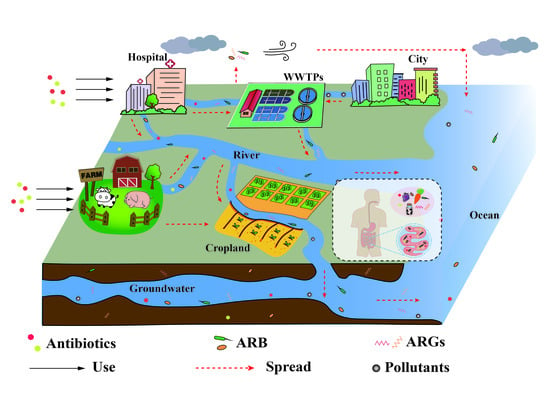
Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies
Abstract
:1. Introduction
2. The Usage and Fate of Antimicrobials in the Environment
2.1. Entry of Antimicrobials into the Environment
2.2. Effects of Antimicrobials on the Environment and Human Health
2.3. The Combined Effects of Environmental Pollutants and Antimicrobials
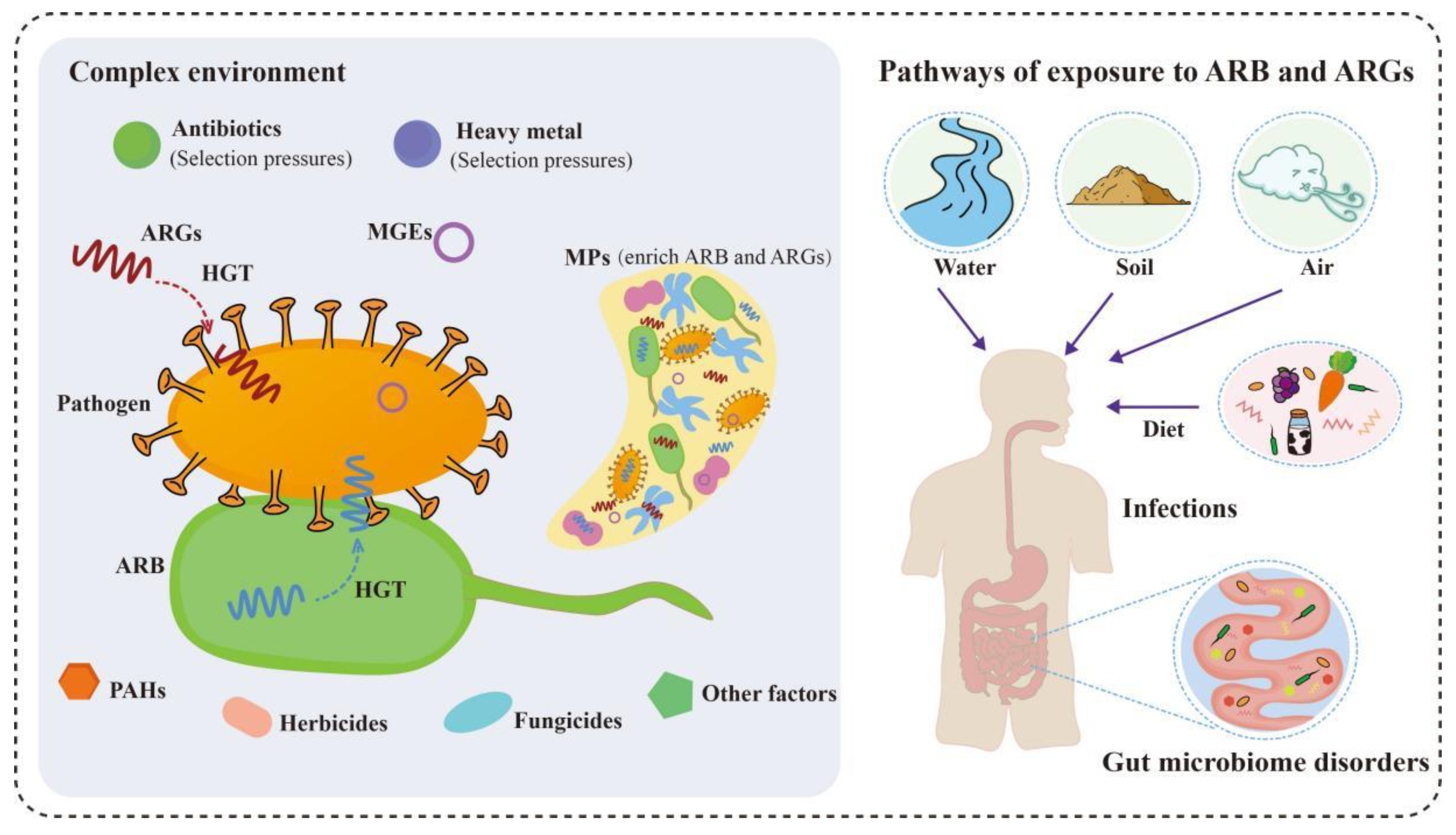
3. The Fate of Antimicrobial Resistance in the Environment
3.1. The Influence of Various Environmental Factors on Antimicrobial Resistance
3.1.1. Heavy Metals
3.1.2. MPs
3.1.3. Other Factors
3.2. The Potential Threat of Antimicrobial Resistance to Humans and the Ecological System
4. Methods to Mitigate Antimicrobial Contamination and Antimicrobial Resistance
4.1. AOP
4.2. CW
4.3. MFC
4.4. Other Technologies
5. Discussion and Prospects
- (1)
- To explore the possible human health risks caused by residual antimicrobials and ARGs in food, which is a topic that has been neglected and deserves attention;
- (2)
- Make an accurate and systematic risk assessment of antimicrobial and antimicrobial resistance based on environmental factors;
- (3)
- Develop fast and real-time monitoring means for antimicrobials and ARGs, and establish uniform standards for the effluent discharge of antimicrobials and ARGs;
- (4)
- Most of the research on the removal methods of antimicrobials and ARGs has been focused on the laboratory scale, which requires the establishment of practical application test sites. Data from the test site is often necessary and reliable;
- (5)
- To develop broad-spectrum, efficient, and safe antimicrobials and ARGs removal technologies is also indispensable.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bacanli, M.; Basaran, N. Importance of antibiotic residues in animal food. Food Chem. Toxicol. 2019, 125, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Danner, M.C.; Robertson, A.; Behrends, V.; Reiss, J. Antibiotic pollution in surface fresh waters: Occurrence and effects. Sci. Total Environ. 2019, 664, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, J.; Feng, H.; Shen, D.; He, S.; Xu, Y. Quantitative study on the fate of antibiotic emissions in China. Environ. Geochem. Health 2020, 42, 3471–3479. [Google Scholar] [CrossRef]
- Bilal, M.; Ashraf, S.S.; Barcelo, D.; Iqbal, H.M.N. Biocatalytic degradation/redefining “removal” fate of pharmaceutically active compounds and antibiotics in the aquatic environment. Sci. Total Environ. 2019, 691, 1190–1211. [Google Scholar] [CrossRef]
- Gothwal, R.; Shashidhar, T. Antibiotic pollution in the environment: A review. Clean-Soil Air Water 2015, 43, 479–489. [Google Scholar] [CrossRef]
- Wei, R.; Ge, F.; Huang, S.; Chen, M.; Wang, R. Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere 2011, 82, 1408–1414. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.G.; Deng, W.J. Antibiotic residues in food: Extraction, analysis, and human health concerns. J. Agric. Food Chem. 2019, 67, 7569–7586. [Google Scholar] [CrossRef]
- Petersen, B.D.; Pereira, T.C.B.; Altenhofen, S.; Nabinger, D.D.; Ferreira, P.M.A.; Bogo, M.R.; Bonan, C.D. Antibiotic drugs alter zebrafish behavior. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2021, 242, 108936. [Google Scholar] [CrossRef]
- Qian, M.; Wang, J.; Ji, X.; Yang, H.; Tang, B.; Zhang, H.; Yang, G.; Bao, Z.; Jin, Y. Sub-chronic exposure to antibiotics doxycycline, oxytetracycline or florfenicol impacts gut barrier and induces gut microbiota dysbiosis in adult zebrafish (Daino rerio). Ecotoxicol. Environ. Saf. 2021, 221, 112464. [Google Scholar] [CrossRef]
- Wang, F.; Gao, J.; Zhai, W.; Cui, J.; Liu, D.; Zhou, Z.; Wang, P. Effects of antibiotic norfloxacin on the degradation and enantioselectivity of the herbicides in aquatic environment. Ecotoxicol. Environ. Saf. 2021, 208, 111717. [Google Scholar] [CrossRef]
- Shen, Y.; Ryser, E.T.; Li, H.; Zhang, W. Bacterial community assembly and antibiotic resistance genes in the lettuce-soil system upon antibiotic exposure. Sci. Total Environ. 2021, 778, 146255. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Engelstadter, J.; Zhang, S.; Ding, P.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J. 2020, 14, 2179–2196. [Google Scholar] [CrossRef]
- Guo, A.; Zhou, Q.; Bao, Y.; Qian, F.; Zhou, X. Prochloraz alone or in combination with nano-CuO promotes the conjugative transfer of antibiotic resistance genes between Escherichia coli in pure water. J. Hazard. Mater. 2022, 424, 127761. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Wang, T.; Xu, N.; Lu, T.; Hong, W.; Penuelas, J.; Gillings, M.; Wang, M.; Gao, W.; et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 2022, 13, 1553. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Lin, Z.; Yuan, T.; Zhou, L.; Cheng, S.; Qu, X.; Lu, P.; Feng, Q. Impact factors of the accumulation, migration and spread of antibiotic resistance in the environment. Environ. Geochem. Health 2021, 43, 1741–1758. [Google Scholar] [CrossRef]
- Cerqueira, F.; Matamoros, V.; Bayona, J.; Pina, B. Antibiotic resistance genes distribution in microbiomes from the soil-plant-fruit continuum in commercial Lycopersicon esculentum fields under different agricultural practices. Sci. Total Environ. 2019, 652, 660–670. [Google Scholar] [CrossRef]
- Li, S.; Zhang, C.; Li, F.; Hua, T.; Zhou, Q.; Ho, S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: A critical review. J. Hazard. Mater. 2021, 411, 125148. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Tian, G.M.; Jin, R.C. The occurrence, maintenance, and proliferation of antibiotic resistance genes (ARGs) in the environment: Influencing factors, mechanisms, and elimination strategies. Appl. Microbiol. Biotechnol. 2018, 102, 8261–8274. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Li, C.; Wang, X.; Cao, Z.; Gao, C.; Su, S.; Xue, B.; Wang, S.; Qiu, Z.; Wang, J.; et al. Monitoring and evaluation of antibiotic resistance genes in three rivers in northeast China. Environ. Sci. Pollut. Res. Int. 2022, 29, 44148–44161. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, C.; Song, L. Distribution of antibiotic resistance genes and bacteria from six atmospheric environments: Exposure risk to human. Sci. Total Environ. 2019, 694, 133750. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jin, L.; He, T.; Chen, B.; Luo, X.; Feng, B.; Huang, W.; Li, J.; Fu, P.; Li, X. Bacteria and antibiotic resistance genes (ARGs) in PM2.5 from China: Implications for human exposure. Environ. Sci. Technol. 2019, 53, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wu, S.; Zeng, Z.; Fu, Z. Effects of environmental pollutants on gut microbiota. Environ. Pollut. 2017, 222, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Pan, Z.; Jin, C.; Ni, Y.; Fu, Z.; Jin, Y. Gut microbiota: An underestimated and unintended recipient for pesticide-induced toxicity. Chemosphere 2019, 227, 425–434. [Google Scholar] [CrossRef]
