Degradation of Benzo[a]pyrene and 2,2′,4,4′-Tebrabrominated Diphenyl Ether in Cultures Originated from an Agricultural Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Soil
2.2. Experimental Setup
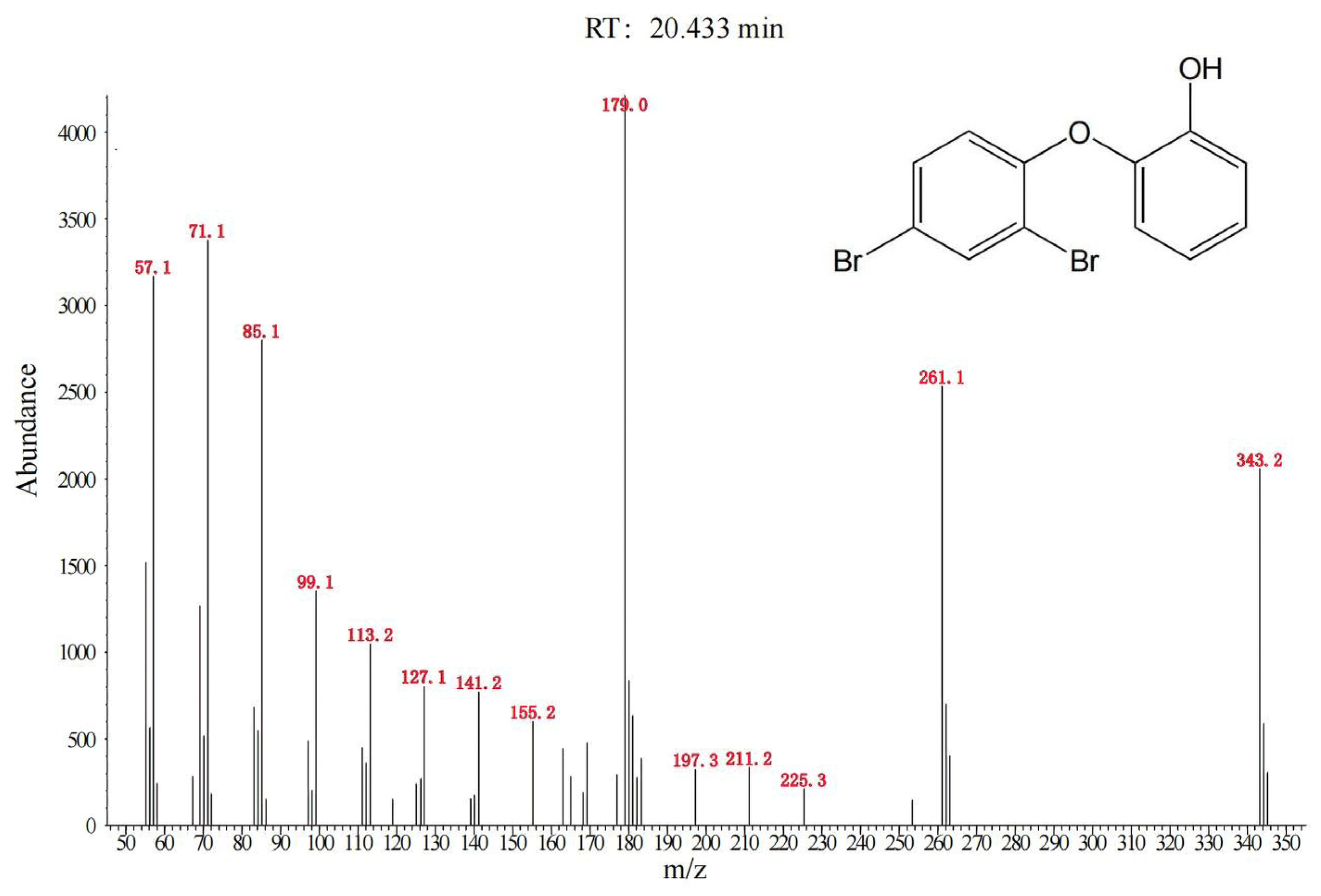
2.3. Analyses of BaP, BDE-47 and Their Degradation Products
2.4. Analyses of Bacterial Community Structure
3. Results and Discussion
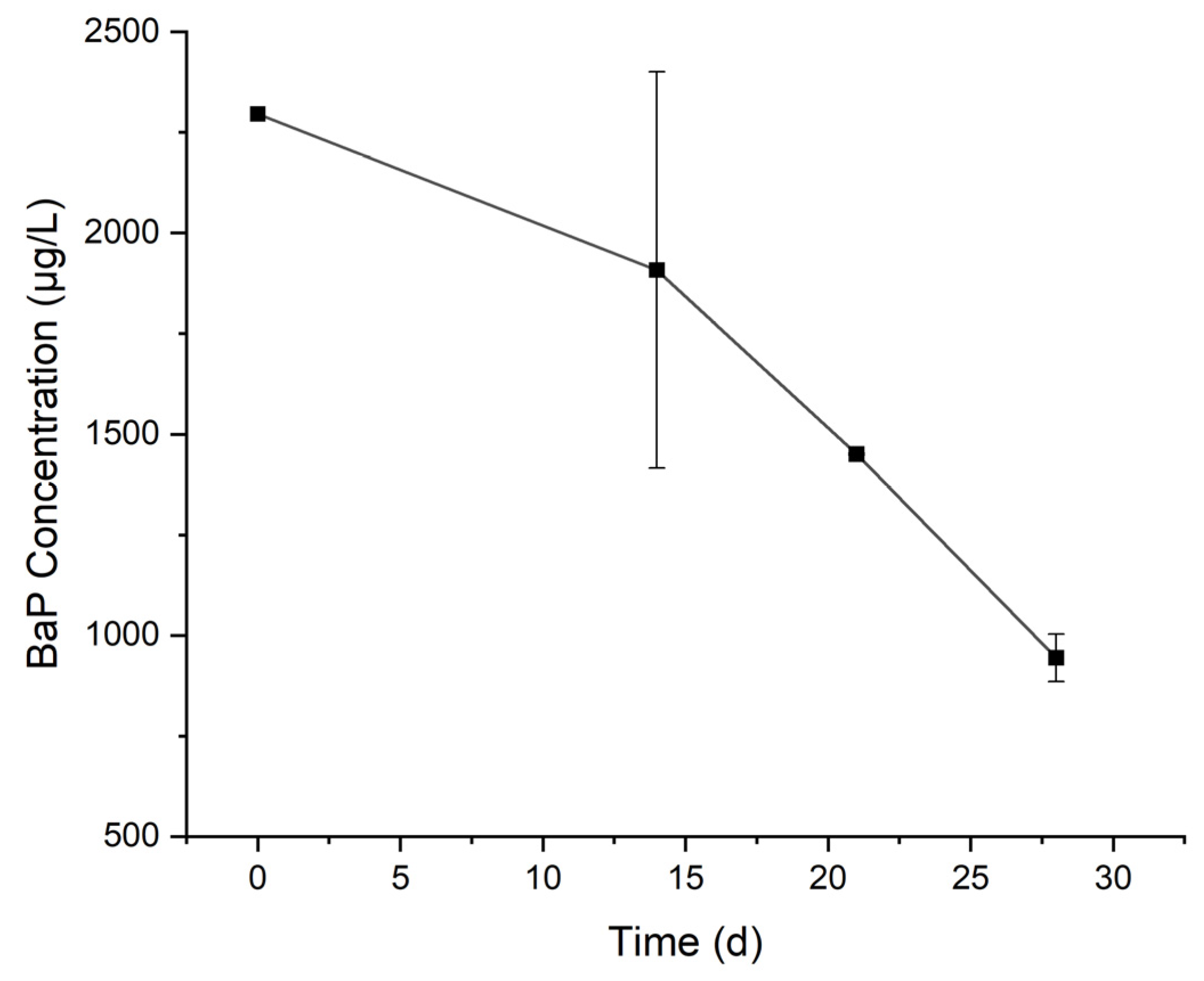
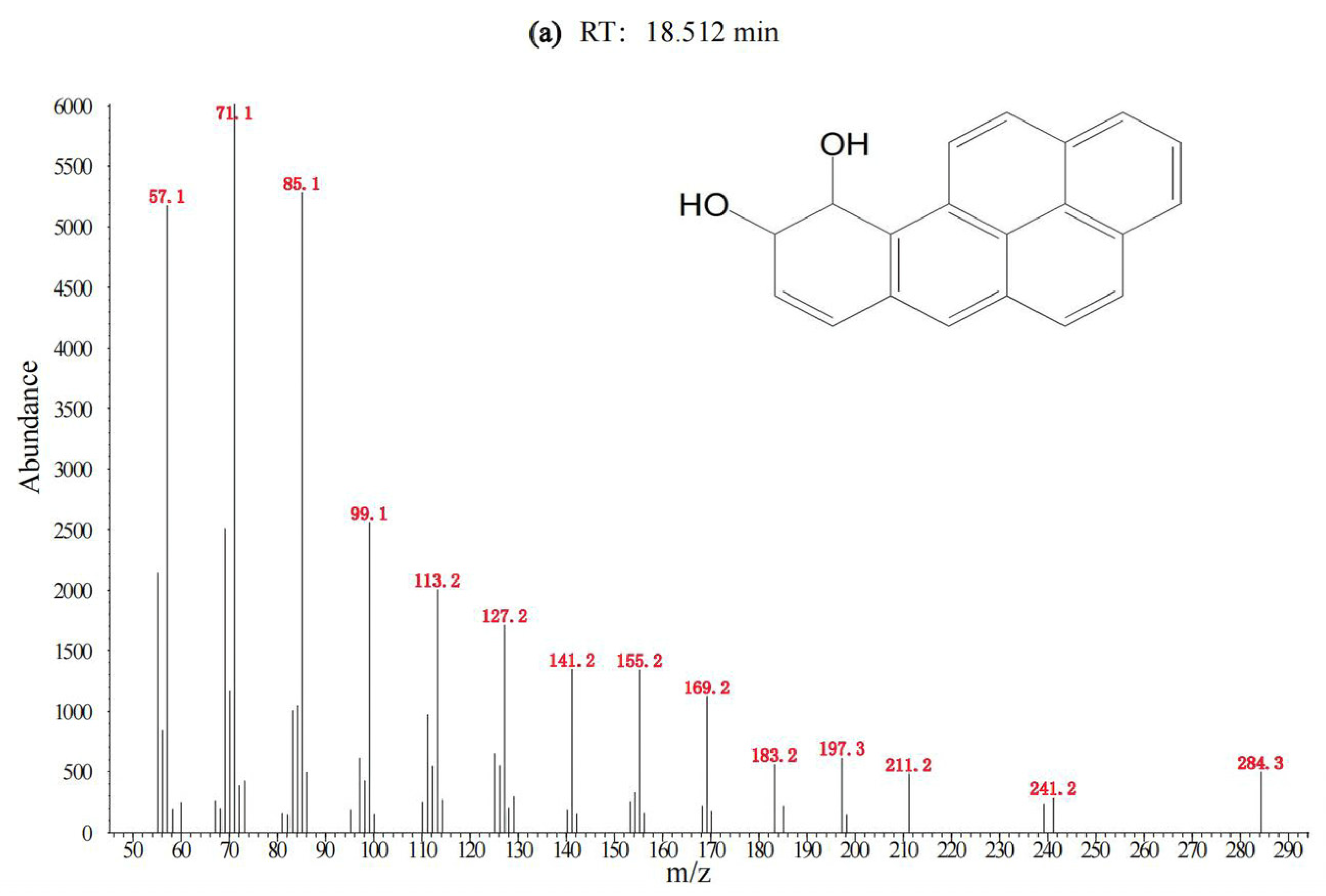
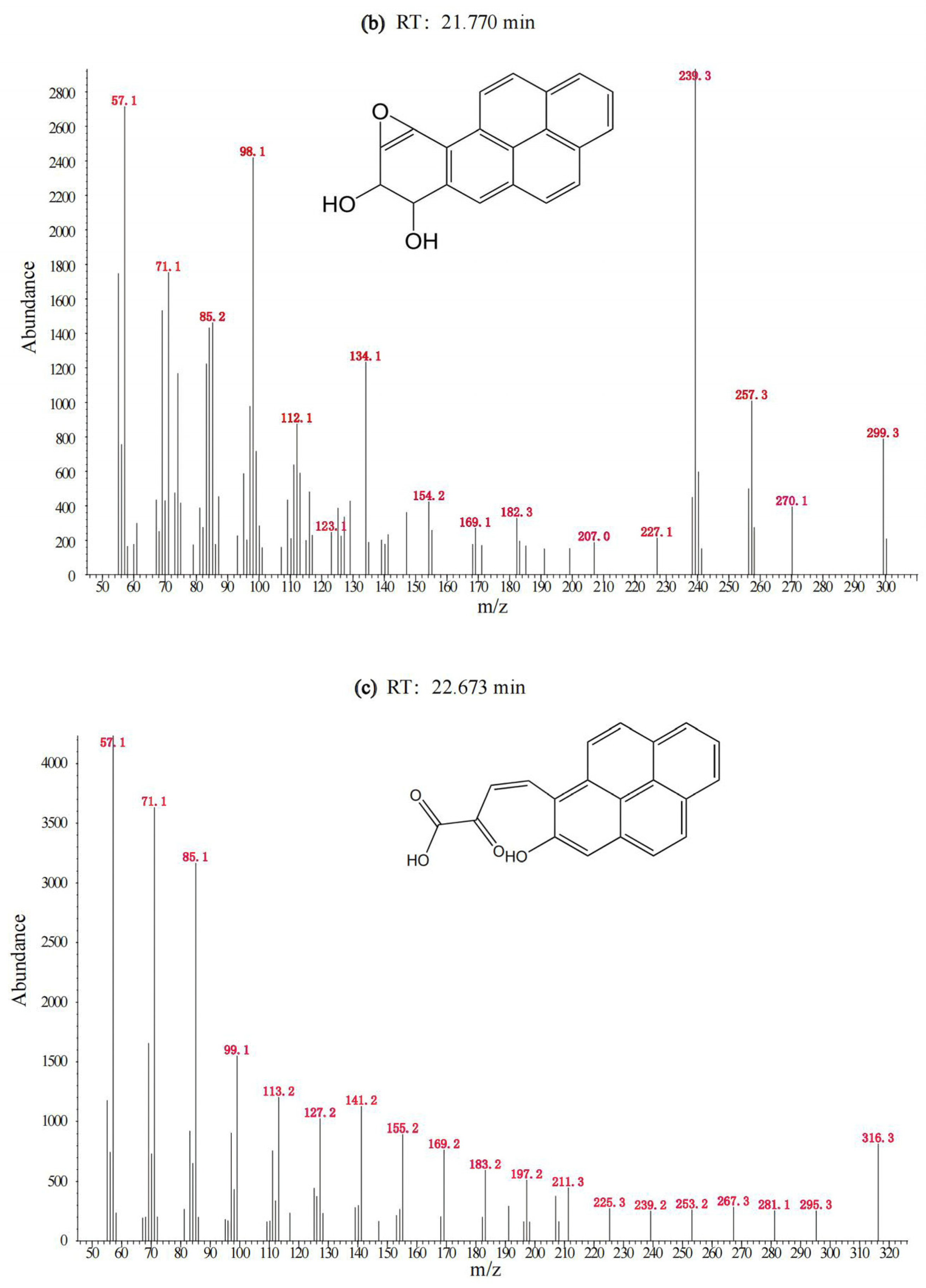
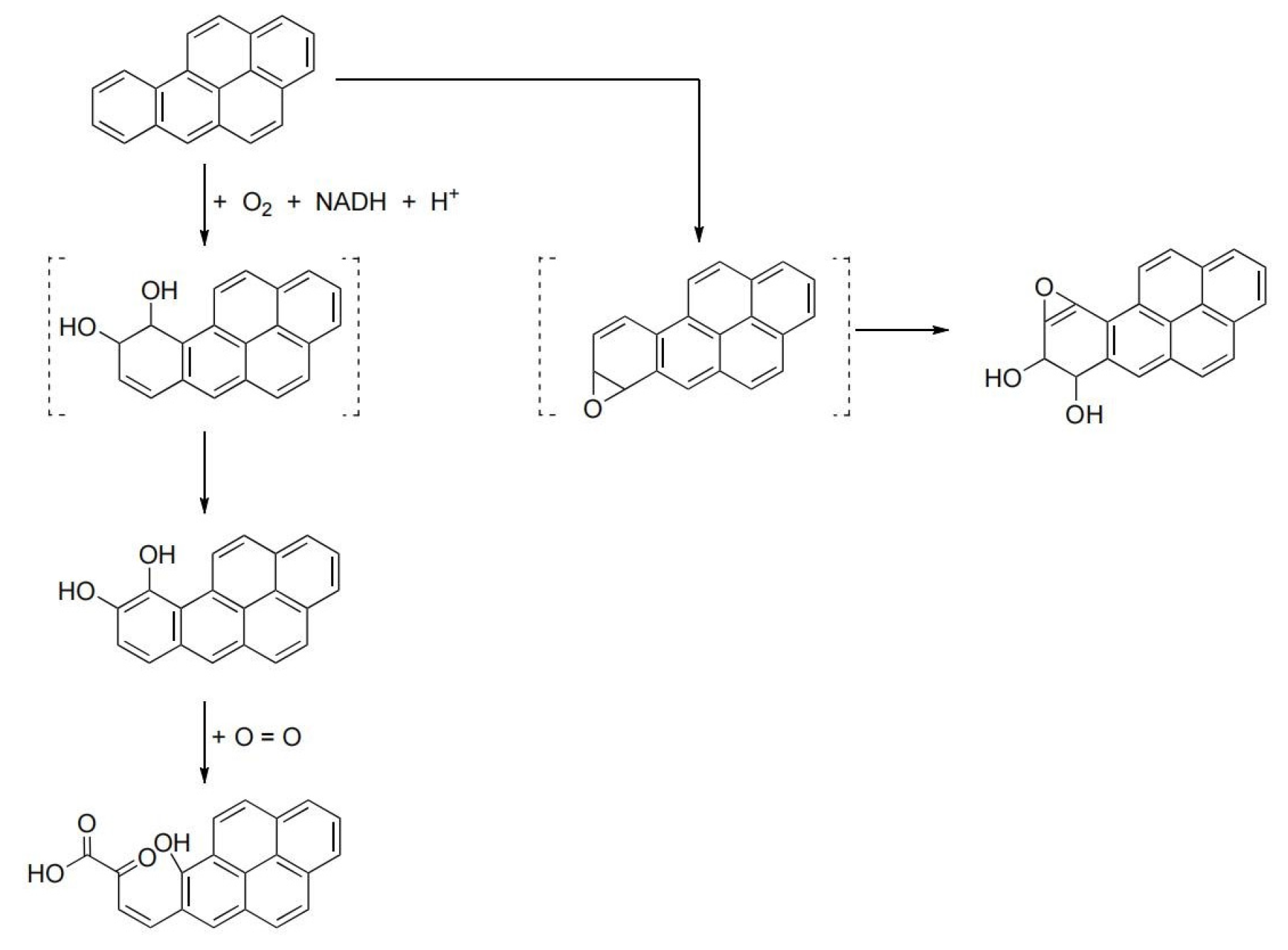
3.1. Degradation of BaP by the Cultured Microorganisms
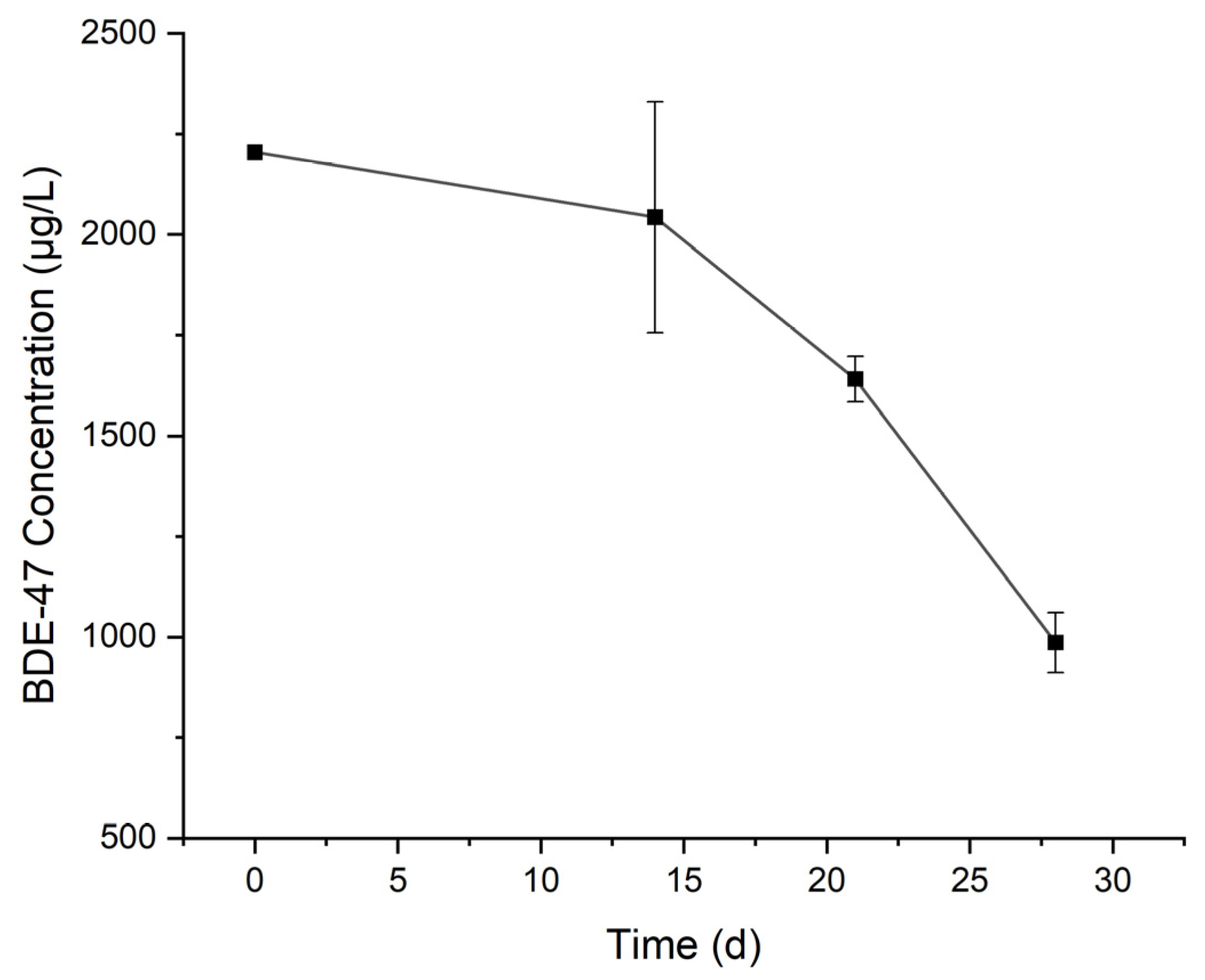
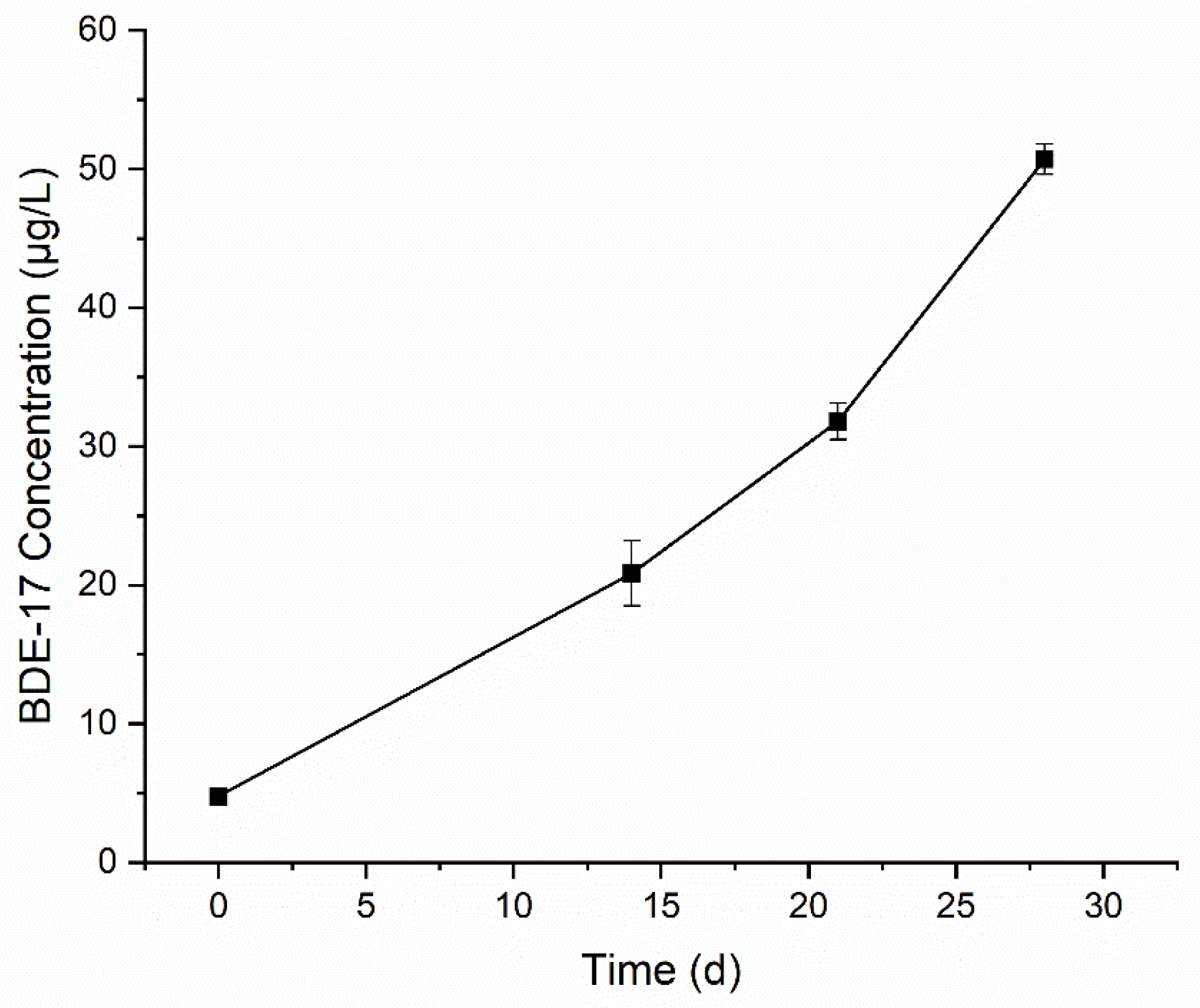
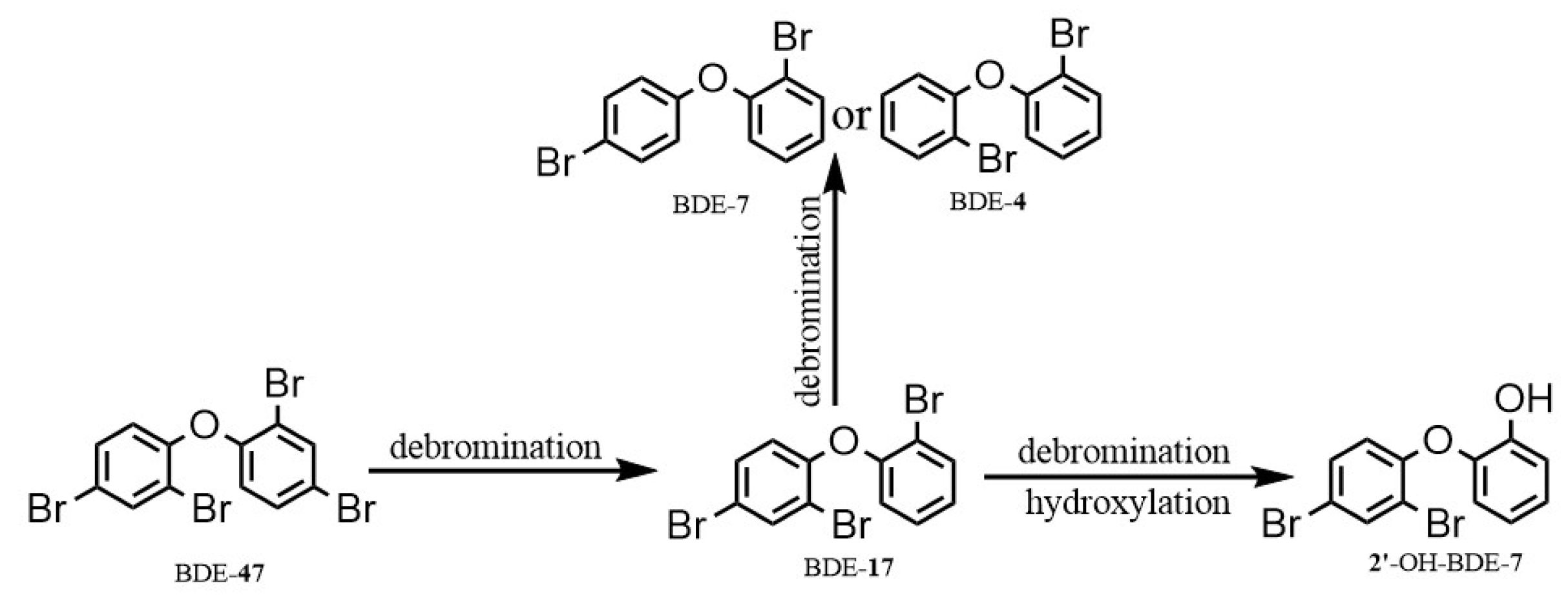
3.2. Degradation of BDE-47 by the Cultured Microorganisms
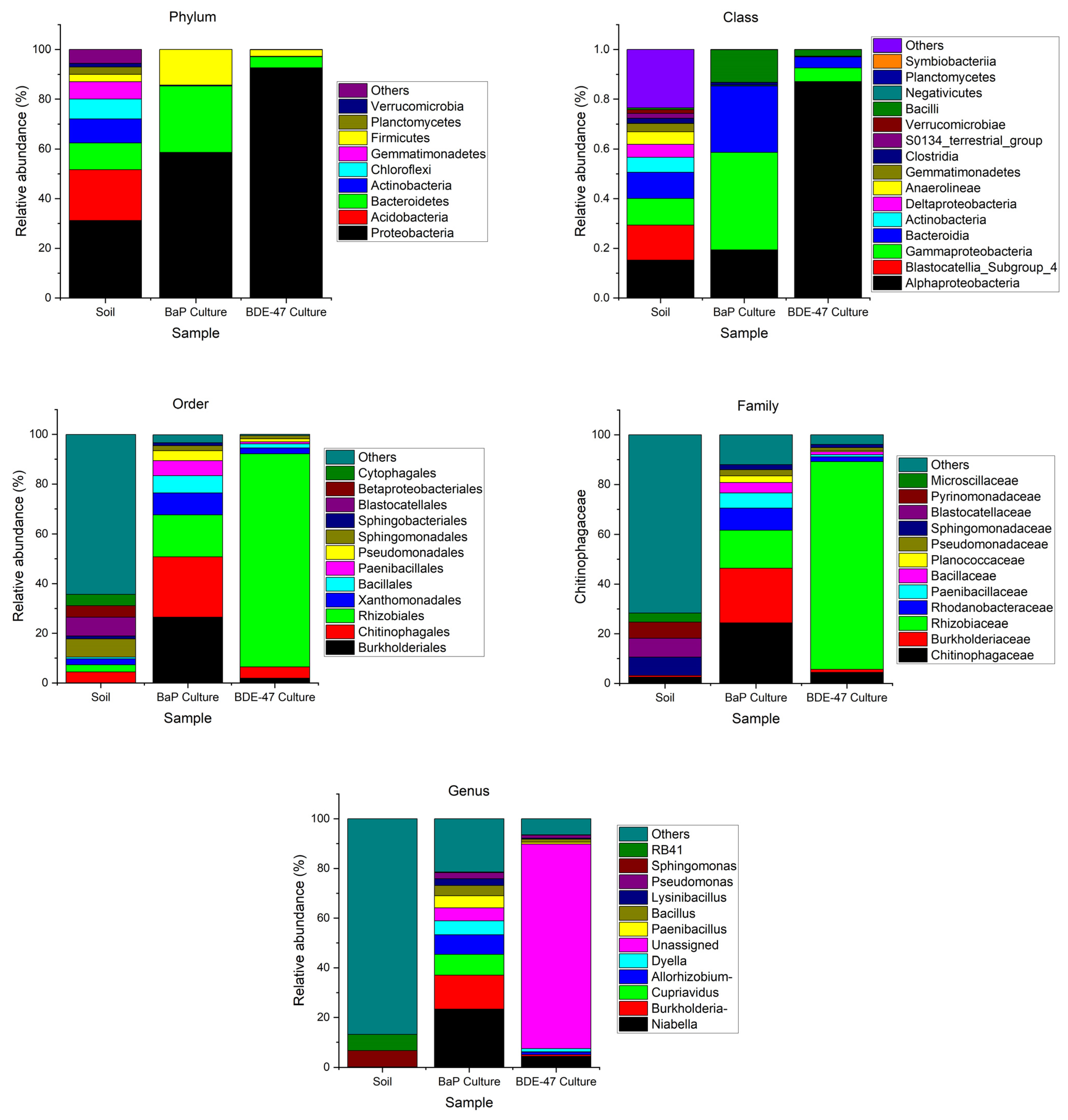
3.3. Characterization of BaP and BDE-47 Degrading Bacteria in the Culture Medium
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dai, C.; Han, Y.; Duan, Y.; Lai, X.; Fu, R.; Liu, S.; Leong, K.H.; Tu, Y.; Zhou, L. Review on the contamination and remediation of polycyclic aromatic hydrocarbons (PAHs) in coastal soil and sediments. Environ. Res. 2022, 205, 112423. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Liang, Y.; Fang, W.; Guan, K.; Huang, C.; Qi, X.; Liang, Z.; Zeng, Y.; Luo, X.; He, Z.; et al. Spatial Distribution, Bioconversion and Ecological Risk of PCBs and PBDEs in the Surface Sediment of Contaminated Urban Rivers: A Nationwide Study in China. Environ. Sci. Technol. 