Molecular Characteristics of Aberrant Gene Mutations and Expression Profiles Induced by Benzo(a)pyrene in Hepatocellular Carcinoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultures and BaP Exposure
2.2. RNA Quantification and Quality Assessment
2.3. Transcriptome Sequencing
2.4. Exosome RNA Analysis
2.5. Analysis of Differential Gene Expression
2.6. Analysis of Gene Ontology and Pathways for the Gene Set
2.7. Enrichment of Joint Pathways for the Integrated Analysis of Gene and Metabolomics Datasets
2.8. Network Construction and Analysis
2.9. SNP and AS Analysis
3. Results
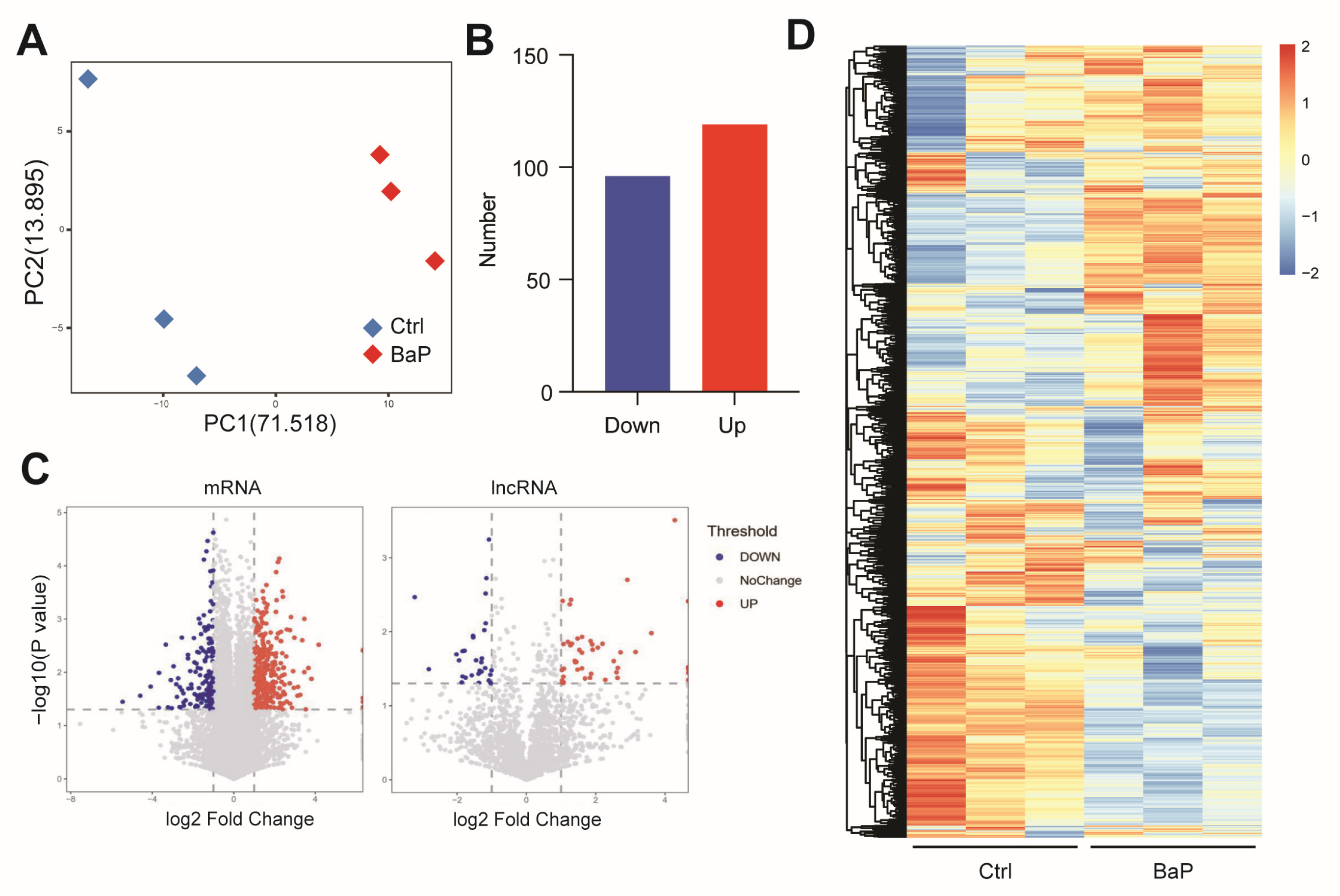
3.1. Long-Term Exposure to Low-Dose BaP Significantly Altered Gene Expressions in HCC Cells
3.2. BaP Exposure Significantly Affects Metastasis-Related Gene Expressions in HCC Cells
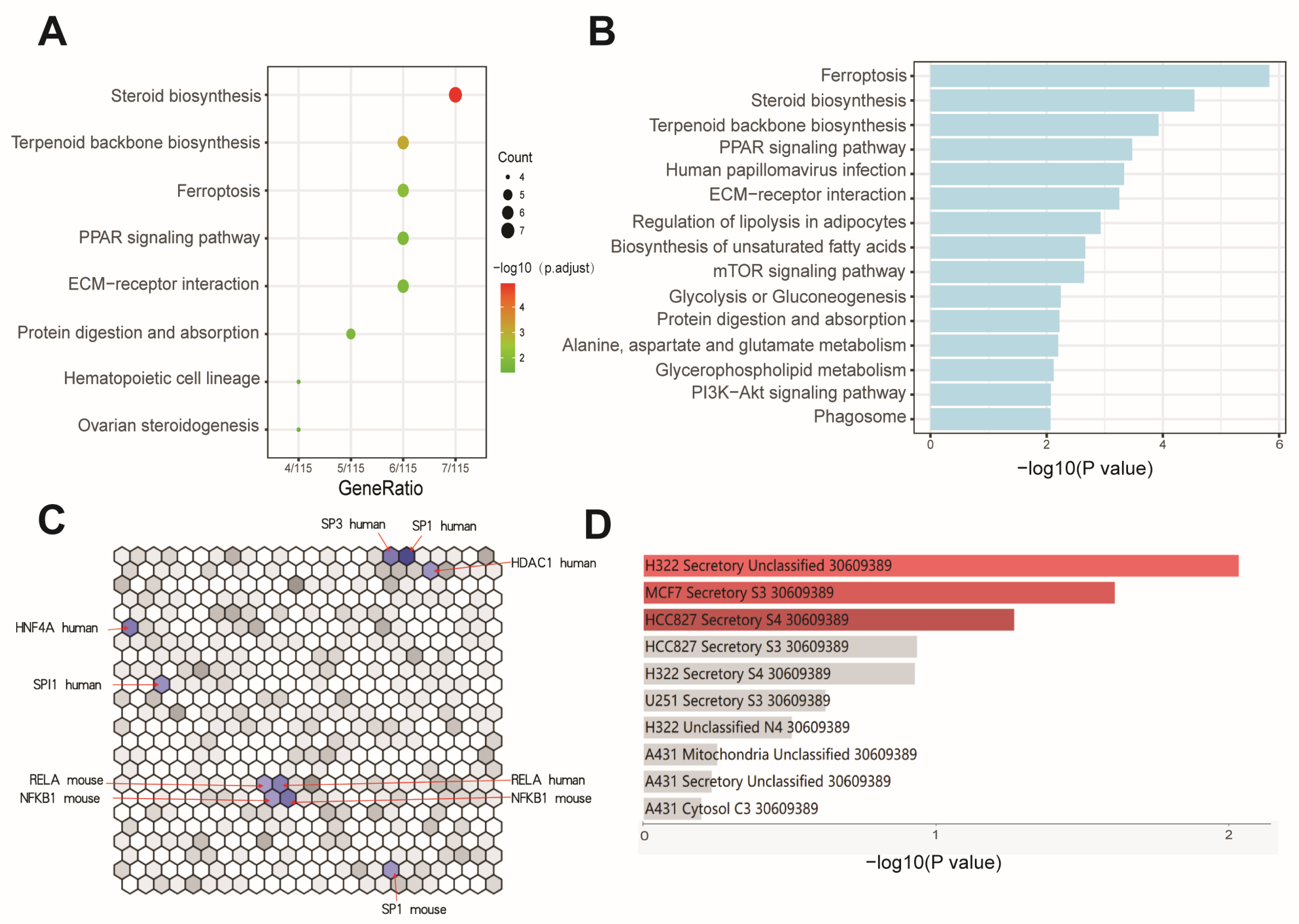
3.3. Effects of BaP Exposure on Metabolic Processes and Key Pathways in HCC Cells
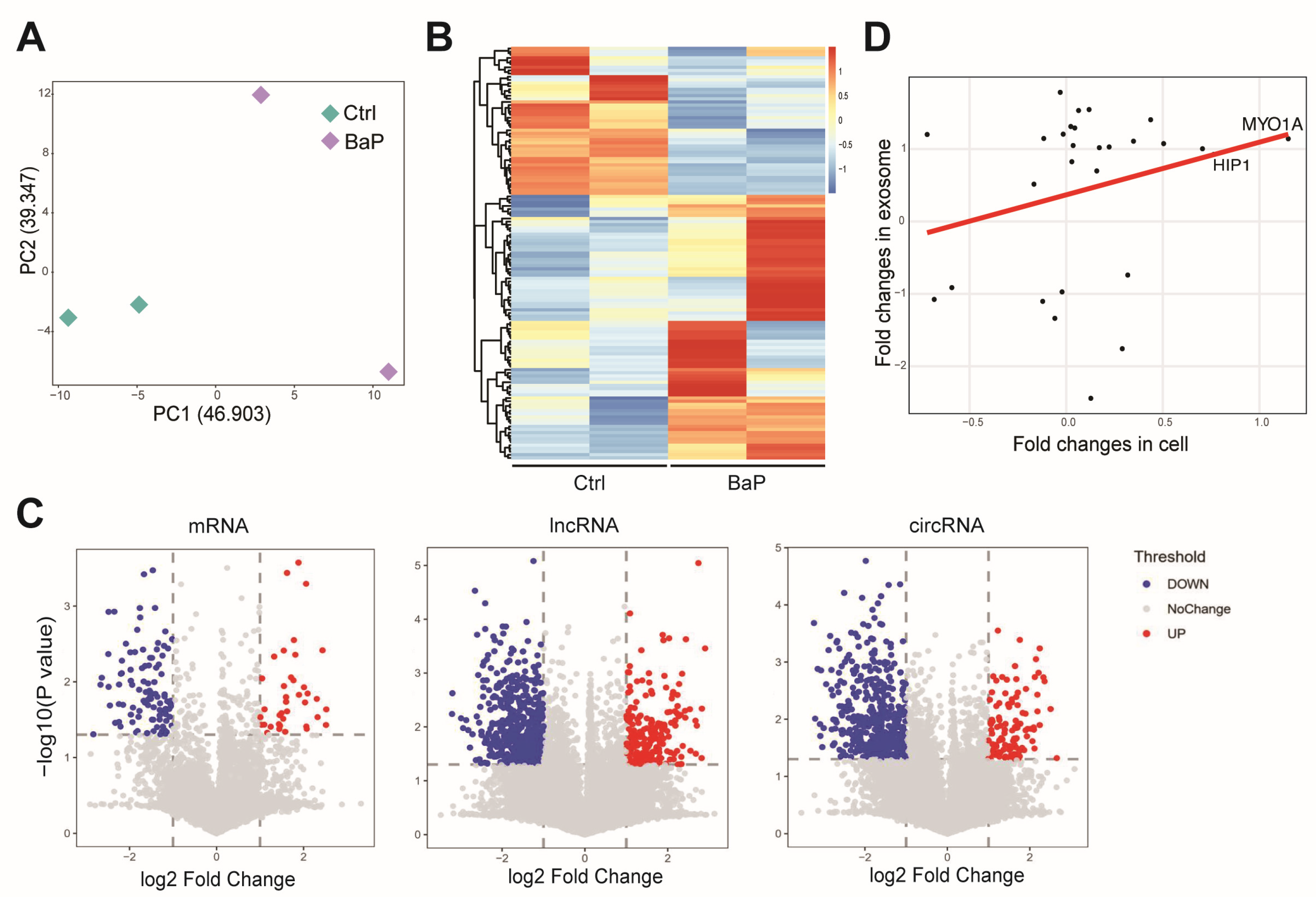
3.4. Effect of BaP Exposure on the RNA Profiling in Exosome of HCC Cells
3.5. BaP-Induced Exosomal circRNA-miRNA-mRNA Interaction Network in HCC Cells
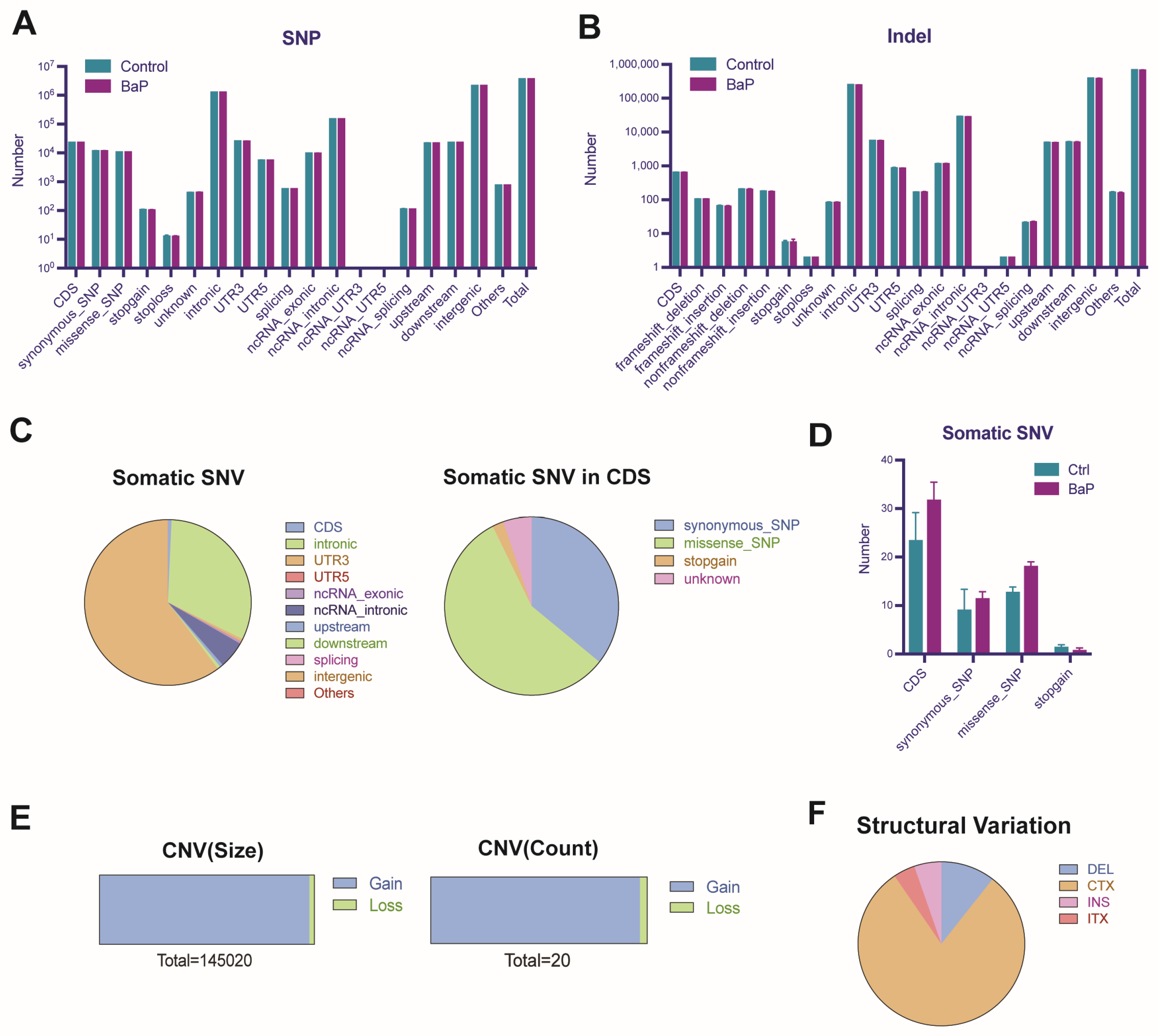
3.6. BaP-Induced Gene Mutation and Genomic Instability in HCC Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bukowska, B.; Mokra, K.; Michałowicz, J. Benzo[a]pyrene-Environmental Occurrence, Human Exposure, and Mechanisms of Toxicity. Int. J. Mol. Sci. 2022, 23, 6348. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, G.R.; Guizellini, G.M.; da Silva, S.A.; de Almeida, A.P.; Pinaffi-Langley, A.C.C.; Rogero, M.M.; de Camargo, A.C.; Torres, E.A.F.S. Polycyclic Aromatic Hydrocarbons in Foods: Biological Effects, Legislation, Occurrence, Analytical Methods, and Strategies to Reduce Their Formation. Int. J. Mol. Sci. 2021, 22, 6010. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-D.; Tang, T.; Tan, Y.-M.; Wang, F.-Y.; Zhang, W.-B.; Li, T.; Xia, M.-Z. Determination of benzo(a)pyrene in fried and baked foods by HPLC combined with vesicular coacervative supramolecular solvent extraction. J. Food Sci. Technol. 2019, 56, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Srogi, K. Monitoring of environmental exposure to polycyclic aromatic hydrocarbons: A review. Environ. Chem. Lett. 2007, 5, 169–195. [Google Scholar] [CrossRef] [PubMed]
- Ribière, C.; Peyret, P.; Parisot, N.; Darcha, C.; Déchelotte, P.J.; Barnich, N.; Peyretaillade, E.; Boucher, D. Oral exposure to environmental pollutant benzo[a]pyrene impacts the intestinal epithelium and induces gut microbial shifts in murine model. Sci. Rep. 2016, 6, 31027. [Google Scholar] [CrossRef] [PubMed]
- Mallah, M.A.; Changxing, L.; Mallah, M.A.; Noreen, S.; Liu, Y.; Saeed, M.; Xi, H.; Ahmed, B.; Feng, F.; Mirjat, A.A.; et al. Polycyclic aromatic hydrocarbon and its effects on human health: An overeview. Chemosphere 2022, 296, 133948. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.L.; Zubair, M.; John, K.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Umannová, L.; Machala, M.; Topinka, J.; Schmuczerová, J.; Krčmář, P.; Neča, J.; Šujanová, K.; Kozubík, A.; Vondráček, J. Benzo[a]pyrene and tumor necrosis factor-α coordinately increase genotoxic damage and the production of proinflammatory mediators in alveolar epithelial type II cells. Toxicol. Lett. 