- Shen, Q.; Huang, Z.; Yao, J.; Jin, Y. Extracellular vesicles-mediated interaction within intestinal microenvironment in inflammatory bowel disease. J. Adv. Res. 2022, 37, 221–233. [Google Scholar] [CrossRef]
- Gerard, P. Gut microbiota and obesity. Cell Mol. Life Sci. 2016, 73, 147–162. [Google Scholar] [CrossRef]
- Nallu, A.; Sharma, S.; Ramezani, A.; Muralidharan, J.; Raj, D. Gut microbiome in chronic kidney disease: Challenges and opportunities. Transl. Res. 2017, 179, 24–37. [Google Scholar] [CrossRef] [Green Version]
- Duan, H.; Yu, L.; Tian, F.; Zhai, Q.; Fan, L.; Chen, W. Antibiotic-induced gut dysbiosis and barrier disruption and the potential protective strategies. Crit. Rev. Food Sci. Nutr. 2022, 62, 1427–1452. [Google Scholar] [CrossRef]
- McInnes, R.S.; McCallum, G.E.; Lamberte, L.E.; van Schaik, W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 2020, 53, 35–43. [Google Scholar] [CrossRef]
- Anthony, W.E.; Burnham, C.-A.D.; Dantas, G.; Kwon, J.H. The gut microbiome as a reservoir for antimicrobial resistance. J. Infect. Dis. 2021, 223, S209–S213. [Google Scholar] [CrossRef]
- Qin, Z.; Wang, W.; Weng, Y.; Bao, Z.; Yang, G.; Jin, Y. Bromuconazole exposure induces cardiotoxicity and lipid transport disorder in larval zebrafish. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2022, 262, 109451. [Google Scholar] [CrossRef]
- Wang, X.; Hu, L.; Jin, C.; Qian, M.; Jin, Y. Effects of maternal exposure to procymidone on hepatic metabolism in the offspring of mice. Environ. Toxicol. 2023. [Google Scholar] [CrossRef]
- Li, N.; Chen, J.; Liu, C.; Yang, J.; Zhu, C.; Li, H. Cu and Zn exert a greater influence on antibiotic resistance and its transfer than doxycycline in agricultural soils. J. Hazard. Mater. 2022, 423, 127042. [Google Scholar] [CrossRef]
- Xia, J.; Sun, H.; Zhang, X.X.; Zhang, T.; Ren, H.; Ye, L. Aromatic compounds lead to increased abundance of antibiotic resistance genes in wastewater treatment bioreactors. Water Res. 2019, 166, 115073. [Google Scholar] [CrossRef]
- Kumar, M.; Jaiswal, S.; Sodhi, K.K.; Shree, P.; Singh, D.K.; Agrawal, P.K.; Shukla, P. Antibiotics bioremediation: Perspectives on its ecotoxicity and resistance. Environ. Int. 2019, 124, 448–461. [Google Scholar] [CrossRef]
- Laws, M.; Shaaban, A.; Rahman, K.M. Antibiotic resistance breakers: Current approaches and future directions. FEMS Microbiol. Rev. 2019, 43, 490–516. [Google Scholar] [CrossRef] [Green Version]
- Tasho, R.P.; Cho, J.Y. Veterinary antibiotics in animal waste, its distribution in soil and uptake by plants: A review. Sci. Total Environ. 2016, 563–564, 366–376. [Google Scholar] [CrossRef]
- Zhou, L.; Limbu, S.M.; Shen, M.; Zhai, W.; Qiao, F.; He, A.; Du, Z.Y.; Zhang, M. Environmental concentrations of antibiotics impair zebrafish gut health. Environ. Pollut. 2018, 235, 245–254. [Google Scholar] [CrossRef]
- Zhang, M.; He, L.Y.; Liu, Y.S.; Zhao, J.L.; Liu, W.R.; Zhang, J.N.; Chen, J.; He, L.K.; Zhang, Q.Q.; Ying, G.G. Fate of veterinary antibiotics during animal manure composting. Sci. Total Environ. 2019, 650, 1363–1370. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhao, T.; Liu, Q.; He, J.; He, D.; Wu, G.; Li, Y.; Jiang, C.; Xu, Z. Residual veterinary antibiotics in pig excreta after oral administration of sulfonamides. Environ. Geochem. Health 2016, 38, 549–556. [Google Scholar] [CrossRef] [PubMed]
- MacLauchlin, C.; Schneider, S.E.; Keedy, K.; Fernandes, P.; Jamieson, B.D. Metabolism, Excretion, and Mass Balance of Solithromycin in Humans. Antimicrob. Agents Chemother. 2018, 62, e01474-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bombaywala, S.; Mandpe, A.; Paliya, S.; Kumar, S. Antibiotic resistance in the environment: A critical insight on its occurrence, fate, and eco-toxicity. Environ. Sci. Pollut. Res. Int. 2021, 28, 24889–24916. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, Q.; Yan, X.; Zhai, Y.; Guo, Z.; Li, N.; Ying, G. Antibiotic pollution in lakes in China: Emission estimation and fate modeling using a temperature-dependent multimedia model. Sci. Total Environ. 2022, 842, 156633. [Google Scholar] [CrossRef]
- Aydin, S.; Aydin, M.E.; Ulvi, A.; Kilic, H. Antibiotics in hospital effluents: Occurrence, contribution to urban wastewater, removal in a wastewater treatment plant, and environmental risk assessment. Environ. Sci. Pollut. Res. Int. 2019, 26, 544–558. [Google Scholar] [CrossRef]
- Behera, S.K.; Kim, H.W.; Oh, J.E.; Park, H.S. Occurrence and removal of antibiotics, hormones and several other pharmaceuticals in wastewater treatment plants of the largest industrial city of Korea. Sci. Total Environ. 2011, 409, 4351–4360. [Google Scholar] [CrossRef]
- Langbehn, R.K.; Michels, C.; Soares, H.M. Antibiotics in wastewater: From its occurrence to the biological removal by environmentally conscious technologies. Environ. Pollut. 2021, 275, 116603. [Google Scholar] [CrossRef]
- Yao, S.; Ye, J.; Yang, Q.; Hu, Y.; Zhang, T.; Jiang, L.; Munezero, S.; Lin, K.; Cui, C. Occurrence and removal of antibiotics, antibiotic resistance genes, and bacterial communities in hospital wastewater. Environ. Sci. Pollut. Res. Int. 2021, 28, 57321–57333. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; Zhang, Z.; Li, P.; Zang, Y.; Liu, X. Antibiotics in aquatic environments of China: A review and meta-analysis. Ecotoxicol. Environ. Saf. 2020, 199, 110668. [Google Scholar] [CrossRef]
- Gu, X.; Zhai, H.; Cheng, S. Fate of antibiotics and antibiotic resistance genes in home water purification systems. Water Res. 2021, 190, 116762. [Google Scholar] [CrossRef]
- Ma, Y.; Li, M.; Wu, M.; Li, Z.; Liu, X. Occurrences and regional distributions of 20 antibiotics in water bodies during groundwater recharge. Sci. Total Environ. 2015, 518–519, 498–506. [Google Scholar] [CrossRef]
- Wei, M.; Lv, D.; Cao, L.; Zhou, K.; Jiang, K. Adsorption behaviours and transfer simulation of levofloxacin in silty clay. Environ. Sci. Pollut. Res. Int. 2021, 28, 46291–46302. [Google Scholar] [CrossRef]
- Kovalakova, P.; Cizmas, L.; McDonald, T.J.; Marsalek, B.; Feng, M.; Sharma, V.K. Occurrence and toxicity of antibiotics in the aquatic environment: A review. Chemosphere 2020, 251, 126351. [Google Scholar] [CrossRef]
- Xu, S.; Jiang, Y.; Liu, Y.; Zhang, J. Antibiotic-accelerated cyanobacterial growth and aquatic community succession towards the formation of cyanobacterial bloom in eutrophic lake water. Environ. Pollut. 2021, 290, 118057. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, Y.; Zhang, J. Mechanisms for the stimulatory effects of a five-component mixture of antibiotics in Microcystis aeruginosa at transcriptomic and proteomic levels. J. Hazard. Mater. 2021, 406, 124722. [Google Scholar] [CrossRef]