2021, 55, 9397–10188. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, G.; Yin, W.; Gong, H.; Zhou, X.; Wang, L.; Li, M.; Chen, B. Pollution characteristics and environment risk assessment of PAHs in agriculture soil around an electronic waste dismantling venues. Environ. Pollut. Control. 2015, 37, 1–5. [Google Scholar]
- Wei, B.K.; Liu, C.; Wang, Y.; Jin, J. Polybrominated diphenyl ether in e-waste dismantling sites in Taizhou City, Zhejiang Province: Concentration, distribution, and migration trend. Environ. Sci. 2020, 41, 4740–4748. [Google Scholar]
- Bukowska, B.; Mokra, K.; Michałowicz, J. Benzo[a]pyrene—Environmental occurrence, human exposure, and mechanisms of toxicity. Int. J. Mol. Sci. 2022, 23, 6348. [Google Scholar] [CrossRef] [PubMed]
- Dingemans, M.M.L.; Berg, M.; Westerink, R.H.S. Neurotoxicity of brominated flame retardants: (In) direct effects of parent and hydroxylated polybrominated diphenyl ethers on the (developing) nervous system. Environ. Health Perspect. 2011, 119, 900–907. [Google Scholar] [CrossRef] [PubMed]
- Punetha, A.; Saraswat, S.; Rai, J.P.N. An insight on microbial degradation of benzo[a]pyrene: Current status and advances in research. World J. Microbiol. Biotechnol. 2022, 38, 61. [Google Scholar] [CrossRef] [PubMed]
