The BDNF–TrkB–CREB Signalling Pathway Is Involved in Bisphenol S-Induced Neurotoxicity in Male Mice by Regulating Methylation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
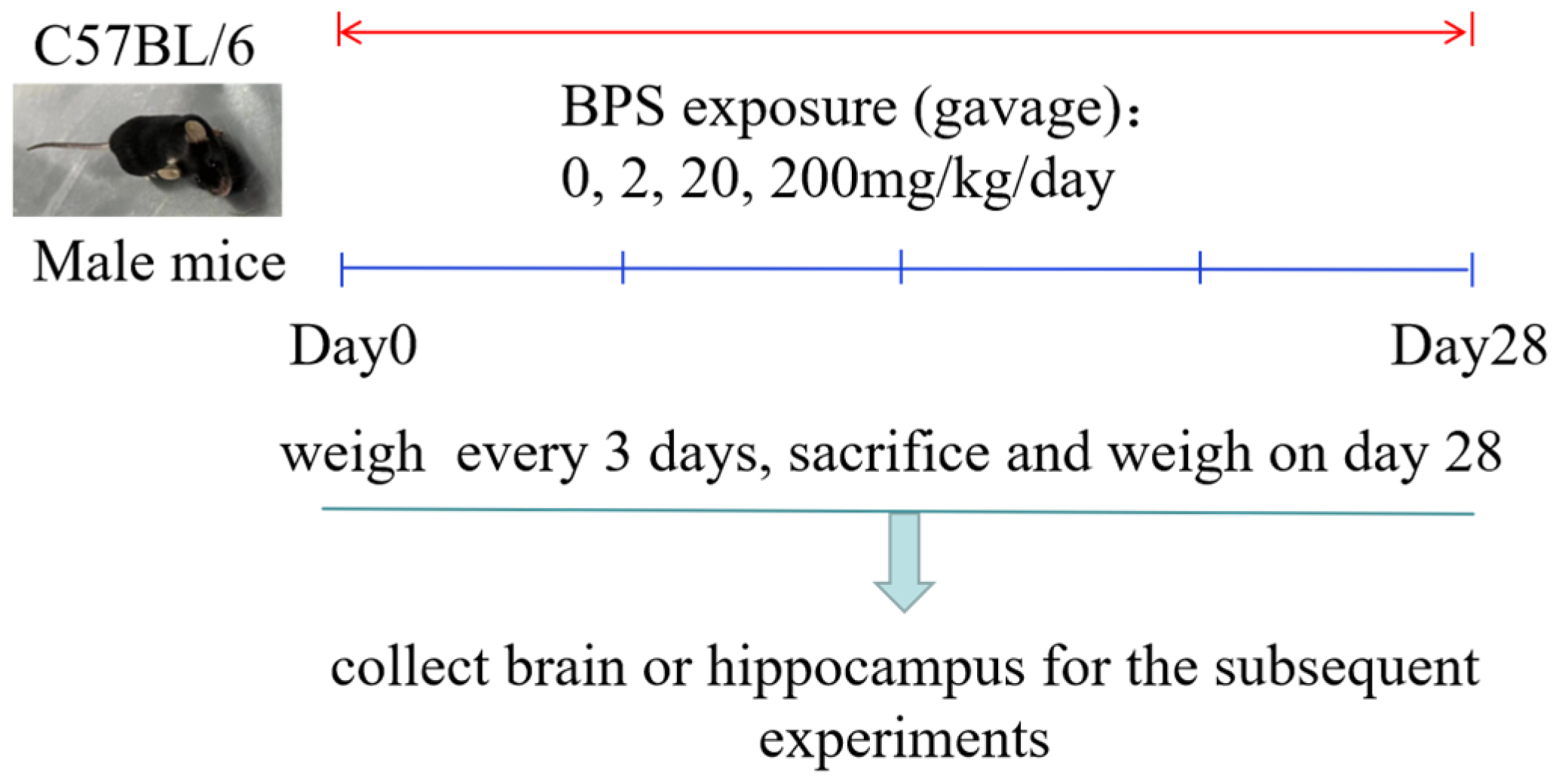
2.2. Animals and Treatment
2.3. Haematoxylin and Eosin (HE) Staining
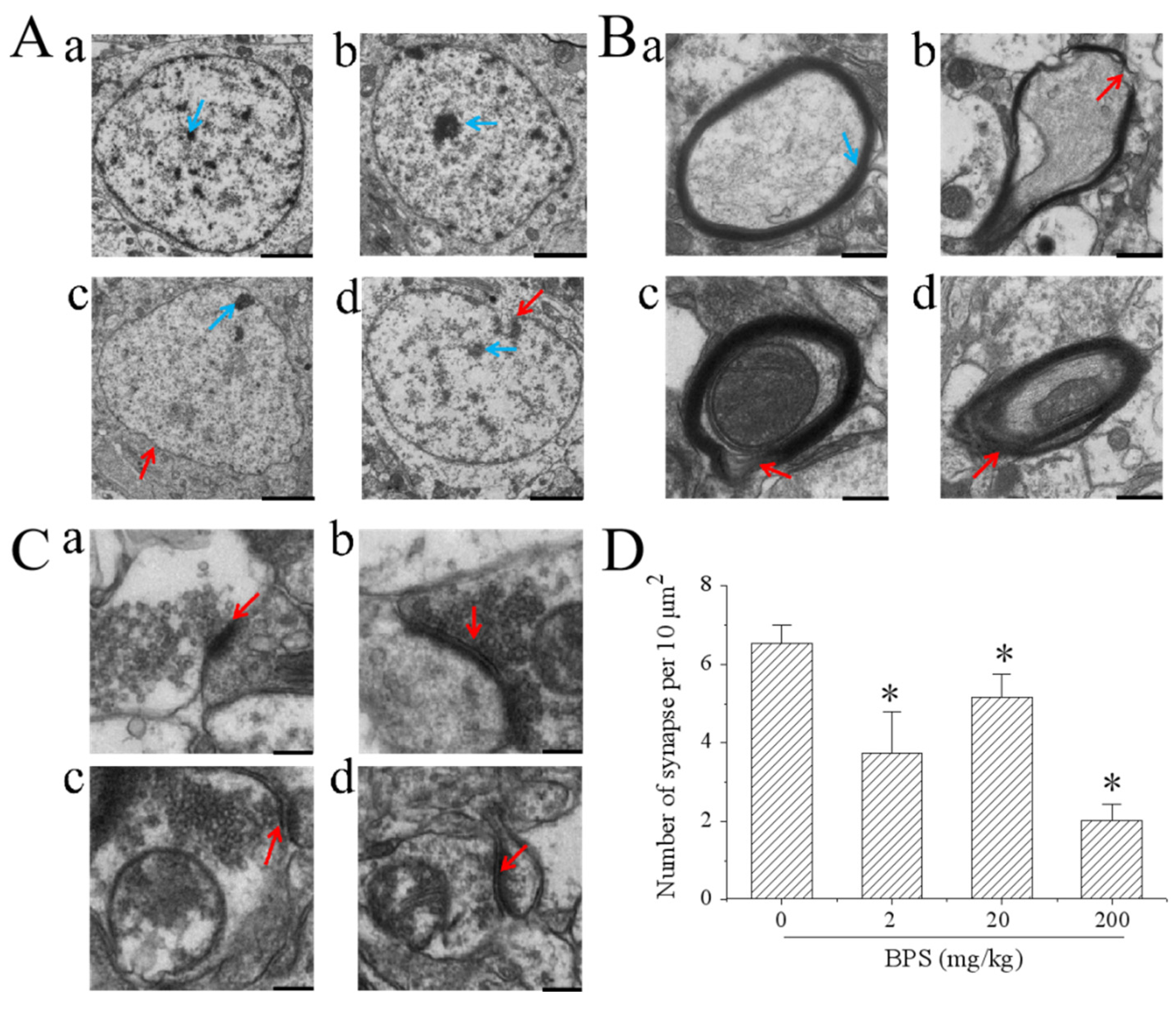
2.4. Transmission Electron Microscopy (TEM)
2.5. Real-Time Quantitative PCR (qRT-PCR)
2.6. Western Blotting
2.7. Bisulfite Sequencing PCR
2.8. Statistical Analysis
3. Results
3.1. The Effect of BPS on Mouse Body and Brain Weight
3.2. The Effect of BPS on Hippocampal Histopathology
3.3. The Effect of BPS on Mouse Hippocampal Ultrastructure
3.4. The Effect of the BDNF–TrkB–CREB Pathway on BPS-Induced Neurotoxicity
3.5. The Effect of BPS on BDNF Promoter IV Methylation in Mouse Hippocampus
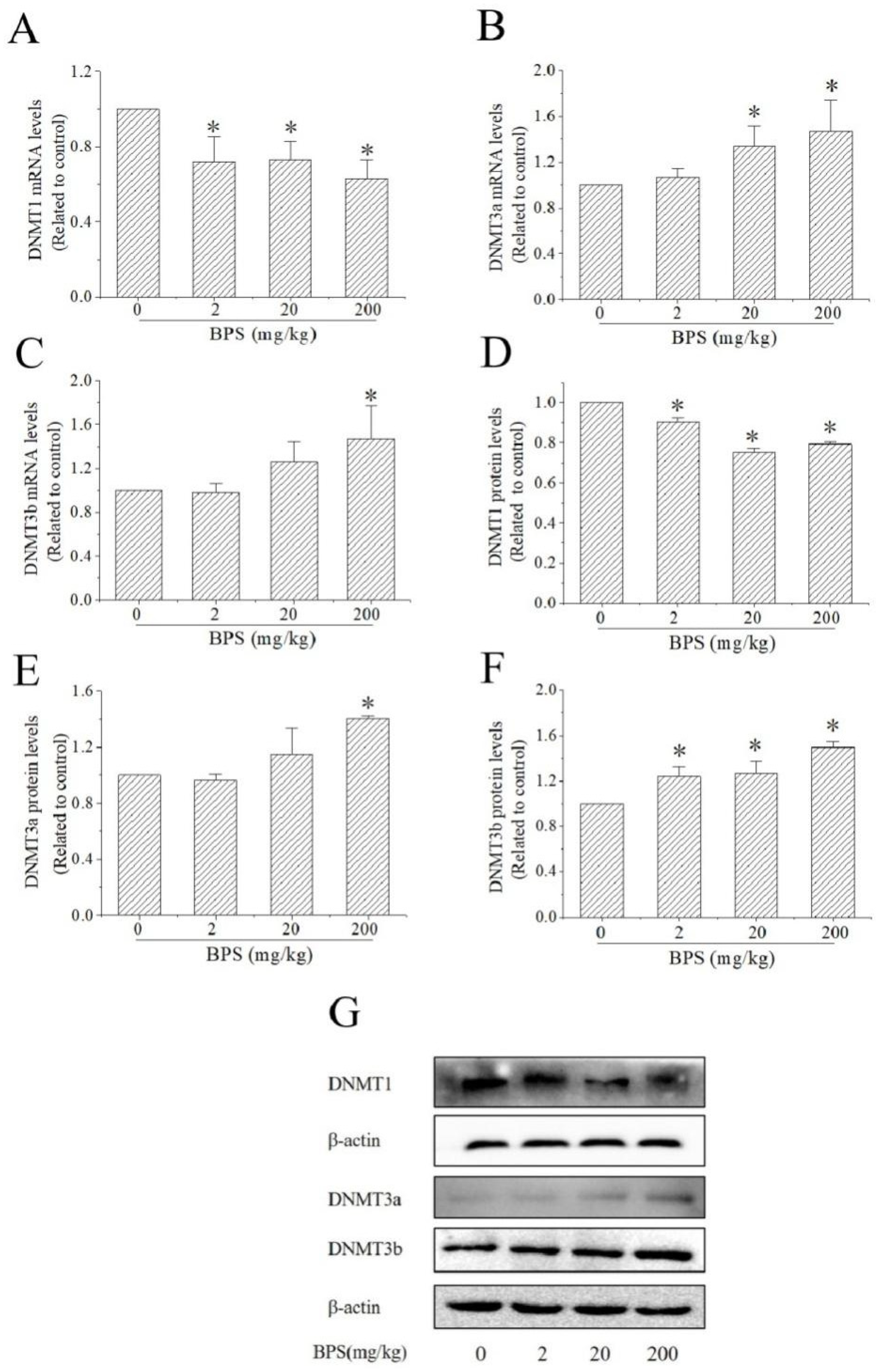
3.6. The Effects of BPS on DNMT Expression in Mouse Hippocampus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cimmino, I.; Fiory, F.; Perruolo, G.; Miele, C.; Beguinot, F.; Formisano, P.; Oriente, F. Potential Mechanisms of Bisphenol A (BPA) Contributing to Human Disease. Int. J. Mol. Sci. 2020, 21, 5761. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Rocha, L.; Ribeiro-Gonçalves, L.; Henriques, B.; Özcan, M.; Tiritan, M.E.; Souza, J.C.M. An integrative review on the toxicity of Bisphenol A (BPA) released from resin composites used in dentistry. J. Biomed. Mater. Res. Part B Appl. Biomater. 2021, 109, 1942–1952. [Google Scholar] [CrossRef] [PubMed]
- Eladak, S.; Grisin, T.; Moison, D.; Guerquin, M.-J.; N’Tumba-Byn, T.; Pozzi-Gaudin, S.; Benachi, A.; Livera, G.; Rouiller-Fabre, V.; Habert, R. A new chapter in the bisphenol A story: Bisphenol S and bisphenol F are not safe alternatives to this compound. Fertil. Steril. 2014, 103, 11–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.; Xia, W.; Yang, S.; Pan, X.; He, Z.; Kannan, K. Spatial distribution of bisphenol S in surface water and human serum from Yangtze River watershed, China: Implications for exposure through drinking water. Chemosphere 2018, 199, 595–602. [Google Scholar] [CrossRef]
