 2013, 85, 7182–7190. [Google Scholar] [CrossRef] [PubMed]
- He, Q.-D.; Huang, D.-P.; Huang, G.; Chen, Z.-G. Advance in research of microfluidic polymerase chain reaction chip. Chin. J. Anal. Chem. 2016, 44, 542–550. [Google Scholar] [CrossRef]
- Jin, U.-H.; Cho, S.-H.; Kim, M.-G.; Ha, S.-D.; Kim, K.-S.; Lee, K.-H.; Kim, K.-Y.; Chung, D.H.; Lee, Y.-C.; Kim, C.-H. PCR method based on the ogdH gene for the detection of Salmonella spp. from chicken meat samples. J. Microbiol. 2004, 42, 216–222. [Google Scholar]
- Guran, H.S.; Oksuztepe, G. Detection and typing of Clostridium perfringens from retail chicken meat parts. Lett. Appl. Microbiol. 2013, 57, 77–82. [Google Scholar] [CrossRef]
- Suo, B.; He, Y.; Tu, S.-I.; Shi, X. A multiplex real-time polymerase chain reaction for simultaneous detection of Salmonella spp., Escherichia coli O157, and Listeria monocytogenes in meat products. Foodborne Pathog. Dis. 2010, 7, 619–628. [Google Scholar] [CrossRef]
- Navas, J.; Ortiz, S.; López, P.; Jantzen, M.M.; Lopez, V.; Martínez-Suárez, J.V. Evaluation of effects of primary and secondary enrichment for the detection of Listeria monocytogenes by real-time PCR in retail ground chicken meat. Foodborne Pathog. Dis. 2006, 3, 347–354. [Google Scholar] [CrossRef]
- Gonzalez, I.; Garcia, T.; Antolin, A.; Hernández, P.E.; Martin, R. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp. in chicken meat. Lett. Appl. Microbiol. 2000, 30, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Lamberti, C.; Cocolin, L. Survey of Campylobacter jejuni in retail chicken meat products by application of a quantitative PCR protocol. Int. J. Food Microbiol. 2010, 141, S75–S79. [Google Scholar] [CrossRef] [PubMed]
- Pentimalli, D.; Pegels, N.; García, T.; Martín, R.; González, I. Specific PCR detection of Arcobacter butzleri, Arcobacter cryaerophilus, Arcobacter skirrowii, and Arcobacter cibarius in chicken meat. J. Food Prot. 2009, 72, 1491–1495. [Google Scholar] [CrossRef] [PubMed]
- Saeki, E.K.; Alves, J.; Bonfante, R.C.; Hirooka, E.Y.; De Oliveira, T.C.R.M. Multiplex PCR (mPCR) for the detection of Salmonella spp. and the differentiation of the Typhimurium and Enteritidis serovars in chicken meat. J. Food Saf. 2012, 33, 25–29. [Google Scholar] [CrossRef]
- Alves, J.; Hirooka, E.Y.; De Oliveira, T.C.R.M. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of Campylobacter spp. and Salmonella spp. in chicken meat. LWT Food Sci. Technol. 2016, 72, 175–181. [Google Scholar] [CrossRef]
- Arunrut, N.; Kiatpathomchai, W.; Ananchaipattana, C. Development and evaluation of real-time loop mediated isothermal amplification assay for rapid and sensitive detection of Salmonella spp. in chicken meat products. J. Food Saf. 2018, 38, e12564. [Google Scholar] [CrossRef]
- Alves, J.; Marques, V.V.; Pereira, L.F.P.; Hirooka, E.Y.; De Oliveira, T.C.R.M. Multiplex PCR for the detection of Campylobacter spp. and Salmonella spp. in chicken meat. J. Food Saf. 2012, 32, 345–350. [Google Scholar] [CrossRef]
- Manguiat, L.S.; Fang, T.J. Evaluation of DAS™ kits for the detection of food-borne pathogens in chicken and meat-based street-vended foods. J. Food Drug Anal. 2013, 21, 198–205. [Google Scholar] [CrossRef] [Green Version]
- Umesha, S.; Manukumar, H.M. Advanced molecular diagnostic techniques for detection of food-borne pathogens: Current applications and future challenges. Crit. Rev. Food Sci. Nutr. 2017, 58, 84–104. [Google Scholar] [CrossRef]
- Kupradit, C.; Rodtong, S.; Ketudat-Cairns, M. Development of a DNA macroarray for simultaneous detection of multiple foodborne pathogenic bacteria in fresh chicken meat. World J. Microbiol. Biotechnol. 2013, 29, 2281–2291. [Google Scholar] [CrossRef]
- Kupradit, C.; Ruamkuson, D.; Rodtong, S.; Ketudat-Cairns, M. Novel multiplex polymerase chain reaction and an oligonucleotide array for specific detection of the dominant foodborne bacterial pathogens in chicken meat. Afr. J. Microbiol. Res. 2013, 7, 3085–3095. [Google Scholar] [CrossRef] [Green Version]
- Tortajada-Genaro, L.A.; Rodrigo, A.; Hevia, E.; Mena, S.; Niñoles, R.; Maquieira, Á. Microarray on digital versatile disc for identification and genotyping of Salmonella and Campylobacter in meat products. Anal. Bioanal. Chem. 2015, 407, 7285–7294. [Google Scholar] [CrossRef] [Green Version]
- Quiñones, B.; Parker, C.T.; Janda, J.M.; Miller, W.G.; Mandrell, R.E. Detection and genotyping of Arcobacter and Campylobacter isolates from retail chicken samples by use of DNA oligonucleotide arrays. Appl. Environ. Microbiol. 2007, 73, 3645–3655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlermroj, R.; Makornwattana, M.; Phuengwas, S.; Meerak, J.; Pichpol, D.; Karoonuthaisiri, N. DNA-based bead array technology for simultaneous identification of eleven foodborne pathogens in chicken meat. Food Control 2019, 101, 81–88. [Google Scholar] [CrossRef]
- Schneid, A.D.S.; Rodrigues, K.L.; Chemello, D.; Tondo, E.C.; Ayub, M.A.Z.; Aleixo, J.A.G. Evaluation of an indirect ELISA for the detection of Salmonella in chicken meat. Braz. J. Microbiol. 2006, 37, 350–355. [Google Scholar] [CrossRef] [Green Version]
- Taha, E.G.; Mohamed, A.; Srivastava, K.K.; Reddy, P.G. Rapid detection of Salmonella in chicken meat using immunomagnetic separation, CHROMagar, ELISA and Real-time polymerase chain reaction (RT-PCR). Int. J. Poult. Sci. 2010, 9, 831–835. [Google Scholar] [CrossRef] [Green Version]
- Lilja, L.; Hänninen, M.-L. Evaluation of a commercial automated ELISA and PCR-method for rapid detection and identification of Campylobacter jejuni and C. coli in poultry products. Food Microbiol. 2001, 18, 205–209. [Google Scholar] [CrossRef]
- Croci, L.; Delibato, E.; Volpe, G.; Palleschi, G. A rapid electrochemical ELISA for the detection of Salmonella in meat samples. Anal. Lett. 2001, 34, 2597–2607. [Google Scholar] [CrossRef]
- Vanderlinde, P.B.; Grau, F.H. Detection of Listeria spp. in Meat and Environmental Samples by an Enzyme-linked Immunosorbent Assay (ELISA). J. Food Prot. 1991, 54, 230–231. [Google Scholar] [CrossRef]
- Charlermroj, R.; Makornwattana, M.; Grant, I.R.; Elliott, C.T.; Karoonuthaisiri, N. Validation of a high-throughput immunobead array technique for multiplex detection of three foodborne pathogens in chicken products. Int. J. Food Microbiol. 2016, 224, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Rozenblum, G.T.; Lopez, V.G.; Vitullo, A.D.; Radrizzani, M. Aptamers: Current challenges and future prospects. Expert Opin. Drug Discov. 2015, 11, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.A.; Zhang, H.; Li, F.; Wang, Z.; Le, X.C.; Li, X.-F. Selection and analytical applications of aptamers binding microbial pathogens. TrAC Trends Anal. Chem. 2011, 30, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef]