- Nzila, A.; Musa, M.M.; Afuecheta, E.; Al-Thukair, A.; Sankaran, S.; Xiang, L.; Li, Q.X. Benzo[a]pyrene biodegradation by multiple and individual mesophilic bacteria under axenic conditions and in soil samples. Int. J. Environ. Res. Public Health 2023, 20, 1855. [Google Scholar] [CrossRef]
- Zhao, Z.; Wong, J.W. Rapid biodegradation of benzo[a]pyrene by Bacillus subtilis BUM under thermophilic conditions. Environ. Eng. Sci. 2009, 27, 939–945. [Google Scholar] [CrossRef]
- Aziz, A.; Agamuthu, P.; Alaribe, F.O.; Fauziah, S.H. Biodegradation of benzo[a]Pyrene by bacterial consortium isolated from mangrove sediment. Environ. Technol. 2018, 39, 527–535. [Google Scholar] [CrossRef]
- Lee, L.K.; Ding, C.; Yang, K.L.; He, J. Complete debromination of tetra and pentabrominated diphenyl ethers by a coculture consisting of Dehalococcoides and Desulfovibrio species. Environ. Sci. Technol. 2011, 45, 8475–8482. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Chow, W.L.; He, J. Isolation of Acetobacterium sp. Strain AG, which reductively debrominates octa- and penta-brominated diphenyl ether technical mixtures. Appl. Environ. Microbiol. 