- Jin, H.; Zhu, J.; Chen, Z.; Zhongjian, C.; Cai, Z. Occurrence and Partitioning of Bisphenol Analogues in Adults’ Blood from China. Environ. Sci. Technol. 2017, 52, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Gayrard, V.; Lacroix, M.; Grandin, F.C.; Collet, S.H.; Mila, H.; Viguié, C.; Gély, C.A.; Rabozzi, B.; Bouchard, M.; Léandri, R.; et al. Oral Systemic Bioavailability of Bisphenol A and Bisphenol S in Pigs. Environ. Health Perspect. 2019, 127, 077005. [Google Scholar] [CrossRef]
- Russo, G.; Barbato, F.; Cardone, E.; Fattore, M.; Albrizio, S.; Grumetto, L. Bisphenol A and Bisphenol S release in milk under household conditions from baby bottles marketed in Italy. J. Environ. Sci. Health Part B 2017, 53, 116–120. [Google Scholar] [CrossRef]
- Fouyet, S.; Olivier, E.; Leproux, P.; Dutot, M.; Rat, P. Bisphenol A, Bisphenol F, and Bisphenol S: The Bad and the Ugly. Where Is the Good? Life 2021, 11, 314. [Google Scholar] [CrossRef]
- da Silva, B.S.; Pietrobon, C.B.; Bertasso, I.M.; Lopes, B.P.; Carvalho, J.C.; Peixoto-Silva, N.; Santos, T.R.; Claudio-Neto, S.; Manhães, A.C.; Oliveira, E.; et al. Short and long-term effects of bisphenol S (BPS) exposure during pregnancy and lactation on plasma lipids, hormones, and behavior in rats. Environ. Pollut. 2019, 250, 312–322. [Google Scholar] [CrossRef]
- Zhang, Z.; Lin, L.; Gai, Y.; Hong, Y.; Li, L.; Weng, L. Subchronic bisphenol S exposure affects liver function in mice involving oxidative damage. Regul. Toxicol. Pharmacol. 2018, 92, 138–144. [Google Scholar] [CrossRef]
- Ji, K.; Hong, S.; Kho, Y.; Choi, K. Effects of Bisphenol S Exposure on Endocrine Functions and Reproduction of Zebrafish. Environ. Sci. Technol. 2013, 47, 8793–8800. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Sekulovski, N.; MacLean, J.A.; Hayashi, K. Effects of bisphenol A analogues on reproductive functions in mice. Reprod. Toxicol. 