- Kim, Y.S.; Raston, N.H.A.; Gu, M.B. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [CrossRef]
- Dong, Y.; Xu, Y.; Yong, W.; Chu, X.; Wang, D. Aptamer and its potential applications for food safety. Crit. Rev. Food Sci. Nutr. 2014, 54, 1548–1561. [Google Scholar] [CrossRef]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Ohk, S.; Koo, O.; Sen, T.; Yamamoto, C.; Bhunia, A.K. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food. J. Appl. Microbiol. 2010, 109, 808–817. [Google Scholar] [CrossRef]
- Muniandy, S.; Dinshaw, I.J.; Teh, S.J.; Lai, C.W.; Ibrahim, F.; Thong, K.L.; Leo, B.F. Graphene-based label-free electrochemical aptasensor for rapid and sensitive detection of foodborne pathogen. Anal. Bioanal. Chem. 2017, 409, 6893–6905. [Google Scholar] [CrossRef]
- Duan, N.; Wu, S.; Dai, S.; Miao, T.; Chen, J.; Wang, Z. Simultaneous detection of pathogenic bacteria using an aptamer based biosensor and dual fluorescence resonance energy transfer from quantum dots to carbon nanoparticles. Microchim. Acta 2014, 182, 917–923. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, X.; Duan, N.; Xia, Y.; Wang, Z.; Che, Z.; Wang, L.; Yang, X.; Chen, X. Selection and characterization, application of a DNA aptamer targeted to Streptococcus pyogenes in cooked chicken. Anal. Biochem. 2018, 551, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Renuka, R.M.; Achuth, J.; Chandan, H.R.; Venkataramana, M.; Kadirvelu, K. Fluorescent dual aptasensor for rapid and sensitive onsite detection of E. coli O157: H7 and its validation onto various food matrices. New J. Chem. 2018, 42, 10807–10817. [Google Scholar] [CrossRef]
- Sundararaj, N.; Kalagatur, N.K.; Mudili, V.; Krishna, K.; Antonysamy, M. Isolation and identification of enterotoxigenic Staphylococcus aureus isolates from Indian food samples: Evaluation of in-house developed aptamer linked sandwich ELISA (ALISA) method. J. Food Sci. Technol. 2019, 56, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Dai, Z.; Tian, X.; Jiang, X. Detection of Listeria monocytogenes based on combined aptamers magnetic capture and loop-mediated isothermal amplification. Food Control 2018, 85, 443–452. [Google Scholar] [CrossRef]
- Kumar, H.; Kuca, K.; Bhatia, S.K.; Saini, K.; Kaushal, A.; Verma, R.; Bhalla, T.C.; Kumar, D. Applications of nanotechnology in sensor-based detection of foodborne pathogens. Sensors 2020, 20, 1966. [Google Scholar] [CrossRef] [Green Version]
- Paniel, N.; Baudart, J.; Hayat, A.; Barthelmebs, L. Aptasensor and genosensor methods for detection of microbes in real world samples. Methods 2013, 64, 229–240. [Google Scholar] [CrossRef]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E.; Stricker, S. Application of electrochemical biosensors for detection of food pathogenic bacteria. Electroanalysis 2000, 12, 317–325. [Google Scholar] [CrossRef]
- Senturk, E.; Aktop, S.; Sanlibaba, P.; Tezel, B.U. Biosensors: A Novel Approach to Detect Food-borne Pathogens. Appl. Microbiol. Open Access 2018, 4, 1–8. [Google Scholar] [CrossRef]
- Poltronieri, P.; Mezzolla, V.; Primiceri, E.; Maruccio, G. Biosensors for the detection of food pathogens. Foods 2014, 3, 511–526. [Google Scholar] [CrossRef]
- Chen, I.-H.; Horikawa, S.; Bryant, K.; Riggs, R.; Chin, B.A.; Barbaree, J.M. Bacterial assessment of phage magnetoelastic sensors for Salmonella enterica Typhimurium detection in chicken meat. Food Control 2017, 71, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Ohk, S.-H.; Bhunia, A.K. Multiplex fiber optic biosensor for detection of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol. 2013, 33, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Valadez, A.M.; Lana, C.A.; Tu, S.-I.; Morgan, M.T.; Bhunia, A.K. Evanescent wave fiber optic biosensor for Salmonella detection in food. Sensors 2009, 9, 5810–5824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Kutsanedzie, F.Y.H.; Sun, H.; Wang, M.; Chen, Q.; Guo, Z.; Wu, J. Rapid Pseudomonas species identification from chicken by integrating colorimetric sensors with near-infrared spectroscopy. Food Anal. Methods 2017, 11, 1199–1208. [Google Scholar] [CrossRef]
- Park, M.-K.; Park, J.W.; Oh, J.-H. Optimization and application of a dithiobis-succinimidyl propionate-modified immunosensor platform to detect Listeria monocytogenes in chicken skin. Sens. Actuators B Chem. 2012, 323–331. [Google Scholar] [CrossRef]
- Chai, G.L.C.; Liu, G.; Chai, C.; Yao, B. Rapid Evaluation of Salmonella pullorum contamination in chicken based on a portable amperometric sensor. J. Biosens. Bioelectron. 2013, 4, 137. [Google Scholar] [CrossRef] [Green Version]
- Abdelhaseib, M.U.; Singh, A.K.; Bhunia, A.K. Simultaneous detection of Salmonella enterica, Escherichia coli and Listeria monocytogenes in food using a light scattering sensor. J. Appl. Microbiol. 2019, 126, 1496–1507. [Google Scholar] [CrossRef]
- Kim, Y.-J.; Kim, H.-S.; Chon, J.-W.; Kim, D.-H.; Hyeon, J.-Y.; Seo, K.-H. New colorimetric aptasensor for rapid on-site detection of Campylobacter jejuni and Campylobacter coli in chicken carcass samples. Anal. Chim. Acta 2018, 1029, 78–85. [Google Scholar] [CrossRef]
- Helali, S.; Alatawi, A.S.E.; AbdelGhani, A. Pathogenic Escherichia coli biosensor detection on chicken food samples. J. Food Saf. 2018, 38, e12510. [Google Scholar] [CrossRef]
- Huang, F.; Xue, L.; Zhang, H.; Guo, R.; Li, Y.; Liao, M.; Wang, M.; Lin, J. An enzyme-free biosensor for sensitive detection of Salmonella using curcumin as signal reporter and click chemistry for signal amplification. Theranostics 2018, 8, 6263–6273. [Google Scholar] [CrossRef]
- Wieser, A.; Schneider, L.; Jung, J.; Schubert, S. MALDI-TOF MS in microbiological diagnostics—Identification of microorganisms and beyond (mini review). Appl. Microbiol. Biotechnol. 2011, 93, 965–974. [Google Scholar] [CrossRef]
- Marvin, L.F.; Roberts, M.A.; Fay, L.B. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in clinical chemistry. Clin. Chim. Acta 2003, 337, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Rychert, J. Benefits and Limitations of MALDI-TOF Mass Spectrometry for the Identification of Microorganisms. J. Infect. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Rasmussen, M.M.; Opintan, J.A.; Frimodt-Møller, N.; Styrishave, B. Beta-lactamase producing Escherichia coli isolates in imported and locally produced chicken meat from Ghana. PLoS ONE 2015, 10, e0139706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdallah, H.M.; Reuland, E.A.; Wintermans, B.B.; Al Naiemi, N.; Koek, A.; Abdelwahab, A.M.; Ammar, A.M.; Mohamed, A.A.; Vandenbroucke-Grauls, C.M.J.E. Extended-spectrum β-lactamases and/or carbapenemases-producing Enterobacteriaceae isolated from retail chicken meat in Zagazig, Egypt. PLoS ONE 2015, 10, e0136052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somsri, A.; Pilasombut, K.; Ngamyeesoon, N.; Rumjuankiat, K. Detection and identification of bacterial contamination in meat by matrix-assisted laser desorption ionization-time of flight -mass spectrometry. Int. J. Agric. Technol. 2017, 13, 1487–1504. [Google Scholar]