2013, 79, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ding, C.; Xu, G.; Rogers, M.J.; Ramaswamy, R.; He, J. Diversity of organohalide respiring bacteria and reductive dehalogenases that detoxify polybrominated diphenyl ethers in E-waste recycling sites. ISME J. 2022, 16, 2123–2131. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Ng, H.L.; Chen, C.; Zhao, S.; He, J. Offshore marine sediment microbiota respire structurally distinct organohalide pollutants. Environ. Sci. Technol. 2022, 56, 8008–8019. [Google Scholar] [CrossRef]
- Xu, G.; Zhao, S.; Chen, C.; Zhao, X.; Ramaswamy, R.; He, J. Dehalogenation of polybrominated diphenyl ethers and polychlorinated biphenyls catalyzed by a reductive dehalogenase in Dehalococcoides mccartyi Strain MB. Environ. Sci. Technol. 2022, 56, 3831–4690. [Google Scholar] [CrossRef]
- Tokarz, J.A., 3rd; Ahn, M.-Y.; Leng, J.; Filley, T.R.; Nies, L. Reductive debromination of polybrominated diphenyl ethers in anaerobic sediment and a biomimetic system. Environ. Sci. Technol. 2008, 42, 1157–1164. [Google Scholar] [CrossRef]
- Huo, L.; Zhao, C.; Gu, T.; Yan, M.; Zhong, H. Aerobic and anaerobic biodegradation of BDE-47 by bacteria isolated from an e-waste-contaminated site and the effect of various additives. Chemosphere 2022, 294, 133739. [Google Scholar] [CrossRef]
- Gu, C.G.; Fan, X.L.; Ti, Q.Q.; Yang, X.L.; Bian, Y.R.; Sun, C.; Jiang, X. Mechanistic insight into hydroxylation of 2,2’,4,4’-tetrabromodiphenyl ether during biodegradation by typical aerobic bacteria: Experimental and computational studies. J. Hazard Mater. 2021, 416, 126132. [Google Scholar] [CrossRef]