2017, 73, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, J.; Xu, T.; Han, H.; Zhu, Z.; Meng, L.; Pang, Q.; Fan, R. Bisphenol A(BPA), BPS and BPB-induced oxidative stress and apoptosis mediated by mitochondria in human neuroblastoma cell lines. Ecotoxicol. Environ. Saf. 2020, 207, 111299. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Zhu, B.; Xu, X.; Zeng, H. Role of the BDNF/TrkB/CREB signaling pathway in the cytotoxicity of bisphenol S in SK-N-SH cells. J. Biochem. Mol. Toxicol. 2021, 35, 1–11. [Google Scholar] [CrossRef]
- Catanese, M.; Vandenberg, L.N. Bisphenol S (BPS) alters maternal behavior and brain in mice exposed during pregnancy/lactation and their daughters. Endocrinology 2016, 158, 516–530. [Google Scholar] [CrossRef] [Green Version]
- Tucker, D.K.; Bouknight, S.H.; Brar, S.S.; Kissling, G.E.; Fenton, S.E. Evaluation of Prenatal Exposure to Bisphenol Analogues on Development and Long-Term Health of the Mammary Gland in Female Mice. Environ. Health Perspect. 2018, 126, 087003. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Li, J.; Xu, S.; Zhou, Y.; Zhao, H.; Li, Y.; Xiong, C.; Sun, X.; Liu, H.; Liu, W.; et al. Prenatal exposure to bisphenol A and its alternatives and child neurodevelopment at 2 years. J. Hazard. Mater. 2019, 388, 121774. [Google Scholar] [CrossRef]
- Salahinejad, A.; Attaran, A.; Naderi, M.; Meuthen, D.; Niyogi, S.; Chivers, D.P. Chronic exposure to bisphenol S induces oxidative stress, abnormal anxiety, and fear responses in adult zebrafish (Danio rerio). Sci. Total Environ. 2020, 750, 141633. [Google Scholar] [CrossRef]
- Castro, B.; Sánchez, P.; Torres, J.M.; Ortega, E. Bisphenol A, bisphenol F and bisphenol S affect differently 5α-reductase expression and dopamine–serotonin systems in the prefrontal cortex of juvenile female rats. Environ. Res. 2015, 142, 281–287. [Google Scholar] [CrossRef]
- Mornagui, B.; Rezg, R.; Repond, C.; Pellerin, L. Effects of bisphenol S, a major substitute of bisphenol A, on neurobehavioral responses and cerebral monocarboxylate transporters expression in mice. Food Chem. Toxicol. 2019, 132, 110670. [Google Scholar] [CrossRef]