- Lei, H.; Song, Y.; Dong, M.; Chen, G.; Cao, Z.; Wu, F.; Chen, C.; Zhang, C.; Liu, C.; Shi, Z.; et al. Metabolomics safety assessments of microcystin exposure via drinking water in rats. Ecotoxicol. Environ. Saf. 2021, 212, 111989. [Google Scholar] [CrossRef]
- Wan, L.; Wu, Y.; Zhang, B.; Yang, W.; Ding, H.; Zhang, W. Effects of moxifloxacin and gatifloxacin stress on growth, photosynthesis, antioxidant responses, and microcystin release in Microcystis aeruginosa. J. Hazard. Mater. 2021, 409, 124518. [Google Scholar] [CrossRef]
- Guo, R.X.; Chen, J.Q. Phytoplankton toxicity of the antibiotic chlortetracycline and its UV light degradation products. Chemosphere 2012, 87, 1254–1259. [Google Scholar] [CrossRef]
- Wan, J.; Guo, P.; Peng, X.; Wen, K. Effect of erythromycin exposure on the growth, antioxidant system and photosynthesis of Microcystis flos-aquae. J. Hazard. Mater. 2015, 283, 778–786. [Google Scholar] [CrossRef]
- Yang, C.; Song, G.; Lim, W. A review of the toxicity in fish exposed to antibiotics. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 2020, 237, 108840. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wang, Y.; Li, J.; Wang, D.; Bao, Z.; Wang, Q.; Jin, Y. Health risks of chlorothalonil, carbendazim, prochloraz, their binary and ternary mixtures on embryonic and larval zebrafish based on metabolomics analysis. J. Hazard. Mater. 2021, 404, 124240. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Huang, X.; Xie, Y.; Song, M.; Zhu, K.; Ding, S. Macrolides induce severe cardiotoxicity and developmental toxicity in zebrafish embryos. Sci. Total Environ. 2019, 649, 1414–1421. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Q.; Chen, B.; Zhang, J.P.; Chen, N.; Liu, C.Z.; Hu, C.Q. Liver toxicity of macrolide antibiotics in zebrafish. Toxicology 2020, 441, 152501. [Google Scholar] [CrossRef]
- Li, J.; Dong, T.; Keerthisinghe, T.P.; Chen, H.; Li, M.; Chu, W.; Yang, J.; Hu, Z.; Snyder, S.A.; Dong, W.; et al. Long-term oxytetracycline exposure potentially alters brain thyroid hormone and serotonin homeostasis in zebrafish. J. Hazard. Mater. 2020, 399, 123061. [Google Scholar] [CrossRef]
- Zhao, X.L.; Li, P.; Zhang, S.Q.; He, S.W.; Xing, S.Y.; Cao, Z.H.; Lu, R.; Li, Z.H. Effects of environmental norfloxacin concentrations on the intestinal health and function of juvenile common carp and potential risk to humans. Environ. Pollut. 2021, 287, 117612. [Google Scholar] [CrossRef]
- Qiu, W.; Fang, M.; Magnuson, J.T.; Greer, J.B.; Chen, Q.; Zheng, Y.; Xiong, Y.; Luo, S.; Zheng, C.; Schlenk, D. Maternal exposure to environmental antibiotic mixture during gravid period predicts gastrointestinal effects in zebrafish offspring. J. Hazard. Mater. 2020, 399, 123009. [Google Scholar] [CrossRef]
- Kwon, H.J.; Mohammed, A.E.; Eltom, K.H.; Albrahim, J.S.; Alburae, N.A. Evaluation of antibiotic-induced behavioral changes in mice. Physiol. Behav. 2020, 223, 113015. [Google Scholar] [CrossRef]
- Peltzer, P.M.; Lajmanovich, R.C.; Attademo, A.M.; Junges, C.M.; Teglia, C.M.; Martinuzzi, C.; Curi, L.; Culzoni, M.J.; Goicoechea, H.C. Ecotoxicity of veterinary enrofloxacin and ciprofloxacin antibiotics on anuran amphibian larvae. Environ. Toxicol. Pharmacol. 2017, 51, 114–123. [Google Scholar] [CrossRef]
- Pan, M.; Chu, L.M. Phytotoxicity of veterinary antibiotics to seed germination and root elongation of crops. Ecotoxicol. Environ. Saf. 2016, 126, 228–237. [Google Scholar] [CrossRef]
- Luo, Y.; Liang, J.; Zeng, G.; Zhang, Y.; Xing, W.; Tang, N. Tetracycline stress disturbs the mobilization of protein bodies in seed storage reserves during radicle elongation after seed germination. Environ. Sci. Pollut. Res. Int. 2020, 27, 42150–42157. [Google Scholar] [CrossRef]
- Cheong, M.; Yoon, Y.; Kim, J.; Hong, Y.; Kim, S.; Lee, Y. Chlortetracycline inhibits seed germination and seedling growth in Brassica campestris by disrupting H2O2 signaling. Appl. Biol. Chem. 2020, 63, 1. [Google Scholar] [CrossRef] [Green Version]
- Bellino, A.; Lofrano, G.; Carotenuto, M.; Libralato, G.; Baldantoni, D. Antibiotic effects on seed germination and root development of tomato (Solanum lycopersicum L.). Ecotoxicol. Environ. Saf. 2018, 148, 135–141. [Google Scholar] [CrossRef]
- Marques, R.; Wistuba, N.; Brito, J.; Bernardoni, V.; Rocha, D.; Gomes, M. Crop irrigation (soybean, bean, and corn) with enrofloxacin-contaminated water leads to yield reductions and antibiotic accumulation. Ecotoxicol. Environ. Saf. 2021, 216, 112193. [Google Scholar] [CrossRef]
- Jones, T.W.; Fino, N.; Olson, J.; Hersh, A.L. The impact of beta-lactam allergy labels on hospitalized children. Infect. Control Hosp. Epidemiol. 2021, 42, 318–324. [Google Scholar] [CrossRef]
- Liu, X.; Steele, J.C.; Meng, X.Z. Usage, residue, and human health risk of antibiotics in Chinese aquaculture: A review. Environ. Pollut. 2017, 223, 161–169. [Google Scholar] [CrossRef]
- Polk, R.E.; Israel, D.; Wang, J.; Venitz, J.; Miller, J.; Stotka, J. Vancomycin skin tests and prediction of “red man syndrome” in healthy volunteers. Antimicrob. Agents Chemother. 1993, 37, 2139–2143. [Google Scholar] [CrossRef] [Green Version]
- Jeffres, M.N. The Whole Price of Vancomycin: Toxicities, Troughs, and Time. Drugs 2017, 77, 1143–1154. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.J.; Liu, A.Y.; Wong, P.H.; Arroyo, A.C. Road Less Traveled: Drug Hypersensitivity to Fluoroquinolones, Vancomycin, Tetracyclines, and Macrolides. Clin. Rev. Allergy Immunol. 2022, 62, 505–518. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Li, D.Y.; Hazen, S.L. Dietary metabolism, the gut microbiome, and heart failure. Nat. Rev. Cardiol. 2019, 16, 137–154. [Google Scholar] [CrossRef]
- Wang, X.; Yang, S.; Li, S.; Zhao, L.; Hao, Y.; Qin, J.; Zhang, L.; Zhang, C.; Bian, W.; Zuo, L.; et al. Aberrant gut microbiota alters host metabolome and impacts renal failure in humans and rodents. Gut 2020, 69, 2131–2142. [Google Scholar] [CrossRef] [PubMed]
- Aversa, Z.; Atkinson, E.J.; Schafer, M.J.; Theiler, R.N.; Rocca, W.A.; Blaser, M.J.; LeBrasseur, N.K. Association of infant antibiotic exposure with childhood health outcomes. Mayo Clin. Proc. 2021, 96, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Nahum, G.G.; Uhl, K.; Kennedy, D.L. Antibiotic use in pregnancy and lactation—What is and is not known about teratogenic and toxic risks. Obstet. Gynecol. 2006, 107, 1120–1138. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishna, K.P.; Hand, T.W. Influence of maternal milk on the neonatal intestinal microbiome. Nutrients 2020, 12, 823. [Google Scholar] [CrossRef] [Green Version]