- Woźniak-Biel, A.; Bugla-Płoskońska, G.; Kielsznia, A.; Korzekwa, K.; Tobiasz, A.; Korzeniowska-Kowal, A.; Wieliczko, A. High Prevalence of resistance to fluoroquinolones and tetracycline Campylobacter spp. isolated from poultry in Poland. Microb. Drug Resist. 2018, 24, 314–322. [Google Scholar] [CrossRef]
- Gardner, J.W.; Bartlett, P.N. A brief history of electronic noses. Sens. Actuators B Chem. 1994, 18, 210–211. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhasti, M.; Mohtasebi, S.S.; Siadat, M.; Balasubramanian, S. Meat Quality Assessment by Electronic Nose (Machine Olfaction Technology). Sensors 2009, 9, 6058–6083. [Google Scholar] [CrossRef]
- Rajamäki, T.; Alakomi, H.-L.; Ritvanen, T.; Skytta, E.; Smolander, M.; Ahvenainen, R. Application of an electronic nose for quality assessment of modified atmosphere packaged poultry meat. Food Control 2006, 17, 5–13. [Google Scholar] [CrossRef]
- Timsorn, K.; Thoopboochagorn, T.; Lertwattanasakul, N.; Wongchoosuk, C. Evaluation of bacterial population on chicken meats using a briefcase electronic nose. Biosyst. Eng. 2016, 151, 116–125. [Google Scholar] [CrossRef]
- Vishnuraj, M.; Kandeepan, G.; Rao, K.; Chand, S.; Kumbhar, V. Occurrence, public health hazards and detection methods of antibiotic residues in foods of animal origin: A comprehensive review. Cogent Food Agric. 2016, 2, 1235458. [Google Scholar] [CrossRef]
- Vermunt, A.E.M.; Stadhouders, J.; Loeffen, G.J.M.; Bakker, R. Improvements of the tube diffusion method for detection of antibiotics and sulfonamides in raw milk. Neth. Milk Dairy J. 1993, 47, 31–40. [Google Scholar]
- Bogaerts, R.; Wolf, F. A standardised method for the detection of residues of antibacterial substances in fresh meat. Fleischwirtschaft 1980, 60, 672–673. [Google Scholar]
- Nouws, J.F.M.; Schothorst, M.; Ziv, G. A critical evaluation of several microbiological test methods for residues of antimicrobial drugs in ruminants. Arch. Lebensm. Hyg. 1979, 30, 4–8. [Google Scholar]
- Elnasri, H.A.; Salman, A.M.; El Rade, S.A. Screening of antibiotic residues in poultry liver, kidney and muscle in Khartoum state, Sudan. J. Appl. Ind. Sci. 2014, 2, 116–122. [Google Scholar]
- Tang, J.S.; Gillevet, P.M. Reclassification of ATCC 9341 from Micrococcus luteus to Kocuria rhizophila. Int. J. Syst. Evol. Microbiol. 2003, 53, 995–997. [Google Scholar] [CrossRef]
- Ezenduka, E.V. Screening of antimicrobial residues in poultry meat in Enugu metropolis, Enugu State, South East Nigeria, Enugu State, South East Nigeria. Vet. Ital. 2019, 55, 143–148. [Google Scholar] [PubMed]
- Sophila, J.R.; Raj, G.D.; Kumanan, K.; Chandra, G.S.; Vairamuthu, S. Microbial inhibition assay for detection of antibiotic residues in chicken meat using vegetative form of Geobacillus stearothermophilus. Pharm. Innov. J. 2018, 7, 753–757. [Google Scholar]
- Shahbazi, Y.; Ahmadi, F.; Karami, N. Screening, determination and confirmation of tetracycline residues in chicken tissues using four-plate test, ELISA and HPLC-UV methods: Comparison between correlation results. Food Agric. Immunol. 2015, 26, 821–834. [Google Scholar] [CrossRef]
- Baazize-Ammi, D.; Dechicha, A.S.; Tassist, A.; Gharbi, I.; Hezil, N.; Kebbal, S.; Morsli, W.; Beldjoudi, S.; Saadaoui, M.R.; Guetarni, D. Screeing and quantification of antibiotic residues in broiler chicken meat and milk in the central region of Algeria. Rev. Sci. Tech. Int. Off. Epiz. 2019, 38, 1–16. [Google Scholar]
- Hussein, M.A.; Khalil, S. Screening of some antibiotics and anabolic steroids residues in broiler fillet marketed in El-Sharkia Governorate. Life Sci. J. 2013, 10, 2111–2118. [Google Scholar]
- Karmi, M. Detection and presumptive identification of antibiotic residues in poultry meat by using FPT. Glob. J. Pharmacol. 2014, 8, 160–165. [Google Scholar]
- Tajik, H.; Malekinejad, H.; Razavi-Rouhani, S.M.; Pajouhi, M.R.; Mahmoudi, R.; Haghnazari, A. Chloramphenicol residues in chicken liver, kidney and muscle: A comparison among the antibacterial residues monitoring methods of Four Plate Test, ELISA and HPLC. Food Chem. Toxicol. 2010, 48, 2464–2468. [Google Scholar] [CrossRef] [PubMed]
- Ramatla, T.A.; Ngoma, L.; Adetunji, M.C.; Mwanza, M. Evaluation of Antibiotic Residues in Raw Meat Using Different Analytical Methods. Antibiotics 2017, 6, 34. [Google Scholar] [CrossRef] [Green Version]
- Kadim, I.; Mahgoub, O.; Al-Marzooqi, W.; Al-Maqbaly, R.; Annamali, K.; Khalaf, S. Enzyme-linked immunosorbent assay for screening antibiotic and hormone residues in broiler chicken meat in the sultanate of Oman. J. Muscle Foods 2010, 21, 243–254. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Z.; Shi, W.; Eremin, S.A.; Shen, J. Development of a Chemiluminescent ELISA for Determining Chloramphenicol in Chicken Muscle. J. Agric. Food Chem. 2006, 54, 5718–5722. [Google Scholar] [CrossRef]
- Prajapati, M.; Ranjit, E.; Shrestha, R.; Shrestha, S.P.; Adhikari, S.K.; Khanal, D.R. Status of antibiotic residues in poultry meat of Nepal. Nepal. Vet. J. 2018, 35, 55–62. [Google Scholar] [CrossRef]
- De Wasch, K.; Okerman, L.; De Brabander, H.; Van Hoof, J.; Croubels, S.; De Backer, P. Detection of residues of tetracycline antibiotics in pork and chicken meat: Correlation between results of screening and confirmatory tests. Analyst 1998, 123, 2737–2741. [Google Scholar] [CrossRef]
- Onyeanu, C.T.; Ezenduka, E.V.; Anaga, A.O. Determination of gentamicin use in poultry farms in Enugu state, Nigeria, and detection of its residue in slaughter commercial broilers. Int. J. One Health 2020, 6, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Mashak, Z.; Langroodi, A.M.; Mehdizadeh, T.; Fathabad, A.E.; Asadi, A.H. Detection of quinolones residues in beef and chicken meat in hypermarkets of Urmia, Iran using ELISA. Iran Agric. Res. 2017, 36, 73–77. [Google Scholar]
- Sherma, J. Thin-layer chromatography in food and agricultural analysis. J. Chromatogr. A 2000, 880, 129–147. [Google Scholar] [CrossRef]
- Khan, A.T. Advantages and Disadvantages of Thin Layer Chromatography. Available online: https://www.biomadam.com/advantages-and-disadvantages-of-thin-layer-chromatography (accessed on 3 October 2020).