- Guo, Z.Y.; Yin, H.; Wei, X.P.; Zhu, M.H.; Lu, G.N.; Dang, Z. Effects of methanol on the performance of a novel BDE-47 degrading bacterial consortium QY2 in the co-metabolism process. J. Hazard Mater. 2021, 415, 125698. [Google Scholar] [CrossRef]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Miec-zkowski, P.; Jones, C.D. Root microbiota drive direct integration of phosphate stress and immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef]
- Zeng, Q.; An, S. Identifying the biogeographic patterns of rare and abundant bacterial communities using different primer sets on the loess plateau. Microorganisms 2021, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Goveas, L.C.; Selvaraj, R.; Vinayagam, R.; Sajankila, S.P.; Pugazhendhi, A. Biodegradation of benzo(a)pyrene by Pseudomonas strains, isolated from petroleum refinery effluent: Degradation, inhibition kinetics and metabolic pathway. Chemosphere 2023, 321, 138066. [Google Scholar] [CrossRef] [PubMed]
- Moody, J.D.; Freeman, J.P.; Fu, P.P.; Cerniglia1, C.E. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 2004, 70, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Shi, Y.; Zhang, B.; Wan, D.; Zhang, Q. An integrated method for studying the biodegradation of benzo[a]pyrene by Citrobacter sp. HJS-1 and interaction mechanism based on the structural model of the initial dioxygenase. Environ. Sci. Pollut. Res. 2023, 30, 85558–85568. [Google Scholar] [CrossRef]
- Qin, W.; Zhu, Y.; Fan, F.Q.; Wang, Y.Y.; Liu, X.; Ding, A.Z.; Dou, J.F. Biodegradation of benzo(a)pyrene by Microbacterium sp. strain under denitrification: Degradation pathway and effects of limiting electron acceptors or carbon source. Biochem. Eng. J. 2017, 121, 131–138. [Google Scholar] [CrossRef]
- Liang, L.; Song, X.; Kong, J.; Shen, C.; Huang, T.; Hu, Z. Anaerobic biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by a facultative anaerobe Pseudomonas sp. JP1. Biodegradation 2014, 25, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, G.; Nong, J.; Xie, Q. Biodegradation of benzo(a)pyrene by a genetically engineered Bacillus licheniformis: Degradation, metabolic pathway and toxicity analysis. Chem. Eng. J. 2023, 478, 147478. [Google Scholar] [CrossRef]
- Johnson, T.O.; Abolaji, A.O.; Omale, S.; Longdet, I.Y.; Kutshik, R.J.; Oyetayo, B.O.; Adegboyega, A.E.; Sagay, A. Benzo[a]pyrene and Benzo[a]pyrene-7,8-dihydrodiol-9,10-epoxide induced locomotor and reproductive senescence and altered biochemical parameters of oxidative damage in Canton-S Drosophila melanogaster. Toxicol. Rep. 2021, 8, 571–580. [Google Scholar] [CrossRef]