- Fonseca, W.; Malinczak, C.A.; Fujimura, K.; Li, D.; McCauley, K.; Li, J.; Best, S.K.K.; Zhu, D.; Rasky, A.J.; Johnson, C.C.; et al. Maternal gut microbiome regulates immunity to RSV infection in offspring. J. Exp. Med. 2021, 218, e20210235. [Google Scholar] [CrossRef]
- Le Roy, T.; Lecuyer, E.; Chassaing, B.; Rhimi, M.; Lhomme, M.; Boudebbouze, S.; Ichou, F.; Haro Barcelo, J.; Huby, T.; Guerin, M.; et al. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biol. 2019, 17, 94. [Google Scholar] [CrossRef] [Green Version]
- San Millan, A. Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 2018, 26, 978–985. [Google Scholar] [CrossRef] [Green Version]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef]
- Dobrijevic, D.; Abraham, A.L.; Jamet, A.; Maguin, E.; van de Guchte, M. Functional comparison of bacteria from the human gut and closely related non-gut bacteria reveals the importance of conjugation and a paucity of motility and chemotaxis functions in the gut environment. PLoS ONE 2016, 11, e0159030. [Google Scholar] [CrossRef]
- Huang, Z.; Weng, Y.; Shen, Q.; Zhao, Y.; Jin, Y. Microplastic: A potential threat to human and animal health by interfering with the intestinal barrier function and changing the intestinal microenvironment. Sci. Total Environ. 2021, 785, 147365. [Google Scholar] [CrossRef]
- Lu, L.; Luo, T.; Zhao, Y.; Cai, C.; Fu, Z.; Jin, Y. Interaction between microplastics and microorganism as well as gut microbiota: A consideration on environmental animal and human health. Sci. Total Environ. 2019, 667, 94–100. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Y.; Liu, X.; Zhao, J.; Liu, R.; Xing, B. Interaction of microplastics with antibiotics in aquatic environment: Distribution, adsorption, and toxicity. Environ. Sci. Technol. 2021, 55, 15579–15595. [Google Scholar] [CrossRef]
- Luo, T.; Wang, C.; Pan, Z.; Jin, C.; Fu, Z.; Jin, Y. Maternal Polystyrene Microplastic Exposure during Gestation and Lactation Altered Metabolic Homeostasis in the Dams and Their F1 and F2 Offspring. Environ. Sci. Technol. 2019, 53, 10978–10992. [Google Scholar] [CrossRef]
- Guo, X.; Chen, C.; Wang, J. Sorption of sulfamethoxazole onto six types of microplastics. Chemosphere 2019, 228, 300–308. [Google Scholar] [CrossRef]
- Guo, X.; Wang, J. Sorption of antibiotics onto aged microplastics in freshwater and seawater. Mar. Pollut. Bull. 2019, 149, 110511. [Google Scholar] [CrossRef]
- Li, J.; Zhang, K.; Zhang, H. Adsorption of antibiotics on microplastics. Environ. Pollut. 2018, 237, 460–467. [Google Scholar] [CrossRef]
- Gonzalez-Pleiter, M.; Pedrouzo-Rodriguez, A.; Verdu, I.; Leganes, F.; Marco, E.; Rosal, R.; Fernandez-Pinas, F. Microplastics as vectors of the antibiotics azithromycin and clarithromycin: Effects towards freshwater microalgae. Chemosphere 2021, 268, 128824. [Google Scholar] [CrossRef]
- Wang, S.; Xue, N.; Li, W.; Zhang, D.; Pan, X.; Luo, Y. Selectively enrichment of antibiotics and ARGs by microplastics in river, estuary and marine waters. Sci. Total Environ. 2020, 708, 134594. [Google Scholar] [CrossRef]
- Zhou, W.; Han, Y.; Tang, Y.; Shi, W.; Du, X.; Sun, S.; Liu, G. Microplastics aggravate the bioaccumulation of two waterborne veterinary antibiotics in an edible bivalve species: Potential mechanisms and implications for human health. Environ. Sci. Technol. 2020, 54, 8115–8122. [Google Scholar] [CrossRef]
- Zhou, W.; Tang, Y.; Du, X.; Han, Y.; Shi, W.; Sun, S.; Zhang, W.; Zheng, H.; Liu, G. Fine polystyrene microplastics render immune responses more vulnerable to two veterinary antibiotics in a bivalve species. Mar. Pollut. Bull. 2021, 164, 111995. [Google Scholar] [CrossRef]
- Liu, J.; Wang, P.; Wang, C.; Qian, J.; Hou, J. Heavy metal pollution status and ecological risks of sediments under the influence of water transfers in Taihu Lake, China. Environ. Sci. Pollut. Res. Int. 2017, 24, 2653–2666. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, Y.; Huang, G.; Yang, C.; Chen, C.; Zhou, T.; Zhao, Y.; Ma, J. Adsorption behavior of the antibiotic levofloxacin on microplastics in the presence of different heavy metals in an aqueous solution. Chemosphere 2020, 260, 127650. [Google Scholar] [CrossRef] [PubMed]
- You, X.; Li, H.; Pan, B.; You, M.; Sun, W. Interactions between antibiotics and heavy metals determine their combined toxicity to Synechocystis sp. J. Hazard. Mater. 2022, 424, 127707. [Google Scholar] [CrossRef] [PubMed]
- Khurana, P.; Pulicharla, R.; Kaur Brar, S. Antibiotic-metal complexes in wastewaters: Fate and treatment trajectory. Environ. Int. 2021, 157, 106863. [Google Scholar] [CrossRef]
- Yang, R.; Wang, J.; Zhu, L.; Wang, J.; Yang, L.; Mao, S.; Conkle, J.L.; Chen, Y.; Kim, Y.M. Effects of interaction between enrofloxacin and copper on soil enzyme activity and evaluation of comprehensive toxicity. Chemosphere 2021, 268, 129208. [Google Scholar] [CrossRef]
- Sciscenko, I.; Arques, A.; Varga, Z.; Bouchonnet, S.; Monfort, O.; Brigante, M.; Mailhot, G. Significant role of iron on the fate and photodegradation of enrofloxacin. Chemosphere 2021, 270, 129791. [Google Scholar] [CrossRef]
- Guo, X.; Wang, J. Projecting the sorption capacity of heavy metal ions onto microplastics in global aquatic environments using artificial neural networks. J. Hazard. Mater. 2021, 402, 123709. [Google Scholar] [CrossRef]
- Tang, S.; Lin, L.; Wang, X.; Yu, A.; Sun, X. Interfacial interactions between collected nylon microplastics and three divalent metal ions (Cu(II), Ni(II), Zn(II)) in aqueous solutions. J. Hazard. Mater. 2021, 403, 123548. [Google Scholar] [CrossRef]
- Jiang, W.; Gao, J.; Cheng, Z.; Wang, P.; Zhou, Z.; Liu, D. The effect of antibiotics on the persistence of herbicides in soil under the combined pollution. Chemosphere 2018, 204, 303–309. [Google Scholar] [CrossRef]
- Xu, S.; Liu, Y.; Zhang, J.; Gao, B. Proteomic mechanisms for the combined stimulatory effects of glyphosate and antibiotic contaminants on Microcystis aeruginosa. Chemosphere 2021, 267, 129244. [Google Scholar] [CrossRef]
- Zhan, J.; Liang, Y.; Liu, D.; Ma, X.; Li, P.; Liu, C.; Liu, X.; Wang, P.; Zhou, Z. Antibiotics may increase triazine herbicide exposure risk via disturbing gut microbiota. Microbiome 2018, 6, 224. [Google Scholar] [CrossRef] [Green Version]
- Santos-Lopez, A.; Marshall, C.W.; Haas, A.L.; Turner, C.; Rasero, J.; Cooper, V.S. The roles of history, chance, and natural selection in the evolution of antibiotic resistance. eLife 2021, 10, e70676. [Google Scholar] [CrossRef]
- Khan, A.; Miller, W.R.; Arias, C.A. Mechanisms of antimicrobial resistance among hospital-associated pathogens. Expert. Rev. Anti. Infect. Ther. 2018, 16, 269–287. [Google Scholar] [CrossRef]