- Chen, F.; Zhou, C.-C.; Yang, Y.; Liu, J.-W.; Yan, C.-H. GM1 Ameliorates Lead-Induced Cognitive Deficits and Brain Damage Through Activating the SIRT1/CREB/BDNF Pathway in the Developing Male Rat Hippocampus. Biol. Trace Element Res. 2018, 190, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, X.; Gao, J.; Liu, Y.; Shi, J.; Gong, Q. Icariside II, a Phosphodiesterase-5 Inhibitor, Attenuates Beta-Amyloid-Induced Cognitive Deficits via BDNF/TrkB/CREB Signaling. Cell. Physiol. Biochem. 2018, 49, 1010–1025. [Google Scholar] [CrossRef] [PubMed]
- Hilakivi-Clarke, L. Maternal exposure to diethylstilbestrol during pregnancy and increased breast cancer risk in daughters. Breast Cancer Res. 2014, 16, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheong, A.; Johnson, S.A.; Howald, E.C.; Ellersieck, M.R.; Camacho, L.; Lewis, S.M.; VanLandingham, M.M.; Ying, J.; Ho, S.-M.; Rosenfeld, C.S. Gene expression and DNA methylation changes in the hypothalamus and hippocampus of adult rats developmentally exposed to bisphenol A or ethinyl estradiol: A CLARITY-BPA consortium study. Epigenetics 2018, 13, 704–720. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhao, C.; Zhong, H.; Zhang, S.; Xia, Y.; Cai, Z. Bisphenol S induced epigenetic and transcriptional changes in human breast cancer cell line MCF-7. Environ. Pollut. 2018, 246, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Roth, T.L.; Lubin, F.D.; Funk, A.J.; Sweatt, J.D. Lasting Epigenetic Influence of Early-Life Adversity on the BDNF Gene. Biol. Psychiatry 2009, 65, 760–769. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.-X.; He, Q.-Z.; Li, W.; Long, D.-X.; Pan, X.-Y.; Chen, C.; Zeng, H. Brain-Derived Neurotrophic Factor Mediated Perfluorooctane Sulfonate Induced-Neurotoxicity via Epigenetics Regulation in SK-N-SH Cells. Int. J. Mol. Sci. 2017, 18, 893. [Google Scholar] [CrossRef] [Green Version]