- Sarker, Y.A.; Hasan, M.M.; Paul, T.K.; Rashid, S.Z.; Alam, M.N.; Sikder, M.H. Screening of antibiotic residues in chicken meat in Bangladesh by thin layer chromatography. J. Adv. Vet. Anim. Res. 2018, 5, 140. [Google Scholar] [CrossRef]
- Tajick, M.A.; Shohreh, B. Detection of antibiotics residue in chicken meat using TLC. Int. J. Poult. Sci. 2006, 5, 611–612. [Google Scholar]
- Billah, M.; Rana, S.M.M.; Hossain, M.S.; Saifuddin, A.K.M.; Islam, S.K.M.A.; Naim, Z.; Barua, S. Determination of the presence and pharmacokinetic profile of ciprofloxacin by TLC and HPLC method respectively in broiler chicken after single oral administration. J. Antibiot. 2014, 67, 745–748. [Google Scholar] [CrossRef]
- Premarathne, J.M.K.J.K.; Satharasinghe, D.A.; Gunasena, A.R.C.; Wanigasekara, A.; Munasinghe, D.M.S.; Abeynayake, P. Thin-layer chromatographic method for quantification of sulfonamides in chicken meat. Food Anal. Methods 2018, 11, 2666–2672. [Google Scholar] [CrossRef]
- Das, S.; Faysal, M.N.A.; Ferdous, J.; Sachi, S.; Islam, M.S.; Sikder, M.H. Detection of oxytetracycline and doxycycline residue in different growth stages of commercial broiler. Bangladesh J. Vet. Med. 2019, 17, 7–14. [Google Scholar]
- Ali, M.R.; Sikder, M.M.; Islam, M.S.; Islam, M.S. Investigation of discriminate and indiscriminate use of doxycycline in broiler: An indoor research on antibiotic doxycycline residue study in edible poultry tissue. Asian J. Med. Biol. Res. 2020, 6, 1–7. [Google Scholar] [CrossRef]
- Mora, L.; Reig, M. Methods for rapid detection of chemical and veterinary drug residues in animal foods. Trends Food Sci. Technol. 2006, 17, 482–489. [Google Scholar] [CrossRef]
- Bergweff, A.A.; Schloesser, J. Residue determination. In Encyclopedia of Food Sciences and Nutrition; Caballero, B., Trugo, L., Finglas, P., Eds.; Elsevier: London, UK, 2003; pp. 254–261. [Google Scholar]
- Chrominfo. Advantages and Disadvantages of HPLC. Available online: https://chrominfo.blogspot.com/2019/03/advantages-and-disadvantages-of-hplc.html (accessed on 3 October 2020).
- Aman, I.M.; Ahmed, H.F.; Mostafa, N.Y.; Kitada, Y.; Kar, G. Detection of tetracycline veterinary drug residues in Egyptian poultry meat by high performance liquid chromatography. J. Vet. Med. Allied Sci. 2017, 1, 52–58. [Google Scholar]
- Shalaby, A.R.; Salama, N.A.; Abou-Raya, S.H.; Emam, W.H.; Mehaya, F.M. Validation of HPLC method for determination of tetracycline residues in chicken meat and liver. Food Chem. 2011, 124, 1660–1666. [Google Scholar] [CrossRef]
- Zhao, S.; Jiang, H.; Li, X.; Mi, T.; Li, C.; Shen, J. Simultaneous Determination of Trace Levels of 10 Quinolones in Swine, Chicken, and Shrimp Muscle Tissues Using HPLC with Programmable Fluorescence Detection. J. Agric. Food Chem. 2007, 55, 3829–3834. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Guo, L.; Xu, F.; Rao, Q.; Xia, X.; Li, X.; Ding, S. Simultaneous Determination of Fluoroquinolones, Tetracyclines and Sulfonamides in Chicken Muscle by UPLC–MS–MS. Chromatographia 2010, 71, 383–388. [Google Scholar] [CrossRef]
- Wang, B.; Pang, M.; Xie, X.; Zhao, M.; Xie, K.; Zhang, Y.; Zhao, X.; Wang, Y.; Wang, R.; Wu, H.; et al. Quantitative analysis of amoxicillin, amoxicillin major metabolites, and ampicillin in chicken tissues via ultra-performance liquid chromatography-electrospray ionization tandem mass spectrometry. Food Anal. Methods 2017, 10, 3292–3305. [Google Scholar] [CrossRef]
- Virolainen, N.E.; Pikkemaat, M.G.; Elferink, J.W.A.; Karp, M.T. Rapid detection of tetracyclines and their 4-epimer derivatives from poultry meat with bioluminescent biosensor bacteria. J. Agric. Food Chem. 2008, 56, 11065–11070. [Google Scholar] [CrossRef]
- Pikkemaat, M.G.; Rapallini, M.L.B.A.; Karp, M.T.; Elferink, J.W.A. Application of a luminescent bacterial biosensor for the detection of tetracyclines in routine analysis of poultry muscle samples. Food Addit. Contam. Part A 2010, 27, 1112–1117. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, C.; Sarpong, V.; Gu, Z. Multisegment nanowire/nanoparticle hybrid arrays as electrochemical biosensors for Simultaneous detection of antibiotics. Biosens. Bioelectron. 2019, 126, 632–639. [Google Scholar] [CrossRef]
- Gan, T.; Shi, Z.; Sun, J.; Liu, Y. Simple and novel electrochemical sensor for the determination of tetracycline based on iron/zinc cations–exchanged montmorillonite catalyst. Talanta 2014, 121, 187–193. [Google Scholar] [CrossRef]
- Mohammad-Razdari, A.; Ghasemi-Varnamkhasti, M.; Izadi, Z.; Rostami, S.; Ensafi, A.A.; Siadat, M.; Losson, E. Detection of sulfadimethoxine in meat samples using a novel electrochemical biosensor as a rapid analysis method. J. Food Compos. Anal. 2019, 82, 103252. [Google Scholar] [CrossRef]
- Kim, D.-M.; Rahman, M.A.; Do, M.H.; Ban, C.; Shim, Y.-B. An amperometric chloramphenicol immunosensor based on cadmium sulfide nanoparticles modified-dendrimer bonded conducting polymer. Biosens. Bioelectron. 2010, 25, 1781–1788. [Google Scholar] [CrossRef]
- Ferguson, J.; Baxter, A.; Young, P.; Kennedy, G.; Elliott, C.; Weigel, S.; Gatermann, R.; Ashwin, H.; Stead, S.; Sharman, M. Detection of chloramphenicol and chloramphenicol glucuronide residues in poultry muscle, honey, prawn and milk using a surface plasmon resonance biosensor and Qflex® kit chloramphenicol. Anal. Chim. Acta 2005, 529, 109–113. [Google Scholar] [CrossRef]
- Huet, A.-C.; Charlier, C.; Singh, G.; Godefroy, S.B.; Leivo, J.; Vehniäinen, M.; Nielen, M.W.F.; Weigel, S.; Delahaut, P. Development of an optical surface plasmon resonance biosensor assay for (fluoro)quinolones in egg, fish, and poultry meat. Anal. Chim. Acta 2008, 623, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ho, B.; Ding, J.L. Future Perspectives on New Approaches in Pathogen Detection. Biomed. Sci. Lett. 2015, 21, 165–171. [Google Scholar] [CrossRef] [Green Version]
- Foddai, A.C.G.; Grant, I.R. Methods for detection of viable foodborne pathogens: Current state-of-art and future prospects. Appl. Microbiol. Biotechnol. 2020, 104, 4281–4288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Country | Year | Source | Pathogen | Disease | Confirmed Cases | Reference |