- Cooper, R.M.; Tsimring, L.; Hasty, J. Inter-species population dynamics enhance microbial horizontal gene transfer and spread of antibiotic resistance. eLife 2017, 6, e25950. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, P.; Yang, Q. Occurrence and diversity of antibiotic resistance in untreated hospital wastewater. Sci. Total Environ. 2018, 621, 990–999. [Google Scholar] [CrossRef]
- Song, L.; Wang, C.; Jiang, G.; Ma, J.; Li, Y.; Chen, H.; Guo, J. Bioaerosol is an important transmission route of antibiotic resistance genes in pig farms. Environ. Int. 2021, 154, 106559. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- Hu, J.; Shi, J.; Chang, H.; Li, D.; Yang, M.; Kamagata, Y. Phenotyping and genotyping of antibiotic-resistant Escherichia coli isolated from a natural river basin. Environ. Sci. Technol. 2008, 42, 3415–3420. [Google Scholar] [CrossRef]
- Pintor-Cora, A.; Alvaro-Llorente, L.; Otero, A.; Rodriguez-Calleja, J.M.; Santos, J.A. Extended-spectrum beta-lactamase-producing Enterobacteriaceae in fresh produce. Foods 2021, 10, 2609. [Google Scholar] [CrossRef]
- Zhou, Z.C.; Feng, W.Q.; Han, Y.; Zheng, J.; Chen, T.; Wei, Y.Y.; Gillings, M.; Zhu, Y.G.; Chen, H. Prevalence and transmission of antibiotic resistance and microbiota between humans and water environments. Environ. Int. 2018, 121, 1155–1161. [Google Scholar] [CrossRef]
- Komijani, M.; Shamabadi, N.S.; Shahin, K.; Eghbalpour, F.; Tahsili, M.R.; Bahram, M. Heavy metal pollution promotes antibiotic resistance potential in the aquatic environment. Environ. Pollut. 2021, 274, 116569. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.Y.; Wu, E.L.; Hartmann, E.M.; Zhao, H.P. Biological mitigation of antibiotic resistance gene dissemination by antioxidant-producing microorganisms in activated sludge Systems. Environ. Sci. Technol. 2021, 55, 15831–15842. [Google Scholar] [CrossRef] [PubMed]
- Bombaywala, S.; Purohit, H.J.; Dafale, N.A. Mobility of antibiotic resistance and its co-occurrence with metal resistance in pathogens under oxidative stress. J. Environ. Manag. 2021, 297, 113315. [Google Scholar] [CrossRef] [PubMed]
- Bergholz, T.M.; Bowen, B.; Wiedmann, M.; Boor, K.J. Listeria monocytogenes shows temperature-dependent and -independent responses to salt stress, including responses that induce cross-protection against other stresses. Appl. Environ. Microbiol. 2012, 78, 2602–2612. [Google Scholar] [CrossRef] [Green Version]
- Zhong, C.; Zhou, Y.; Fu, J.; Qi, X.; Wang, Z.; Li, J.; Zhang, P.; Zong, G.; Cao, G. Cadmium stress efficiently enhanced meropenem degradation by the meropenem- and cadmium-resistant strain Pseudomonas putida R51. J. Hazard. Mater. 2022, 429, 128354. [Google Scholar] [CrossRef]
- Imran, M.; Das, K.R.; Naik, M.M. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: An emerging health threat. Chemosphere 2019, 215, 846–857. [Google Scholar] [CrossRef]
- Buledi, J.A.; Amin, S.; Haider, S.I.; Bhanger, M.I.; Solangi, A.R. A review on detection of heavy metals from aqueous media using nanomaterial-based sensors. Environ. Sci. Pollut. Res. Int. 2021, 28, 58994–59002. [Google Scholar] [CrossRef]
- Feng, G.; Huang, H.; Chen, Y. Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: A review. J. Hazard. Mater. 2021, 420, 126602. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, J.; Wu, J.; Wang, J.; Luo, Y. Potential risks of microplastics combined with superbugs: Enrichment of antibiotic resistant bacteria on the surface of microplastics in mariculture system. Ecotoxicol. Environ. Saf. 2020, 187, 109852. [Google Scholar] [CrossRef]
- Li, R.; Zhu, L.; Yang, K.; Li, H.; Zhu, Y.G.; Cui, L. Impact of Urbanization on Antibiotic Resistome in Different Microplastics: Evidence from a Large-Scale Whole River Analysis. Environ. Sci. Technol. 2021, 55, 8760–8770. [Google Scholar] [CrossRef]
- Wang, Z.; Su, Y.; Zhu, J.; Wu, D.; Xie, B. Size-dependent effects of microplastics on antibiotic resistance genes fate in wastewater treatment systems: The role of changed surface property and microbial assemblages in a continuous exposure mode. Sci. Total Environ. 2022, 851, 158264. [Google Scholar] [CrossRef]
- Guo, X.; Pang, J.; Chen, S.; Jia, H. Sorption properties of tylosin on four different microplastics. Chemosphere 2018, 209, 240–245. [Google Scholar] [CrossRef]
- Wang, J.; Guo, X.; Xue, J. Biofilm-Developed Microplastics As Vectors of Pollutants in Aquatic Environments. Environ. Sci. Technol. 2021, 55, 12780–12790. [Google Scholar] [CrossRef]
- Maurya, A.P.; Rajkumari, J.; Pandey, P. Enrichment of antibiotic resistance genes (ARGs) in polyaromatic hydrocarbon-contaminated soils: A major challenge for environmental health. Environ. Sci. Pollut. Res. Int. 2021, 28, 12178–12189. [Google Scholar] [CrossRef]
- Liao, H.; Li, X.; Yang, Q.; Bai, Y.; Cui, P.; Wen, C.; Liu, C.; Chen, Z.; Tang, J.; Che, J.; et al. Herbicide selection promotes antibiotic resistance in soil microbiomes. Mol. Biol. Evol. 2021, 38, 2337–2350. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhu, D.; Ding, J.; Zheng, F.; Zhou, S.; Lu, T.; Zhu, Y.G.; Qian, H. The fungicide azoxystrobin perturbs the gut microbiota community and enriches antibiotic resistance genes in Enchytraeus crypticus. Environ. Int. 2019, 131, 104965. [Google Scholar] [CrossRef]
- Jin, M.; Liu, L.; Wang, D.N.; Yang, D.; Liu, W.L.; Yin, J.; Yang, Z.W.; Wang, H.R.; Qiu, Z.G.; Shen, Z.Q.; et al. Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J. 2020, 14, 1847–1856. [Google Scholar] [CrossRef]
- Liu, X.; Wang, D.; Tang, J.; Liu, F.; Wang, L. Effect of dissolved biochar on the transfer of antibiotic resistance genes between bacteria. Environ. Pollut. 2021, 288, 117718. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Y.; Jin, M.; Yuan, Z.; Bond, P.; Guo, J. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 2020, 169, 115229. [Google Scholar] [CrossRef]
- Fang, H.; Han, L.; Zhang, H.; Long, Z.; Cai, L.; Yu, Y. Dissemination of antibiotic resistance genes and human pathogenic bacteria from a pig feedlot to the surrounding stream and agricultural soils. J. Hazard. Mater. 2018, 357, 53–62. [Google Scholar] [CrossRef]
- Kampouris, I.D.; Agrawal, S.; Orschler, L.; Cacace, D.; Kunze, S.; Berendonk, T.U.; Klumper, U. Antibiotic resistance gene load and irrigation intensity determine the impact of wastewater irrigation on antimicrobial resistance in the soil microbiome. Water Res. 2021, 193, 116818. [Google Scholar] [CrossRef] [PubMed]
- Han, X.M.; Hu, H.W.; Chen, Q.L.; Yang, L.Y.; Li, H.L.; Zhu, Y.G.; Li, X.Z.; Ma, Y.B. Antibiotic resistance genes and associated bacterial communities in agricultural soils amended with different sources of animal manures. Soil Biol. Biochem. 2018, 126, 91–102. [Google Scholar] [CrossRef]