- Lu, X.; Chen, C.; Zhang, S.; Ou, Y.; Yin, L.; Wu, W. Microbial Degradation of 2, 2’, 4,4’-tetrabrominated diphenyl ether under anaerobic condition. Environ. Sci. 2012, 33, 1000–1007. [Google Scholar]
- Cao, Y.; Yin, H.; Peng, H.; Tang, S.; Lu, G.; Dang, Z. Biodegradation of 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) by Phanerochaete chrysosporium in the presence of Cd2+. Environ. Sci. Pollut. Res. 2017, 24, 11415–11424. [Google Scholar] [CrossRef]
- Li, Z.; Wang, J. Analysis of the functional gene of degrading BDE-47 by Acinetobacter pittii GB-2 based on transcriptome sequencing. Gene 2022, 844, 146826. [Google Scholar] [CrossRef] [PubMed]
- Aitken, M.D.; Stringfellow, W.T.; Nagel, R.D.; Kazunga, C.; Chen, S.H. Characteristics of phenanthrene-degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can. J. Microbiol. 1998, 44, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Walter, U.; Beyer, M.; Klein, J.; Rehm, H.J. Degradation of pyrene by Rhodococcus sp. UW1. Appl. Microbiol. Biotechnol. 1991, 34, 671–676. [Google Scholar] [CrossRef]
- Schneider, J.; Grosser, R.; Jayasimhulu, K.; Xue, W.; Warshawsky, D. Degradation of pyrene, benz[a]anthracene and benzo[a]pyrene by Mycobacterium sp. strain RJGII-135, isolated from a former coal gasification site. Appl. Environ. Microbiol. 1996, 62, 13–19. [Google Scholar] [CrossRef]