|---|---|---|---|---|---|---|
| Canada | 2015–2019 | Frozen raw breaded chicken products | Salmonella enteritidis | Salmonellosis | 584 | [5] |
| United Kingdom | 2017 | Chicken liver dishes | Campylobacter spp. | Campylobacteriosis | 7 | [6] |
| India | 2016 | Cooked chicken | Clostridium perfringens or Bacillus cereus | Food poisoning | 68 | [7] |
| Zimbabwe | 2014 | Stewed chicken | Staphylococcus aureus | Food poisoning | 53 | [8] |
| United States | 2013–2014 | Chicken dishes | Salmonella Heidelberg | Salmonellosis | 634 | [9] |
| Australia | 2012 | Chicken liver pâté | Campylobacter spp. | Campylobacteriosis | 15 | [10] |
| United States | 2012 | Undercooked chicken liver | Campylobacter jejuni | Campylobacteriosis | 6 | [11] |
| United Kingdom | 2011 | Undercooked chicken liver pâté | Campylobacter coli, Campylobacter jejuni | Campylobacteriosis | 22 | [12] |
| United Kingdom | 2011 | Chicken liver parfait | Campylobacter spp. | Campylobacteriosis | 3 | [13] |
| Australia | 2009 | Chicken wraps | Listeria monocytogenes | Listeriosis | 36 | [14] |
| United Kingdom | 2009 | Chicken liver pâté | Salmonella typhimurium DT8, Campylobacter spp. | Campylobacteriosis, Salmonellosis | 14 | [15] |
| United Kingdom | 2007 | Lemon-and-coriander chicken wraps | Verotoxin-producing Escherichia coli O157 | Diarrhoea | 12 | [16] |
| Australia | 2005 | Chicken dishes | Campylobacter spp. | Campylobacteriosis | 11 | [17] |
| United Kingdom | 1984–1985 | Live chicken | Campylobacter jejuni | Campylobacteriosis | 19 | [18] |
| Name of Antibiotic Class | Types of Antibiotics | Mode of Administration | Biological Effect | Reference |
|---|---|---|---|---|
| Tetracyclines | Tetracycline; Oxytetracycline; Doxycycline; Chlortetracycline | Oral and intramuscular | Bacteriostatic activity against a wide array of Gram-positive and-negative bacteria, mycoplasmas, some mycobacteria, as well as several protozoa and filariae | [22] |
| Macrolides | Tylosin; Tilmicosin | Oral | Antibacterial activity against pathogens such as Gram-positive and-negative bacteria | [23] |
| Lipopeptides | Polymyxins | Oral | Antibacterial activity against Gram-negative bacteria | [24] |
| Penicillins | Penicillin | Oral | Growth promoter | [25,26] |
| Folate Pathway Inhibitors | Trimethoprim | Oral | Treatment of respiratory, gastrointestinal infections | [27] |
| Quinolones | Enrofloxacin; Ciprofloxacin; Danofloxacin | Oral | Growth promoter and antibacterial activity against pathogens such as Gram-positive and-negative bacteria | [28,29,30] |
| Aminoglycosides | Neomycin; Streptomycin | Oral | Antibacterial activity against Gram-negative bacteria | [31] |
| Lincosamides | Lincomycin | Oral and intramuscular | Antibacterial activity against Gram-positive bacteria | [32] |
| Source of Isolation | Site of Isolation | Types of Medium | Incubation Temperature/Time | Types of Bacteria | Antibiogram Assay | Antibiotics Resistance | Reference |
|---|---|---|---|---|---|---|---|
| Egg | Shell surface, yolk, albumin | MacConkey agar, Eosin methylene blue (EMB) agar, Bismuth sulphite agar, Salmonella Shigella agar, Xylose lysine deoxycholate agar | 37 °C/24–48 h | Citrobacter spp., Enterobacter spp., Escherichia spp., Klebsiella spp., Proteus spp., Shigella spp., Serratia spp. | Disk diffusion | Cefixime, amoxicillin, amoxyclave | [45] |
| Whole egg content | Xylose Lysine Deoxycholate agar, MacConkey agar | 37 °C/24–48 h | Salmonella typhi, Salmonella enteritidis | Disk diffusion | Co-trimoxazole, nalidixic acid, ampicillin, tetracycline, kanamycin | [46] | |
| Shell surface, interior | Xylose lysine deoxycholate agar, Salmonella Shigella agar | 37 °C/24 h | Salmonella spp. | Disk diffusion | Tetracyclin, ampicillin, amoxicillin | [47] | |
| Shell surface | Eosin methylene blue (EMB) agar | 37 °C/24 h | Escherichia coli | Disk diffusion | Penicillin, ciprofloxacin, rifampicin, kanamycin, streptomycin, cefixime, erythromycin, ampicillin, tetracycline | [48] | |
| Shell surface | Salmonella Shigella agar | 37 °C/24 h | Salmonella typhimurium, Salmonella enteritidis | Disk diffusion | Erythromycin, ampicillin, penicillin, tetracycline | [49] | |
| Shell surface, yolk, albumin | Salmonella Shigella agar, Xylose lysine deoxycholate agar, Bismuth sulphite agar | 35–37 °C/24 h | Salmonella enterica subsp. salamae, Salmonella enterica subsp. indica, Salmonella paratyphi-A, Salmonella bongori, Salmonella choleraesuis | Disk diffusion | Amoxicillin, ampicillin | [50] | |
| Yolk | Blood agar, McConkey agar | 37 °C/24 h | Escherichia coli | Disk diffusion | Ampicillin | [51] | |
| Interior content | Xylose lysine deoxycholate agar, Bismuth sulphite agar | 37 °C/24 h | Salmonella enteritidis | MIC | Ampicillin, nalidixic acid, tetracycline | [52] | |
| Shell surface, interior | McConkey agar | 37 °C/24 h | Escherichia coli, Salmonella spp., Campylobacter spp. and Listeria spp. Enterobacter spp. Klebsiella spp. | Disk diffusion | Streptomycin, tetracycline, kanamycin | [53] | |
| Shell surface | Eosin methylene blue (EMB) agar | 37 °C/24 h | Escherichia coli | Disk diffusion | Ampicillin, streptomycin, tetracycline | [54] | |
| Shell surface, interior | Brilliant green agar, McConkey agar, Salmonella Shigella agar | 37 °C/24 h | Salmonella enteritidis | Disk diffusion | Bacitracin, erythromycin, novobiocin | [55] | |
| Shell surface, yolk | Blood agar, Mannitol salt agar | 37 °C/24–48 h | Staphylococcus aureus | Disk diffusion | Erythromycin, tetracycline | [56] | |
| Shell surface, yolk, albumin | Hektoen enteric agar | 37 °C/24 h | Salmonella typhimurium | Disk diffusion | Bacitracin, polymyxin-B, colistin | [57] | |
| Shell surface, yolk | Hektoen enteric agar | 37 °C/24 h | Salmonella typhimurium | Disk diffusion | Clindamycin, oxacillin, penicillin, vancomycin | [58] | |
| ealthy chicken | Skin, feather, nasal, cloaca | Mannitol salt agar, McConkey agar, Brilliant green agar, Blood agar | ND | Staphylococcus aureus, Escherichia coli, Pasteurella spp., Salmonella spp. | Disk diffusion | Tetracycline | [59] |