- Xu, H.; Chen, Z.; Huang, R.; Cui, Y.; Li, Q.; Zhao, Y.; Wang, X.; Mao, D.; Luo, Y.; Ren, H. Antibiotic resistance gene-carrying plasmid spreads into the plant endophytic bacteria using soil bacteria as carriers. Environ. Sci. Technol. 2021, 55, 10462–10470. [Google Scholar] [CrossRef] [PubMed]
- Scaccia, N.; Vaz-Moreira, I.; Manaia, C.M. The risk of transmitting antibiotic resistance through endophytic bacteria. Trends Plant Sci. 2021, 26, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.J.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef]
- Heintz-Buschart, A.; Wilmes, P. Human gut microbiome: Function matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef]
- Launay, A.; Ballard, S.A.; Johnson, P.D.; Grayson, M.L.; Lambert, T. Transfer of vancomycin resistance transposon Tn1549 from Clostridium symbiosum to Enterococcus spp. in the gut of gnotobiotic mice. Antimicrob. Agents Chemother. 2006, 50, 1054–1062. [Google Scholar] [CrossRef] [Green Version]
- Parnanen, K.; Karkman, A.; Hultman, J.; Lyra, C.; Bengtsson-Palme, J.; Larsson, D.G.J.; Rautava, S.; Isolauri, E.; Salminen, S.; Kumar, H.; et al. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 2018, 9, 3891. [Google Scholar] [CrossRef] [Green Version]
- Gasparrini, A.J.; Wang, B.; Sun, X.; Kennedy, E.A.; Hernandez-Leyva, A.; Ndao, I.M.; Tarr, P.I.; Warner, B.B.; Dantas, G. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 2019, 4, 2285–2297. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269–281. [Google Scholar]
- Diawara, I.; Zerouali, K.; Katfy, K.; Barguigua, A.; Belabbes, H.; Timinouni, M.; Elmdaghri, N. Phenotypic and genotypic characterization of Streptococcus pneumoniae resistant to macrolide in Casablanca, Morocco. Infect. Genet. Evol. 2016, 40, 200–204. [Google Scholar] [CrossRef]
- Garcia-Solache, M.; Rice, L.B. The enterococcus: A model of adaptability to its environment. Clin. Microbiol. Rev. 2019, 32, e00058-18. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Shen, P.; Liang, W.; Jin, J.; Jiang, X. A putative multi-replicon plasmid co-harboring beta-lactamase genes blaKPC-2, blaCTX-M-14 and blaTEM-1 and trimethoprim resistance gene dfrA25 from a Klebsiella pneumoniae sequence type (ST) 11 strain in China. PLoS ONE 2017, 12, e0171339. [Google Scholar] [CrossRef] [Green Version]
- Breidenstein, E.B.; de la Fuente-Nunez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Kim, M.; Park, J.; Park, W. Genomic and phenotypic analyses of multidrug-resistant Acinetobacter baumannii NCCP 16007 isolated from a patient with a urinary tract infection. Virulence 2021, 12, 150–164. [Google Scholar] [CrossRef]
- Zhu, Z.; Jiang, S.; Qi, M.; Liu, H.; Zhang, S.; Liu, H.; Zhou, Z.; Wang, L.; Wang, C.; Luo, Y.; et al. Prevalence and characterization of antibiotic resistance genes and integrons in Escherichia coli isolates from captive non-human primates of 13 zoos in China. Sci. Total Environ. 2021, 798, 149268. [Google Scholar] [CrossRef]
- Hassoun-Kheir, N.; Stabholz, Y.; Kreft, J.U.; de la Cruz, R.; Romalde, J.L.; Nesme, J.; Sorensen, S.J.; Smets, B.F.; Graham, D.; Paul, M. Comparison of antibiotic-resistant bacteria and antibiotic resistance genes abundance in hospital and community wastewater: A systematic review. Sci. Total Environ. 2020, 743, 140804. [Google Scholar] [CrossRef]
- Stange, C.; Sidhu, J.P.S.; Toze, S.; Tiehm, A. Comparative removal of antibiotic resistance genes during chlorination, ozonation, and UV treatment. Int. J. Hyg. Environ. Health 2019, 222, 541–548. [Google Scholar] [CrossRef]
- Wang, J.; Chu, L.; Wojnarovits, L.; Takacs, E. Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: An overview. Sci. Total Environ. 2020, 744, 140997. [Google Scholar] [CrossRef]
- Ahmed, Y.; Zhong, J.; Yuan, Z.; Guo, J. Simultaneous removal of antibiotic resistant bacteria, antibiotic resistance genes, and micropollutants by a modified photo-Fenton process. Water Res. 2021, 197, 117075. [Google Scholar] [CrossRef]
- Liang, C.; Wei, D.; Zhang, S.; Ren, Q.; Shi, J.; Liu, L. Removal of antibiotic resistance genes from swine wastewater by membrane filtration treatment. Ecotoxicol. Environ. Saf. 2021, 210, 111885. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xin, Y.; Huang, X.; Liu, C. Response of antibiotic resistance genes in constructed wetlands during treatment of livestock wastewater with different exogenous inducers: Antibiotic and antibiotic-resistant bacteria. Bioresour. Technol. 2020, 314, 123779. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.S.; Wu, J.W.; Dong, L.L.; Liu, B.F.; Xing, D.F.; Yang, S.S.; Wu, X.K.; Wang, Q.; Fan, J.N.; Feng, L.P.; et al. Removal of antibiotic resistant bacteria and antibiotic resistance genes in wastewater effluent by UV-activated persulfate. J. Hazard. Mater. 2020, 388, 122070. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhuan, R. Degradation of antibiotics by advanced oxidation processes: An overview. Sci. Total Environ. 2020, 701, 135023. [Google Scholar] [CrossRef]
- Li, S.; Wu, Y.; Zheng, H.; Li, H.; Zheng, Y.; Nan, J.; Ma, J.; Nagarajan, D.; Chang, J.S. Antibiotics degradation by advanced oxidation process (AOPs): Recent advances in ecotoxicity and antibiotic-resistance genes induction of degradation products. Chemosphere 2023, 311, 136977. [Google Scholar] [CrossRef]
- Leng, L.; Wei, L.; Xiong, Q.; Xu, S.; Li, W.; Lv, S.; Lu, Q.; Wan, L.; Wen, Z.; Zhou, W. Use of microalgae based technology for the removal of antibiotics from wastewater: A review. Chemosphere 2020, 238, 124680. [Google Scholar] [CrossRef]
- Phoon, B.L.; Ong, C.C.; Mohamed Saheed, M.S.; Show, P.L.; Chang, J.S.; Ling, T.C.; Lam, S.S.; Juan, J.C. Conventional and emerging technologies for removal of antibiotics from wastewater. J. Hazard. Mater. 2020, 400, 122961. [Google Scholar] [CrossRef]
- Pelalak, R.; Alizadeh, R.; Ghareshabani, E.; Heidari, Z. Degradation of sulfonamide antibiotics using ozone-based advanced oxidation process: Experimental, modeling, transformation mechanism and DFT study. Sci. Total Environ. 2020, 734, 139446. [Google Scholar] [CrossRef]
- Luo, L.; Wang, G.; Wang, Z.; Ma, J.; He, Y.; He, J.; Wang, L.; Liu, Y.; Xiao, H.; Xiao, Y.; et al. Optimization of Fenton process on removing antibiotic resistance genes from excess sludge by single-factor experiment and response surface methodology. Sci. Total Environ. 2021, 788, 147889. [Google Scholar] [CrossRef]
- Vilela, P.B.; Mendonca Neto, R.P.; Starling, M.; da Martins, S.A.; Pires, G.F.F.; Souza, F.A.R.; Amorim, C.C. Metagenomic analysis of MWWTP effluent treated via solar photo-Fenton at neutral pH: Effects upon microbial community, priority pathogens, and antibiotic resistance genes. Sci. Total Environ. 2021, 801, 149599. [Google Scholar] [CrossRef]