- Naderi, M.; Kwong, R.W. A comprehensive review of the neurobehavioral effects of bisphenol S and the mechanisms of action: New insights from in vitro and in vivo models. Environ. Int. 2020, 145, 106078. [Google Scholar] [CrossRef]
- Buuse, M.V.D.; Buret, L.; Hill, R. Involvement of brain-derived neurotrophic factor (BDNF) in the long-term memory effects of glucocorticoid stimulation during adolescence/young adulthood. Behav. Brain Res. 2019, 377, 112223. [Google Scholar] [CrossRef]
- Mersha, M.D.; Patel, B.M.; Patel, D.; Richardson, B.N.; Dhillon, H.S. Effects of BPA and BPS exposure limited to early embryogenesis persist to impair non-associative learning in adults. Behav. Brain Funct. 2015, 11, 27. [Google Scholar] [CrossRef] [Green Version]
- Teleanu, D.M.; Niculescu, A.-G.; Lungu, I.I.; Radu, C.I.; Vladâcenco, O.; Roza, E.; Costăchescu, B.; Grumezescu, A.M.; Teleanu, R.I. An Overview of Oxidative Stress, Neuroinflammation, and Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 5938. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.; Kanski, J.; Varadarajan, S.; Tsoras, M.; Butterfield, D.A. Elevation of brain glutathione by gamma-glutamylcysteine ethyl ester protects against peroxynitrite-induced oxidative stress. J. Neurosci. Res. 2002, 68, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Van de Vijver, K.I.; Holsbeek, L.; Das, K.; Blust, R.; Joiris, C.; De Coen, W. Occurrence of Perfluorooctane Sulfonate and Other Perfluorinated Alkylated Substances in Harbor Porpoises from the Black Sea. Environ. Sci. Technol. 2006, 41, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Zhang, Y.; Huang, L.; Xu, S.; Li, J.; Yang, L.; Wang, L.; Xing, C.; Wang, X.; Peng, Y. L-3-n-Butylphthalide Regulates Proliferation, Migration, and Differentiation of Neural Stem Cell In Vitro and Promotes Neurogenesis in APP/PS1 Mouse Model by Regulating BDNF/TrkB/CREB/Akt Pathway. Neurotox. Res. 2018, 34, 477–488. [Google Scholar] [CrossRef]
- Odaira, T.; Nakagawasai, O.; Takahashi, K.; Nemoto, W.; Sakuma, W.; Lin, J.-R.; Tan-No, K. Mechanisms underpinning AMP-activated protein kinase-related effects on behavior and hippocampal neurogenesis in an animal model of depression. Neuropharmacology 2019, 150, 121–133. [Google Scholar] [CrossRef]