| Cloaca | Eosin methylene blue (EMB) agar | ND | Escherichia coli | Disk diffusion | Gentamycin, erythromycin, penicillin, cephalexin, amoxicillin, nalidixic acid | [60] | |
| Cloaca | Xylose lysine deoxycholate agar, Brilliant green agar | 37 °C/24 h | Salmonella spp. | Disk diffusion | Kanamycin, sulfamethoxazole-trimethoprim, nalidixic acid, ampicillin, cefoxitin, streptomycin, tetracycline, chloramphenicol | [61] | |
| Cloaca | Xylose lysine deoxycholate agar | ND | Salmonella spp. | Disk diffusion | Tetracycline, chloramphenicol, ampicillin, streptomycin | [62] | |
| Cloaca | McConkey agar, Eosin methylene blue (EMB) agar | 37 °C/18–24 h | Escherichia coli | Disk diffusion | Ampicillin, tetracycline, sulfamethoxazole-trimethoprim, nalidixic acid | [63] | |
| Meat | Drumsticks, gizzards, liver | Xylose lysine deoxycholate agar, Brilliant green agar | 37 °C/24 h | Salmonella spp. | Disk diffusion | Erythromycin, penicillin, amoxicillin | [64] |
| Liver, gizzards, hearts | Enterococcosel agar | 37 °C/48 h | Enterococcus faecalis | Disk diffusion | Oxytetracycline, dihydrostreptomycin | [65] | |
| Brest | Enterococcosel agar | 35 °C/24 h | Enterococcus faecium | MIC | Quinupristin-dalfopristin | [66] | |
| Brest, muscle | McConkey agar supplemented with 5% sheep blood | 37 °C/18–24 h | Escherichia coli | Disk diffusion | Tetracycline, chloramphenicol, nitrofurantoin | [67] |
| Type of PCR | Sample Used | Target Site of Bacterial Pathogen | Primers | Probe | Detection Chemistry | Limit of Detection | Reference |
|---|---|---|---|---|---|---|---|
| Simple | Meat (PND) | Spiked Salmonella typhimurium:ogdh gene | Forward 5′-GCCTTCCTGAAACGTGACCTA-3′ and reverse 5′-ACCATCTCTTTCAGCATGGGT3′ | NA | NA | 102 cfu/mL | [73] |
| Multiplex | Meat (Breasts, wings, drumsticks, legs) | Clostridium perfringens:cpa, cpb, etx, iA, cpe and cpb2 genes | Forward 5′-GCTAATGTTACTGCCGTTGA-3′ and reverse 5′-CCTCTGATACATCGTGTAAG-3′; Forward 5′- GCGAATATGCTGAATCATCTA-3′ and reverse 5′-GCAGGAACATTAGTATATCTTC-3′; Forward 5′-GCGGTGATATCCATCTATTC-3′ and reverse 5′-CCACTTACTTGTCCTACTAAC-3′; Forward 5′-ACTACTCTCAGACAAGACAG-3′ and reverse 5′-CTTTCCTTCTATTACTATACG-3′; Forward 5′-GGAGATGGTTGGATATTAGG-3′ and reverse 5′-GGACCAGCAGTTGTAGATA-3′; Forward 5′-AGATTTTAAATATGATCCTAACC-3′ and reverse 5′-CAATACCCTTCACCAAATACTC-3′ | NA | NA | NA | [74] |
| Multiplex Real-Time | Meat (PND) | Salmonella spp.: invA; Escherichia coli O157: rfbE; Listeria monocytogenes: hlyA gene | Forward 5′-GTTGAGGATGTTATTCGCAAAGG-3′ and reverse 5′-GGAGGCTTCCGGGTCAAG-3′; Forward 5′-TGTTCCAACACTGACATATATAGCATCA-3′ and reverse 5′-TGCCAAGTTTCATTATCTGAATCAA-3′; Forward 5′-ACTGAAGCAAAGGATGCATCTG-3′ 3′ and reverse 5′-TTTTCGATTGGCGTCTTAGGA-3′ | 5′-CCGTCAGACCTCTGGCAGTACCTTCCTC-3′; 5′-ATGCTATAAAATACACAGGAGCCACCCCCA-3′; 5′-CACCACCAGCATCTCCGCCTGC-3′ | TaqMan probes labelled with fluorescent dyes CAL Fluor Orange 560, Quasar 670, Fluorescein amidite (FAM), and 5-Carboxytetramethylrhodamine (TAMRA), respectively | NA | [75] |
| Real-Time | Meat (PND) | Spiked Listeria monocytogenes: ilyA gene | Forward 5′-GGCTTTCAGCTGGGCATAACCAA-3′ and reverse 5′-GCGGTCAGTGTAAAAAGTGGCACA-3′ | NA | Brilliant SYBR Green QPCR Master Mix | 1 cfu/g | [76] |
| Simple | Meat (Breasts, drumsticks, legs) | Arcobacter spp.: 16S rRNA | Forward 5′-AGAACGGGTTATAGCTTGCTAT-3′ and reverse 5′-GATACAATACAGGCTAATCTCT-3′ | NA | NA | NA | [77] |
| Real-Time Quantitative | Meat (Breasts, wings, legs) | Campylobacter jejuni: rpoB gene | Forward 5′-GAGTAAGCTTGGTAAGATTAAAG-3′ and reverse 5′-AAGAAGTTTTAGAGTTTCTCC-3′ | NA | FluoCycle SYBR GreenMix | 10 cfu/g | [78] |
| Simple | Meat (PND) | Arcobacter, butzleri: 16S rRNA, A. cryaerophilus, A. skirrowii, A. cibarius, gyrA gene | Forward 5′-AGTTGTTGTGAGGCTCCAC-3′ and reverse 5′-GCAGACACTAATCTATCTCTAAATCA-3′; Forward 5′-TGCTAAAATTGCAGATGTACCA-3′; and reverse 5′- AATTCCTTTTTCAGAAACTGTACG-3′; Forward 5′- GAGACAACTTTTGGAACTATTCTATGA-3′ and reverse 5′-GAAGATAGATTAACTTTTGCTTGTTG-3′; Forward 5′- TGGAAATATTGTTGGTGAAGTTCAG-3′ and reverse 5′- ATCTACATTTACAATACTTACTCCCGAA-3′ | NA | NA | NA | [79] |
| Multiplex | Meat (PND) | Spiked Salmonella spp. invA, sdf, STM4492 genes | Forward 5′-AAA CGT TGA AAA ACT GAG GA-3′ and reverse 5′-TCG TCA TTC CAT TAC CTA CC-3′; Forward 5′-AAA TGT GTT TTA TCT GAT GCA AGA GG-3′ and reverse 5′-GTT CGT TCT TCT GGT ACT TAC GAT GAC-3′; Forward 5′-ACA GCT TGG CCT ACG CGA G-3′ and reverse 5′-AGC AAC CGT TCG GCC TGA C-3′ | NA | NA | 105 cfu/mL | [80] |
| Multiplex Real-Time | Meat (Skin) | Spiked Salmonella spp.: invA, Campylobacter spp.: 16S rRNA | Forward 5′-TCGTCATTCCATTACCTACC-3′ and reverse 5′-AAACGTTGAAAAACTGAGGA-3′; Forward 5′-CTGCTTAACACAAGTTGAGTAGG-3′ and reverse 5′-TTCCTTAGGTACCGTCAGAA-3′ | 5′-TCTGGTTGATTTCCTGATCGCA-3′; 5;′- TGTCATCCTCCACGCGGCGTTGCTGC-3′ | Cyanines, Fluorescein amidite and VIC fluorophores | 1 and 106 cfu/mL | [81] |
| Real-Time Loop-mediated isothermal amplification | Meat (PND) | Spiked Salmonella spp.: gene62181533 | Forward 5′-TGA TACTGT GTC TGC GTC CC-3′ and reverse 5′-CGG AGC GGA TAAACG GAG TT-3′ | NA | NA | 7 cfu/mL | [82] |
| Multiplex Real-Time | Meat (Skin) | Spiked Salmonella spp.: invA, Campylobacter spp.: 16S rRNA | Forward 5′-TCGTCATTCCATTACCTACC-3′ and reverse 5′-AAACGTTGAAAAACTGAGGA-3′; Forward 5′-CTGCTTAACACAAGTTGAGTAGG-3′ and reverse 5′-TTCCTTAGGTACCGTCAGAA-3′ | 5′-TCTGGTTGATTTCCTGATCGCA-3′; 5′-TGTCATCCTCCACGCGGCGTTGCTGC-3′ | Labeled with Fluorescein amidite (FAM), Cyanines, and VIC fluorophores | 1; 102 cfu/mL | [83] |
| Sample Used | Target Site of Bacterial Pathogen | Probe | Array Matrix | Limit of Detection | Reference |
|---|---|---|---|---|---|
| Meat (Breast, wings, thighs) | Spiked Salmonella spp.: fimY, Shigella spp.: ipaH, Listeria monocytogenes: prfA, Escherichia coli: uspA genes | FY5′-GCCTCAATACAGGAGACAGGTAGCGCC-3′; 5′-ATATCGCTTTGTTGCCAACTGAGCGC-3′; 5′-AAATAAGTAGTGACTCAATGAATAGCCGAG-3′; 5′-AGTTGTAATTATTGCCTGAGAAATGATAC-3′, IH5′-GGGAGTGACAGCAAATGACCTCCGC-3′; 5′-CGGCACTGGTTCTCCCTCTGGGGACCA-3′; 5′-TGTGGATGAGATAGAAGTCTACCTGG-3′; 5′-AGAATGAGTACTCTCAGAGGGTGGCTGAC-3′; 5′-AGAAACTTCAGCTCTCCACTGCCGTGA-3′, PA5′-ACGGGAAGCTTGGCTCTATTTTGCGG-3′; 5′-AGCTTACAAGTATTAGCGAGAACGGGACCA-3′; 5′-ACAAAGGTGCTTTCGTTATAATGTCTGGCT-3′; 5′-AATTTAGAAGTCATTAGCGAACAGGCT-3′; 5′-CATACAGCCTAGCTAAATTTAATGAT-3′; 5′-AAACATCGGTTGGCTATTATAAGTTTAG-3′, UA5′-AAGAGACACATCATGCGCTGACCGAGCT-3; 5′-GGTAGAGAAAGCAGTCTCTATGGCTCGCCC-3′; 5′-ACCGTTCACGTTGATATGCTGATTGTTCCG-3′; 5′-TTGTTTATCTAACGAGTAAGCAAG-3′; 5′-AAGGTAAGGATGGTCTTAACACTGAAT-3′; 5′-GGTGACGTAACGGCACAAGAAACGCTAGCT-3′ | Nylon membrane | 103 cfu/mL | [86] |