- Ahmed, Y.; Lu, J.; Yuan, Z.; Bond, P.L.; Guo, J. Efficient inactivation of antibiotic resistant bacteria and antibiotic resistance genes by photo-Fenton process under visible LED light and neutral pH. Water Res. 2020, 179, 115878. [Google Scholar] [CrossRef]
- Tang, H.; Shang, Q.; Tang, Y.; Liu, H.; Zhang, D.; Du, Y.; Liu, C. Filter-membrane treatment of flowing antibiotic-containing wastewater through peroxydisulfate-coupled photocatalysis to reduce resistance gene and microbial inhibition during biological treatment. Water Res. 2021, 207, 117819. [Google Scholar] [CrossRef]
- Fang, H.; Zhang, Q.; Nie, X.; Chen, B.; Xiao, Y.; Zhou, Q.; Liao, W.; Liang, X. Occurrence and elimination of antibiotic resistance genes in a long-term operation integrated surface flow constructed wetland. Chemosphere 2017, 173, 99–106. [Google Scholar] [CrossRef]
- Sharma, V.K.; Johnson, N.; Cizmas, L.; McDonald, T.J.; Kim, H. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 2016, 150, 702–714. [Google Scholar] [CrossRef]
- Lv, M.; Zhang, D.; Niu, X.; Ma, J.; Lin, Z.; Fu, M. Insights into the fate of antibiotics in constructed wetland systems: Removal performance and mechanisms. J. Environ. Manag. 2022, 321, 116028. [Google Scholar] [CrossRef]
- Chen, J.; Ying, G.G.; Wei, X.D.; Liu, Y.S.; Liu, S.S.; Hu, L.X.; He, L.Y.; Chen, Z.F.; Chen, F.R.; Yang, Y.Q. Removal of antibiotics and antibiotic resistance genes from domestic sewage by constructed wetlands: Effect of flow configuration and plant species. Sci. Total Environ. 2016, 571, 974–982. [Google Scholar] [CrossRef]
- Sakurai, K.; Pompei, C.; Tomita, I.; Santos-Neto, A.; Silva, G. Hybrid constructed wetlands as post-treatment of blackwater: An assessment of the removal of antibiotics. J. Environ. Manag. 2021, 278, 111552. [Google Scholar] [CrossRef]
- Tang, X.Y.; Liu, H.P.; Naila, R.S.L.; Dai, Y.; Zhang, X.M.; Tam, N.F.Y.; Xiong, C.H.; Yang, Y. Irrigation using hybrid constructed wetland treated domestic sewage: Uptake of phthalic acid esters and antibiotics by Ipomoea aquatica forssk. J. Hazard. Mater. 2021, 405, 124025. [Google Scholar] [CrossRef]
- Sabri, N.A.; Schmitt, H.; van der Zaan, B.M.; Gerritsen, H.W.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Performance of full scale constructed wetlands in removing antibiotics and antibiotic resistance genes. Sci. Total Environ. 2021, 786, 147368. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Zheng, J.; Huang, X.; Wang, Z.; Liu, Y.; Zhu, G. Elimination of veterinary antibiotics and antibiotic resistance genes from swine wastewater in the vertical flow constructed wetlands. Chemosphere 2013, 91, 1088–1093. [Google Scholar] [CrossRef]
- Avila, C.; Garcia-Galan, M.J.; Borrego, C.M.; Rodriguez-Mozaz, S.; Garcia, J.; Barcelo, D. New insights on the combined removal of antibiotics and ARGs in urban wastewater through the use of two configurations of vertical subsurface flow constructed wetlands. Sci. Total Environ. 2021, 755, 142554. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Ngo, H.H.; Guo, W.; Lee, D.; Nghiem, D.L.; Zhang, J.; Liang, S.; Varjani, S.; Wang, J. Performance of microbial fuel cell for treating swine wastewater containing sulfonamide antibiotics. Bioresour. Technol. 2020, 311, 123588. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Zhao, L.; Chen, J.; Kim, J.; Huang, C.H.; Pavlostathis, S.G. Tetracycline inhibition and transformation in microbial fuel cell systems: Performance, transformation intermediates, and microbial community structure. Bioresour. Technol. 2021, 322, 124534. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, H.; Zhang, S.; Li, S.; Zeng, W.; Li, F. The influence of external resistance on the performance of microbial fuel cell and the removal of sulfamethoxazole wastewater. Bioresour. Technol. 2021, 336, 125308. [Google Scholar] [CrossRef]
- Yang, X.L.; Wang, Q.; Li, T.; Xu, H.; Song, H.L. Antibiotic removal and antibiotic resistance genes fate by regulating bioelectrochemical characteristics in microbial fuel cells. Bioresour. Technol. 2022, 348, 126752. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, T.; Chen, J.; Guo, J.; Luo, H.; Chen, W.; Mo, Y.; Wei, Z.; Huang, X. The reduction and fate of antibiotic resistance genes (ARGs) and mobile genetic elements (MGEs) in microbial fuel cell (MFC) during treatment of livestock wastewater. J. Contam. Hydrol. 2022, 247, 103981. [Google Scholar] [CrossRef]
- Xu, H.; Song, H.L.; Singh, R.P.; Yang, Y.L.; Xu, J.Y.; Yang, X.L. Simultaneous reduction of antibiotics leakage and methane emission from constructed wetland by integrating microbial fuel cell. Bioresour. Technol. 2021, 320, 124285. [Google Scholar] [CrossRef]
- Dai, M.; Wu, Y.; Wang, J.; Lv, Z.; Li, F.; Zhang, Y.; Kong, Q. Constructed wetland-microbial fuel cells enhanced with iron carbon fillers for ciprofloxacin wastewater treatment and power generation. Chemosphere 2022, 305, 135377. [Google Scholar] [CrossRef]
- Song, H.-L.; Zhang, C.; Lu, Y.-X.; Li, H.; Shao, Y.; Yang, Y.-L. Enhanced removal of antibiotics and antibiotic resistance genes in a soil microbial fuel cell via in situ remediation of agricultural soils with multiple antibiotics. Sci. Total Environ. 2022, 829, 154406. [Google Scholar] [CrossRef]
- Guo, R.; Chen, J. Application of alga-activated sludge combined system (AASCS) as a novel treatment to remove cephalosporins. Chem. Eng. J. 2015, 260, 550–556. [Google Scholar] [CrossRef]
- Pan, M.; Lyu, T.; Zhan, L.; Matamoros, V.; Angelidaki, I.; Cooper, M.; Pan, G. Mitigating antibiotic pollution using cyanobacteria: Removal efficiency, pathways and metabolism. Water Res. 2021, 190, 116735. [Google Scholar] [CrossRef]
- Song, C.; Wei, Y.; Qiu, Y.; Qi, Y.; Li, Y.; Kitamura, Y. Biodegradability and mechanism of florfenicol via Chlorella sp. UTEX1602 and L38: Experimental study. Bioresour. Technol. 2019, 272, 529–534. [Google Scholar] [CrossRef]
- Yang, K.; Lu, J.; Jiang, W.; Jiang, C.; Chen, J.; Wang, Z.; Guo, R. An integrated view of the intimate coupling UV irradiation and algal treatment on antibiotic: Compatibility, efficiency and microbic impact assessment. J. Environ. Chem. Eng. 2017, 5, 4262–4268. [Google Scholar] [CrossRef]
- Bhattacharya, S.S.; Yadav, J.S. Microbial P450 enzymes in bioremediation and drug discovery: Emerging potentials and challenges. Curr. Protein Pept. Sci. 2018, 19, 75–86. [Google Scholar] [CrossRef]
- Ezzariai, A.; Hafidi, M.; Khadra, A.; Aemig, Q.; El Fels, L.; Barret, M.; Merlina, G.; Patureau, D.; Pinelli, E. Human and veterinary antibiotics during composting of sludge or manure: Global perspectives on persistence, degradation, and resistance genes. J. Hazard. Mater. 2018, 359, 465–481. [Google Scholar] [CrossRef]
- Lin, H.; Sun, W.; Yu, Y.; Ding, Y.; Yang, Y.; Zhang, Z.; Ma, J. Simultaneous reductions in antibiotics and heavy metal pollution during manure composting. Sci. Total Environ. 2021, 788, 147830. [Google Scholar] [CrossRef]