- Yan, T.; Xu, M.; Wan, S.; Wang, M.; Wu, B.; Xiao, F.; Bi, K.; Jia, Y. Schisandra chinensis produces the antidepressant-like effects in repeated corticosterone-induced mice via the BDNF/TrkB/CREB signaling pathway. Psychiatry Res. 2016, 243, 135–142. [Google Scholar] [CrossRef]
- Kaur, G.; Rathod, S.S.S.; Ghoneim, M.M.; Alshehri, S.; Ahmad, J.; Mishra, A.; Alhakamy, N.A. DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders. Biology 2022, 11, 90. [Google Scholar] [CrossRef]
- Kundakovic, M.; Gudsnuk, K.; Herbstman, J.B.; Tang, D.; Perera, F.P.; Champagne, F.A. DNA methylation of BDNF as a biomarker of early-life adversity. Proc. Natl. Acad. Sci. USA 2014, 112, 6807–6813. [Google Scholar] [CrossRef] [Green Version]
- Toledo-Rodriguez, M.; Lotfipour, S.; Leonard, G.; Perron, M.; Richer, L.; Veillette, S.; Pausova, Z.; Paus, T. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010, 153B, 1350–1354. [Google Scholar] [CrossRef]
- Sui, L.; Wang, Y.; Ju, L.-H.; Chen, M. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiol. Learn. Mem. 2012, 97, 425–440. [Google Scholar] [CrossRef]
- Mishima, Y.; Brueckner, L.; Takahashi, S.; Kawakami, T.; Otani, J.; Shinohara, A.; Takeshita, K.; Garvilles, R.G.; Watanabe, M.; Sakai, N.; et al. Enhanced processivity of Dnmt1 by monoubiquitinated histone H3. Genes Cells 2019, 25, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.-X.; Koirala, P.; Zhang, W.; Ni, C.; Wang, Z.; Yang, L.; Mo, Y.-Y. lncRNA RMST Enhances DNMT3 Expression through Interaction with HuR. Mol. Ther. 2019, 28, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Chedin, F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog. Mol. Biol. Transl. Sci. 2011, 101, 255–285. [Google Scholar] [CrossRef] [PubMed]



| Gene | Primer | Product Length (bp) |
|---|---|---|
| BDNF | Forward: GCCCATGAAAGAAGTAAACGTCC | 136 |
| Reverse: AGTGTCAGCCAGTGATGTCGTC | ||
| TrkB | Forward: AACGGAGACTACACCCTGATGG | 251 |