2011, 206, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Skupińska, K.; Misiewicz, I.; Kasprzycka-Guttman, T. Polycyclic aromatic hydrocarbons: Physicochemical properties, environmental appearance and impact on living organisms. Acta Pol. Pharm. 2004, 61, 233–240. [Google Scholar]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some non-heterocyclic polycyclic aromatic hydrocarbons and some related exposures. IARC Monogr. Eval. Carcinog. Risks Hum. 2010, 92, 1–853. [Google Scholar]
- Rota, M.; Bosetti, C.; Boccia, S.; Boffetta, P.; La Vecchia, C. Occupational exposures to polycyclic aromatic hydrocarbons and respiratory and urinary tract cancers: An updated systematic review and a meta-analysis to 2014. Arch. Toxicol. 2014, 88, 1479–1490. [Google Scholar] [CrossRef] [PubMed]
- Benford, D.; Dinovi, M.; Setzer, R.W. Application of the margin-of-exposure (MoE) approach to substances in food that are genotoxic and carcinogenic e.g.,: Benzo[a]pyrene and polycyclic aromatic hydrocarbons. Food Chem. Toxicol. 2010, 48 (Suppl. 1), S42–S48. [Google Scholar] [CrossRef] [PubMed]
- Petit, P.; Maître, A.; Persoons, R.; Bicout, D.J. Lung cancer risk assessment for workers exposed to polycyclic aromatic hydrocarbons in various industries. Environ. Int. 2019, 124, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Erkekoglu, P.; Oral, D.; Chao, M.-W.; Kocer-Gumusel, B. Hepatocellular Carcinoma and Possible Chemical and Biological Causes: A Review. J. Environ. Pathol. Toxicol. Oncol. 2017, 36, 171–190. [Google Scholar] [CrossRef] [PubMed]
- Santella, R.M.; Wu, H.-C. Environmental Exposures and Hepatocellular Carcinoma. J. Clin. Transl. Hepatol. 2013, 1, 138–143. [Google Scholar] [PubMed]