| Meat (Breast, wings, thighs) | Spiked Salmonella serotype enteritidis: fimY, Listeria monocytogenes: prfA, Shigella boydii: ipaH genes | FY5′-GCCTCAATACAGGAGACAGGTAGCGCC-3′; 5′-ATATCGCTTTGTTGCCAACTGAGCGC-3′; 5′-AAATAAGTAGTGACTCAATGAATAGCCGAG-3′; 5′-AGTTGTAATTATTGCCTGAGAAATGATAC-3′, PA5′-ACGGGAAGCTTGGCTCTATTTTGCGG-3′; 5′-AGCTTACAAGTATTAGCGAGAACGGGACCA-3′; 5′-ACAAAGGTGCTTTCGTTATAATGTCTGGCT-3′; 5′-AATTTAGAAGTCATTAGCGAACAGGCT-3′; 5′-AAACATCGGTTGGCTATTATAAGTTTAG-3′, IH5′-GGGAGTGACAGCAAATGACCTCCGC-3′; 5′-CGGCACTGGTTCTCCCTCTGGGGACCA-3′; 5′-TGTGGATGAGATAGAAGTCTACCTGG-3′; 5′-AGAATGAGTACTCTCAGAGGGTGGCTGAC-3′; 5′-AGAAACTTCAGCTCTCCACTGCCGTGA-3′ | Nylon membrane | 104–106 cfu/mL | [87] |
| Meat (PND) | Spiked Salmonella enteritidis: sdf, Salmonella typhimurium: STM4497 gene, Campylobacter jejuni: hipO, Campylobacter coli: ceuE gene | Btn-TG-T10-AATCAGCCTGTTGTCTGCTCACCATTC-3′; Btn-TG-T10-AGATCATCGTCGACATGCTCAC-3′, Btn-TG-T10-CATTGCGAGATACTATGCTTTG-3′, Btn-TG-T10-CTGTAAGTATTTTGGCAAGTTT-3′ | DVD chips | 0.2 pg genomic DNA | [88] |
| Type of ELISA | Sample Used | Target Site of Bacterial Pathogen | Sensitivity | Limit of Detection | Reference |
|---|---|---|---|---|---|
| Indirect | Meat (Thighs, legs) | Outer membrane protein of Salmonella enterica serovar Enteritidis | 94% | NA | [91] |
| Sandwich | Spiked wings | Salmonella spp. | 75% | 1.6 × 103 cfu/mL | [92] |
| Sandwich | Meat (PND) | Campylobacter spp. | ND | NA | [93] |
| Sandwich | Spiked meat (PND) and naturally contaminated | Salmonella spp. | ND | 5 × 103 cfu/mL | [94] |
| Sample Used | Target Site of Bacterial Pathogen | Aptamer Name and Sequence | Detection Format | Limit of Detection | Reference |
|---|---|---|---|---|---|
| Spiked meat | Listeria monocytogenes: InlA gene | A8, 5′-ATC CAT GGG GCG GAGATG AGG GGG AGG AGG GCG GGT ACC CGG TTGAT-3′, A610.2, 5′- GGT TACTGA AGC ATA TGT CCG GGG GAT TGC CAA GCCTTC CC-3′ | Sandwich ELISA | 103 cfu/mL | [104] |
| Spiked meat | Whole-cell of Salmonella enterica serovar Typhimurium | ND, 5′-TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG-3′ | Electrochemical | 101 cfu/mL | [105] |
| Spiked meat (Breast) | Whole cell of Salmonella typhimurium | ND, 5′-NH2-ATAGGAGTCACGACGACCAGAAAGTAATGCCCGGTAGTTATTCAAAGATGAGTAGGAAAAGATATGTGCGTCTACCTCTTGACTAAT-3′ | FRET | 35 cfu/mL | [106] |
| Spiked cooked meat | Whole-cell of Streptococcus pyogenes | S2, 5′-GTTCGGGGTCGGGGTGAGTGGGGCCTAGGAGTGGGGGCGC-3′, S8, 5′-ATGGGGGGCGGGGAGGTGGGTACAGGGTCGGGGATGGCAG-3′, S10, 5′-CGGGCGGGGCGTGGGGTGTTGGAGTGGAGGGCGGGGCGGC-3′, S12, 5′-GCGGGCGGGGGGAGGGCGGCCGTGGGCTGCGAGTGGGAGG-3′, S15, 5′-CAGGGTGCGGGAGGGCCAAAGGGGGGAGGGCCCGGGGGGA-3′ | FRET | 70 cfu/mL | [107] |
| Spiked chicken | E. coli O157: H7 | F1N, 5′-ATAGGAGTCACGACGACCAGAA, R1N, ATTAGTCAAGAGGTAGACGCACATA, Bio Rev, 5Biosg/ATTAGTCAAGAGGTAGACGCACATA | Quantum dots | 102 cfu/mL | [108] |
| Biosensor Type | Sensing Platform | Chicken Matrix | Pathogens/Toxins | Limit of Detection | Analysis Time | Reference |
|---|---|---|---|---|---|---|
| Phage magnetoelastic | Gold electrode | Boneless and skinless breast fillets | Spiked Salmonella enterica serovar Typhimurium | 7.86 × 103 cfu/mm2 | 2–10 min | [116] |
| Multiplex fiber optic | Polystyrene waveguides | Breast | Spiked Listeria monocytogenes, Escherichia coli O157:H7, Salmonella enterica | 103 cfu/mL | <24 h | [117] |
| Fiber optic | Polystyrene waveguides | Breast | Spiked Salmonella enteritidis | 102 cfu/mL | <8 h | [118] |
| Colorimetric | C2 reverse-phase silica gel plates with sensitive dyes | Breast fillets | Spiked Pseudomonas gessardii, Pseudomonas psychrophila, Pseudomonas fragi, Pseudomonas fluorescens | NA | ND | [119] |
| Dithiobis-succinimidyl propionate-modifiedimmunosensor | Gold electrode | Skin | Spiked Listeria monocytogene | 103 cfu/25 g | 45 min | [120] |
| Amperometric | Screen-printed electrode | NS | Salmonella pullorum | 100 cfu/mL | 1.5–2 h | [121] |
| Optical scattering | SELA plates | Breast | Spiked Listeria monocytogenes, Escherichia coli O157: H7, Salmonella enteritidis | ND | 29–40 h | [122] |
| Colorimetric | Aptamer | NS | Campylobacter coli, Campylobacter jejuni | 7.2 × 105 cfu/mL | 30 min | [123] |
| Meat Sample | Method Type | Types of Antibiotics Residue | Microbial Test Strains | Reference |
|---|---|---|---|---|
| Muscles, kidney, liver, gizzard | Three-Plate test | Tetracycline, β-lactams, sulphonamides, aminoglycosides | Bacillus subtilis | [143] |
| Spiked liver, kidney, breast, thigh muscle, skin | ND | Enrofloxacin, ciprofloxacin, oxytetracycline | Geobacillus stearothermophilus | [144] |
| Breast, liver, thigh tissue | Four-Plate test | Tetracycline | Bacillus subtilis | [145] |
| Breast | Four-Plate test | Tetracycline, β-lactams, sulphonamides, aminoglycosides | Bacillus subtilis, Micrococcus luteus | [146] |
| Fillet | ND | Oxytetracycline, enrofloxacin | Bacillus subtilis | [147] |
| Breast, thighs | Four-Plate test | Tetracycline, β-lactams, sulphonamides, aminoglycosides, macrolides, quinolones | Bacillus subtlis spore, Micrococcus luteus, Escherichia coli | [148] |
| Liver, kidney, muscle | Four-Plate test | Chloramphenicol | Bacillus subtilis, Staphylococcus aureus | [149] |
| Type of ELISA | Sample Used | Target Antibiotic | Limit of Detection | Reference |
|---|---|---|---|---|
| Competitive | Breast, liver, thigh tissue | Tetracycline | 0.05 µg/Kg | [145] |
| Competitive | Liver, kidney | Ciprofloxacin, streptomycin, sulphanilamide, tetracycline | 10 ppb | [150] |
| ND | Breast | Enrofl oxacin, ciprofloxacin, streptomycin, chloramphenicol | ND | [153] |
| Competitive | Breast | Tetracycline, streptomycin, chloramphenicol, sulfamethazine | ND | [151] |
| ND | Breast | Tetracycline | ND | [154] |
| Indirect competitive | Spiked muscles | Chloramphenicol | 6 ng/L | [152] |