- Cui, E.; Cui, B.; Fan, X.; Li, S.; Gao, F. Ryegrass (Lolium multiflorum L.) and Indian mustard (Brassica juncea L.) intercropping can improve the phytoremediation of antibiotics and antibiotic resistance genes but not heavy metals. Sci. Total Environ. 2021, 784, 147093. [Google Scholar] [CrossRef]
- Marti, E.; Monclus, H.; Jofre, J.; Rodriguez-Roda, I.; Comas, J.; Balcazar, J.L. Removal of microbial indicators from municipal wastewater by a membrane bioreactor (MBR). Bioresour. Technol. 2011, 102, 5004–5009. [Google Scholar] [CrossRef]
- Le, T.H.; Ng, C.; Tran, N.H.; Chen, H.; Gin, K.Y. Removal of antibiotic residues, antibiotic resistant bacteria and antibiotic resistance genes in municipal wastewater by membrane bioreactor systems. Water Res. 2018, 145, 498–508. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, Z.; Zhou, S.; Jin, M.; Lu, T.; Cui, L.; Qian, H. Macleaya cordata extract, an antibiotic alternative, does not contribute to antibiotic resistance gene dissemination. J. Hazard. Mater. 2021, 412, 125272. [Google Scholar] [CrossRef]
- Jia, Y.; Yang, B.; Shi, J.; Fang, D.; Wang, Z.; Liu, Y. Melatonin prevents conjugative transfer of plasmid-mediated antibiotic resistance genes by disrupting proton motive force. Pharmacol. Res. 2022, 175, 105978. [Google Scholar] [CrossRef] [PubMed]
- Carfrae, L.A.; MacNair, C.R.; Brown, C.M.; Tsai, C.N.; Weber, B.S.; Zlitni, S.; Rao, V.N.; Chun, J.; Junop, M.S.; Coombes, B.K.; et al. Mimicking the human environment in mice reveals that inhibiting biotin biosynthesis is effective against antibiotic-resistant pathogens. Nat. Microbiol. 2020, 5, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.K.; Alhhazmi, A.; DeCoteau, J.F.; Luo, Y.; Geyer, C.R. RecA inhibitors potentiate antibiotic activity and block evolution of antibiotic resistance. Cell Chem. Biol. 2016, 23, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.L.; Chen, Z.G.; Yang, T.C.; Jiang, M.; Wang, J.; Cheng, Z.X.; Yang, M.J.; Zhu, J.X.; Zhang, T.T.; Li, H.; et al. Glutamine promotes antibiotic uptake to kill multidrug-resistant uropathogenic bacteria. Sci. Transl. Med. 2021, 13, eabj0716. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schaberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]

| Antimicrobials | Organism | Antimicrobial Concentration | Effects | Reference |
|---|---|---|---|---|
| A five-component mixture (amoxicillin, ciprofloxacin, spiramycin, sulfamethoxazole, and tetracycline) | Microcystis aeruginosa NIES-843 | 50–500 ng/L | increased the concentration of microcystin, promoted the growth | [56] |
| Erythromycin | Microcystis flos-aquae | >10 μg/L | inhibited the growth and photosynthesis | [60] |
| Tilmicosin | Danio rerio | >2 mg/L | induced deformities and lethality | [63] |
| Oxytetracycline | Danio rerio | 1 μg/L, 100 μg/L | affected the thyroid hormone homeostasis, reduced tryptophan hydroxylase | [64] |
| Norfloxacin | Carp | 100 ng/L | induced the oxidative stress, damaged the barrier function of the intestine | [65] |
| Amoxicillin | BALB/c mice | 50 mg/kg/day | reduced recognition memory, increased depression | [68] |
| Enrofloxacin and ciprofloxacin | Rhinella arenarum | >10 μg/L | affected the development, growth | [69] |
| Chlortetracycline | Brassica campestris | 1 mg/L | shortened primary root length, decreased chlorophyll level | [72] |
| Enrofloxacin | Soybean | 10 μg/L | reduced yield | [74] |
| Pathogens | Diseases | Treatment Antimicrobials | ARGs | Reference |
|---|---|---|---|---|
| Staphylococcus aureus | endocarditis, pneumonia | Lincomycin, Vancomycin | mecA, VanA | [150] |
| Streptococcus pneumoniae | pneumonia, bacteremia | Amoxicillin, Piperacillin | Mef, emr(B) | [151] |
| Enterococcus | urinary tract infection, endocarditis | Ampicillin, Piperacillin | VanA, ermB, tet(L), cat, parC, Cfr | [152] |
| Klebsiella pneumoniae | pneumonia, pulmonary abscess, endocarditis | Gentamicin, Kanamycin | blaKPC-2, blaCTX-M-14, blaTEM-1, dfrA25 | [153] |
| Pseudomonas aeruginosa | cystic fibrosis, ventilator-associated pneumonia | Gentamicin, Meropenem, Ceftazidime | crc, lon, psrA, ampD, gyrA, nalA, nfxB, cbrA | [154] |
| Acinetobacter baumannii | respiratory infections, urinary tract infection, meningitis | Imipenem, Polymyxin B | pmrC, blaADC-25, aadA, macA, gyrA, oprD, rpoB | [155] |
| Escherichia coli | gastrointestinal infection, urinary tract infection, arthritis, meningitis | Ampicillin, Amoxicillin | cmlA, flor, aadA, sul1, sul2, tetA, blaCTX-M, blaTEM, aphA3, qnrA, qnrS, OqxAB | [156] |
| Medicine | Object of Study | Effects | Application | Reference |
|---|---|---|---|---|
| Macleaya cordata extract | Enchytraeus crypticus | Decreases ARGs abundance | Anti-inflammatory effects, as a feed supplement | [200] |
| Melatonin | Escherichia coli DH5α | Prevents plasmid-mediated binding transfer of antimicrobial resistance genes by disrupting proton dynamics | As inhibitors of ARGs transmission | [201] |
| Biotin biosynthesis inhibitors (MAC13772) | Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Acinetobacter baumannii | Inhibits biotin synthesis to kill the pathogenic bacterium | Infection treatment | [202] |
| RecA Inhibitors | Escherichia coli ATCC25922 | Enhance the activity of bactericidal antimicrobials, reduce the acquisition of antimicrobial resistance mutations, block the horizontal transfer of mobile genetic elements | As an adjunct to antimicrobials | [203] |
| Glutamine | multidrug resistant Escherichia coli | Promotes bacterial uptake of antimicrobials to kill multidrug-resistant uropathogenic bacteria | As an adjunct to antimicrobials | [204] |
| Teixobactin | Staphylococcus aureus, Mycobacterium tuberculosis, Eleftheria terrae | Inhibits cell wall synthesis to kill pathogens without detectable resistance | A new antimicrobial | [205] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Weng, Y.; Luo, T.; Wang, Q.; Yang, G.; Jin, Y. Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies. Toxics 2023, 11, 185. https://doi.org/10.3390/toxics11020185
Wang W, Weng Y, Luo T, Wang Q, Yang G, Jin Y. Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies. Toxics. 2023; 11(2):185. https://doi.org/10.3390/toxics11020185
Chicago/Turabian StyleWang, Weitao, You Weng, Ting Luo, Qiang Wang, Guiling Yang, and Yuanxiang Jin. 2023. "Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies" Toxics 11, no. 2: 185. https://doi.org/10.3390/toxics11020185
APA StyleWang, W., Weng, Y., Luo, T., Wang, Q., Yang, G., & Jin, Y. (2023). Antimicrobial and the Resistances in the Environment: Ecological and Health Risks, Influencing Factors, and Mitigation Strategies. Toxics, 11(2), 185. https://doi.org/10.3390/toxics11020185