| Reverse: GCAATCACCACCACGGCATA | ||
| CREB | Forward: TGGCTAACAATGGTACGGATGG | 195 |
| Reverse: GTGCTGTGCGGATCTGGTATGT | ||
| DNMT 1 | Forward: AATGGTGTTGTCTACCGACTGG | 158 |
| Reverse: TTGATGTAGTCAGAATACTTGCGG | ||
| DNMT 3a | Forward: TTGATGTAGTCAGAATACTTGCGG | 154 |
| Reverse: AAGCCAAACACCCTTTCCAT | ||
| DNMT 3b | Forward: CCTGCCCGCAAAGGTTTATA | 101 |
| Reverse: AATGGACGGTTGTCGCCCT | ||
| GAPDH | Forward: CCTCGTCCCGTAGACAAAATG | 133 |
| Reverse: TGAGGTCAATGAAGGGGTCGT |
| Group | n | Body Weight (g) | Body Weight Gain (g) | Brain Weight (g) | Brain Index (%) |
|---|---|---|---|---|---|
| Control | 10 | 21.83 ± 1.35 | 1.12 ± 0.86 | 0.437 ± 0.013 | 2.01 ± 0.09 |
| 2 mg/kg BPS | 10 | 22.10 ± 1.14 | 1.16 ± 0.87 | 0.436 ± 0.015 | 1.97 ± 0.11 |
| 20 mg/kg BPS | 10 | 22.18 ± 1.27 | 0.99 ± 0.66 | 0.443 ± 0.009 | 2.00 ± 0.10 |
| 200 mg/kg BPS | 10 | 21.65 ± 1.26 | 1.05 ± 0.68 | 0.427 ±0.009 | 1.98 ± 0.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-Z.; Wu, Z.-Y.; Zhu, B.-Q.; Wang, Y.-X.; Kan, Y.-Q.; Zeng, H.-C. The BDNF–TrkB–CREB Signalling Pathway Is Involved in Bisphenol S-Induced Neurotoxicity in Male Mice by Regulating Methylation. Toxics 2022, 10, 413. https://doi.org/10.3390/toxics10080413
Li Y-Z, Wu Z-Y, Zhu B-Q, Wang Y-X, Kan Y-Q, Zeng H-C. The BDNF–TrkB–CREB Signalling Pathway Is Involved in Bisphenol S-Induced Neurotoxicity in Male Mice by Regulating Methylation. Toxics. 2022; 10(8):413. https://doi.org/10.3390/toxics10080413
Chicago/Turabian StyleLi, Yi-Zhou, Zi-Yao Wu, Bi-Qi Zhu, Yu-Xiao Wang, Ya-Qi Kan, and Huai-Cai Zeng. 2022. "The BDNF–TrkB–CREB Signalling Pathway Is Involved in Bisphenol S-Induced Neurotoxicity in Male Mice by Regulating Methylation" Toxics 10, no. 8: 413. https://doi.org/10.3390/toxics10080413
APA StyleLi, Y.-Z., Wu, Z.-Y., Zhu, B.-Q., Wang, Y.-X., Kan, Y.-Q., & Zeng, H.-C. (2022). The BDNF–TrkB–CREB Signalling Pathway Is Involved in Bisphenol S-Induced Neurotoxicity in Male Mice by Regulating Methylation. Toxics, 10(8), 413. https://doi.org/10.3390/toxics10080413