- Trzesicka-Mlynarz, D.; Ward, O.P. Degradation of polycyclic aromatic hydrocarbons (PAHs) by a mixed culture and its component pure cultures, obtained from PAH-contaminated soil. Can. J. Microbiol. 1995, 41, 470–476. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, S.; Zhang, H.; Zhang, S.; Yi, L.; Yeerkenbieke, G.; Lu, X. Degradation of Benzo[a]pyrene and 2,2′,4,4′-Tebrabrominated Diphenyl Ether in Cultures Originated from an Agricultural Soil. Toxics 2024, 12, 33. https://doi.org/10.3390/toxics12010033
Shi S, Zhang H, Zhang S, Yi L, Yeerkenbieke G, Lu X. Degradation of Benzo[a]pyrene and 2,2′,4,4′-Tebrabrominated Diphenyl Ether in Cultures Originated from an Agricultural Soil. Toxics. 2024; 12(1):33. https://doi.org/10.3390/toxics12010033
Chicago/Turabian StyleShi, Shuai, Huiqian Zhang, Shuai Zhang, Lijin Yi, Gulijiazi Yeerkenbieke, and Xiaoxia Lu. 2024. "Degradation of Benzo[a]pyrene and 2,2′,4,4′-Tebrabrominated Diphenyl Ether in Cultures Originated from an Agricultural Soil" Toxics 12, no. 1: 33. https://doi.org/10.3390/toxics12010033
APA StyleShi, S., Zhang, H., Zhang, S., Yi, L., Yeerkenbieke, G., & Lu, X. (2024). Degradation of Benzo[a]pyrene and 2,2′,4,4′-Tebrabrominated Diphenyl Ether in Cultures Originated from an Agricultural Soil. Toxics, 12(1), 33. https://doi.org/10.3390/toxics12010033