| ND | Muscles, liver, kidney | Gentamicin | 0.05 µg/Kg | [155] |
| Competitive | Breast | Quinolone | 0.05 µg/Kg | [28] |
| Competitive | Breast | Quinolone | 0.05 µg/Kg | [156] |
| Sample Used | Stationary Phase | Mobile Phase | Target Antibiotic | Reference |
|---|---|---|---|---|
| Breast, thigh muscle, liver | Silica | Acetone and Methanol: 1:1 | Ciprofloxacin, enrofloxacin, oxytetracycline, doxycycline, amoxicillin | [159] |
| Liver | Silica | Acetone and Methanol: 1:1 | ND | [160] |
| Liver, kidney | Silica | Acetone and Methanol: 1:1 | Sulphanilamide, streptomycin, ciprofloxacin, tetracycline | [150] |
| Oral administration of chicken blood | Silica | Acetone and Methanol: 1:1 | Ciprofloxacin | [161] |
| Spiked muscles | Silica | Chloroform and n-Butanol: 90:10 | Sulfadiazine, sulfadoxine, sulfamethazine, sulfathiazole, sulfaquinoxaline | [162] |
| MPND | Silica | Acetone and Methanol: 1:1 | Doxycycline, oxytetracycline | [163] |
| Breast, thigh muscle, liver, kidney | Silica | Acetone and Methanol: 1:1 | Doxycycline | [164] |
| Sample Used | Types of Antibiotic | Method Used | Chromatography Conditions Used | Limit of Detection | Reference | |||
|---|---|---|---|---|---|---|---|---|
| Model | Column | Solvent | Flow Rate | |||||
| Breast, liver, thigh tissue | Tetracycline | HPLC-UV | KNAUER liquid chromatography system, Berlin, Germany | Eurospher RP-C18 column (250 × 4.6 mm i.d.). A guard column (Eurospher 100-5 C18) was used to protect the analytical column | Mobile phase was a gradient elution using MeOH; acetonitrile; 0.03 M oxalic acid buffer pH 2.5; water | 0.9 mL/min | 25 µg/Kg | [145] |
| Spiked breast, thigh, liver, kidney | Oxytetracycline, tetracycline | HPLC-UV | HPLC (Shimadzu Corporation, Tokyo, Japan) | Inertsil ODS-3 column | Mobile phase consisting of methanol:acetonitrile: 0.01 M oxalic acid dihydrate (5:18:77 v/v/v) | 1 mL/min | 50 ng/mL | [168] |
| Spiked meat, liver | Oxytetracycline, tetracycline, chlortetracycline, doxycycline | HPLC-DAD | The HPLC system of a HP 1100 chromatograph (Agilent Technologies, Palo Alto, CA, USA) | The analytical column was reversed-phase (Nuclosil 100 C18, 25 cm × 4.6 mm I.D., 5 µm, Germany) | The mixture of acetonitrile/0.03 M oxalic acid (40:60, v/v); The mixture of methanol/acetonitrile/0.03 M oxalic acid (10:30:60, v/v/v); The mixture of methanol/acetonitrile/0.03 M oxalic acid (20:20:60, v/v/v) | 1. 1 mL/min; 2. 1 mL/min; 3. 1 mL/min; 4. NS; 5. NS | 4.4, 5, 13 and 10 ng/g | [169] |
| Spiked muscle | Marbofloxacin, ciprofloxacin, norfloxacin, lomefloxacin, danofloxacin, enrofloxacin, sarafloxacin, difloxacin, oxolinic acid, flumequine | HPLC-FAD | HPLC system (Waters, Milford, MA, USA) | The reverse phase analytical column was a Symmetry C18 (250 mm × 4.5 mm i.d., 5 µ m) from Waters | Mobile phase consisted of aqueous formic acid solution (0.02%, pH 2.8) and acetonitrile | 1.0 mL/min | 0.3–1.0 ng/g | [170] |
| Spiked muscle | Ofloxacin, norfloxacin, ciprofloxacin, enrofloxacin, oxytetracycline, tetracycline, chlortetracycline, doxycycline, sulfadiazine, sulfamethazine, sulfadimethoxydiazine, sulfamonomethoxine, sulfamethoxazole, sulfaquinoxaline | UPLC-MS-MS | UPLC system (Waters, Milford, MA, USA) | UPLC BEH C18 column(50 mm 9 2.1 mm i.d., 1.7 lm) from Waters | Mobile phase, consisting of methanol (solvent A) and 0.01% formic acid in water (solvent B) | 0.3 mL/min | 0.3 µg/Kg | [171] |
| Spiked muscle, liver, kidney | Amoxicillin, amoxicillin metabolites, ampicillin | UPLC-MS-MS | UPLC system (Waters, Milford, MA, USA) | UPLC HSS T3 column (100 × 2.1 mm, internal diameter (i.d.) 1.8 μm) | A (0.15% formic acid) and B (acetonitrile) | 0.5 mL/min | 0.01–1.36 µg/Kg | [172] |
| Biosensor Type | Sensing Platform | Chicken Matrix | Antibiotic | Limit of Detection | Analysis Time | Reference |
|---|---|---|---|---|---|---|
| Bioluminescent biosensor | Bacteria E. coli K12 | Spiked breast fillet | Tetracycline | 100 ng/g | 4 h | [173] |
| Electrochemical | Gold and platinum nanowire | Spiked breast | Penicillin and tetracycline | 41.2 μA μM−1 cm−2 and 26.4 μA μM−1 cm−2 | ND | [175] |
| Electrochemical | Glassy carbon electrode | PND | Tetracycline | 0.10 µM | ND | [176] |
| Electrochemical | Pencil graphite electrode | Spiked PND | Sulfadimethoxine | 3.7 × 10−16 M | ND | [177] |
| Amperometric | Glassy carbon electrode | PND | Chloramphenicol | 45 pg/mL | ND | [178] |
| Surface plasmon resonance | NS | Spiked muscle | Chloramphenicol and chloramphenicol glucuronide | ND | ND | [179] |
| Bioluminescent biosensor bacteria | Bacteria E. coli | Spiked muscle | Tetracycline | ND | ND | [175] |
| Surface plasmon resonance | NS | Spiked breast | Norfloxacin, sarafloxacin, difloxacin, ciprofloxacin, enrofloxacin, flumequine, danofloxacin, marbofloxacin, pefloxacin, enoxacin, lomefloxacin, ofloxacin, oxolinic acid | ND | ND | [180] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, H.; Bhardwaj, K.; Kaur, T.; Nepovimova, E.; Kuča, K.; Kumar, V.; Bhatia, S.K.; Dhanjal, D.S.; Chopra, C.; Singh, R.; et al. Detection of Bacterial Pathogens and Antibiotic Residues in Chicken Meat: A Review. Foods 2020, 9, 1504. https://doi.org/10.3390/foods9101504
Kumar H, Bhardwaj K, Kaur T, Nepovimova E, Kuča K, Kumar V, Bhatia SK, Dhanjal DS, Chopra C, Singh R, et al. Detection of Bacterial Pathogens and Antibiotic Residues in Chicken Meat: A Review. Foods. 2020; 9(10):1504. https://doi.org/10.3390/foods9101504
Chicago/Turabian StyleKumar, Harsh, Kanchan Bhardwaj, Talwinder Kaur, Eugenie Nepovimova, Kamil Kuča, Vinod Kumar, Shashi Kant Bhatia, Daljeet Singh Dhanjal, Chirag Chopra, Reena Singh, and et al. 2020. "Detection of Bacterial Pathogens and Antibiotic Residues in Chicken Meat: A Review" Foods 9, no. 10: 1504. https://doi.org/10.3390/foods9101504
APA StyleKumar, H., Bhardwaj, K., Kaur, T., Nepovimova, E., Kuča, K., Kumar, V., Bhatia, S. K., Dhanjal, D. S., Chopra, C., Singh, R., Guleria, S., Bhalla, T. C., Verma, R., & Kumar, D. (2020). Detection of Bacterial Pathogens and Antibiotic Residues in Chicken Meat: A Review. Foods, 9(10), 1504. https://doi.org/10.3390/foods9101504