- Ba, Q.; Li, J.; Huang, C.; Qiu, H.; Li, J.; Chu, R.; Zhang, W.; Xie, D.; Wu, Y.; Wang, H. Effects of benzo[a]pyrene exposure on human hepatocellular carcinoma cell angiogenesis, metastasis, and NF-κB signaling. Environ. Health Perspect. 2015, 123, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Ba, Q.; Huang, C.; Fu, Y.; Li, J.; Li, J.; Chu, R.; Jia, X.; Wang, H. Cumulative metabolic effects of low-dose benzo(a)pyrene exposure on human cells. Toxicol. Res. 2016, 5, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Maan, K.; Baghel, R.; Dhariwal, S.; Sharma, A.; Bakhshi, R.; Rana, P. Metabolomics and transcriptomics based multi-omics integration reveals radiation-induced altered pathway networking and underlying mechanism. NPJ Syst. Biol. Appl. 2023, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Ueng, T.-H.; Chang, Y.-L.; Tsai, Y.-Y.; Su, J.-L.; Chan, P.-K.; Shih, J.-Y.; Lee, Y.-C.; Ma, Y.-C.; Kuo, M.-L. Potential roles of fibroblast growth factor-9 in the benzo(a)pyrene-induced invasion in vitro and the metastasis of human lung adenocarcinoma. Arch. Toxicol. 2010, 84, 651–660. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, W.; Wang, H.; Xia, X.; Zhang, C. Benzo(a)pyrene promotes A549 cell migration and invasion through up-regulating Twist. Arch. Toxicol. 2015, 89, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.E.; Holloway, A.C.; Foster, W.G. Benzo-[a]-pyrene increases invasion in MDA-MB-231 breast cancer cells via increased COX-II expression and prostaglandin E2 (PGE2) output. Clin. Exp. Metastasis 2005, 22, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, T.; Li, L.; Wang, H.; Zhang, D.; Yang, H. Benzo(a)pyrene promotes Hep-G2 cell migration and invasion by upregulating phosphorylated extracellular signal-regulated kinase expression. Oncol. Lett. 2018, 15, 8325–8332. [Google Scholar] [CrossRef]
- Brown, K.K.; Toker, A. The phosphoinositide 3-kinase pathway and therapy resistance in cancer. F1000Prime Rep. 2015, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.E.; Apsel, B.; Uotila, A.; Loewith, R.; Knight, Z.A.; Ruggero, D.; Shokat, K.M. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009, 7, e38. [Google Scholar] [CrossRef] [PubMed]
- Sze, K.M.-F.; Wong, K.L.-T.; Chu, G.K.-Y.; Lee, J.M.-F.; Yau, T.-O.; Ng, I.O.-L. Loss of phosphatase and tensin homolog enhances cell invasion and migration through AKT/Sp-1 transcription factor/matrix metalloproteinase 2 activation in hepatocellular carcinoma and has clinicopathologic significance. Hepatology 2011, 53, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Zhang, H.; Matei, I.R.; Costa-Silva, B.; Hoshino, A.; Rodrigues, G.; Psaila, B.; Kaplan, R.N.; Bromberg, J.F.; Kang, Y.; et al. Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 2017, 17, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.-N.; Tsuruga, T.; Yeung, T.-L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.-K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.-H.; Niu, J.-J.; Guo, F.; Liu, S.; Pan, L.-L. The study on the interactions between polycyclic aromatic hydrocarbon-albumin adducts and various risk factors to primary hepatocellular carcinoma. Zhonghua Yu Fang Yi Xue Za Zhi 2010, 44, 427–432. [Google Scholar]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- García-Silva, S.; Peinado, H. Melanosomes foster a tumour niche by activating CAFs. Nat. Cell Biol. 2016, 18, 911–913. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Dong, J.; Li, S.; Wang, C.; Ding, H.; Li, H.; Su, X.; Ge, X.; Sun, L.; Bai, C.; et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int. J. Biol. Sci. 2015, 11, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Conigliaro, A.; Costa, V.; Dico, A.L.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef] [PubMed]
- Plebanek, M.P.; Angeloni, N.L.; Vinokour, E.; Li, J.; Henkin, A.; Martinez-Marin, D.; Filleur, S.; Bhowmick, R.; Henkin, J.; Miller, S.D.; et al. Pre-metastatic cancer exosomes induce immune surveillance by patrolling monocytes at the metastatic niche. Nat. Commun. 2017, 8, 1319. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Zhu, Y.; Cheng, S.; Zhang, K.; Wang, H.; Ba, Q. Molecular Characteristics of Aberrant Gene Mutations and Expression Profiles Induced by Benzo(a)pyrene in Hepatocellular Carcinoma Cells. Toxics 2024, 12, 499. https://doi.org/10.3390/toxics12070499
Cao X, Zhu Y, Cheng S, Zhang K, Wang H, Ba Q. Molecular Characteristics of Aberrant Gene Mutations and Expression Profiles Induced by Benzo(a)pyrene in Hepatocellular Carcinoma Cells. Toxics. 2024; 12(7):499. https://doi.org/10.3390/toxics12070499
Chicago/Turabian StyleCao, Xinyi, Ying Zhu, Shujun Cheng, Kunxiao Zhang, Hui Wang, and Qian Ba. 2024. "Molecular Characteristics of Aberrant Gene Mutations and Expression Profiles Induced by Benzo(a)pyrene in Hepatocellular Carcinoma Cells" Toxics 12, no. 7: 499. https://doi.org/10.3390/toxics12070499





