TAS2R Receptor Response Helps Design New Antimicrobial Molecules for the 21st Century
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
Preliminary Experimental Validation
2.2. Model Development
3. Results and Discussion
3.1. Model Outcomes Meet Standard Quality Criteria
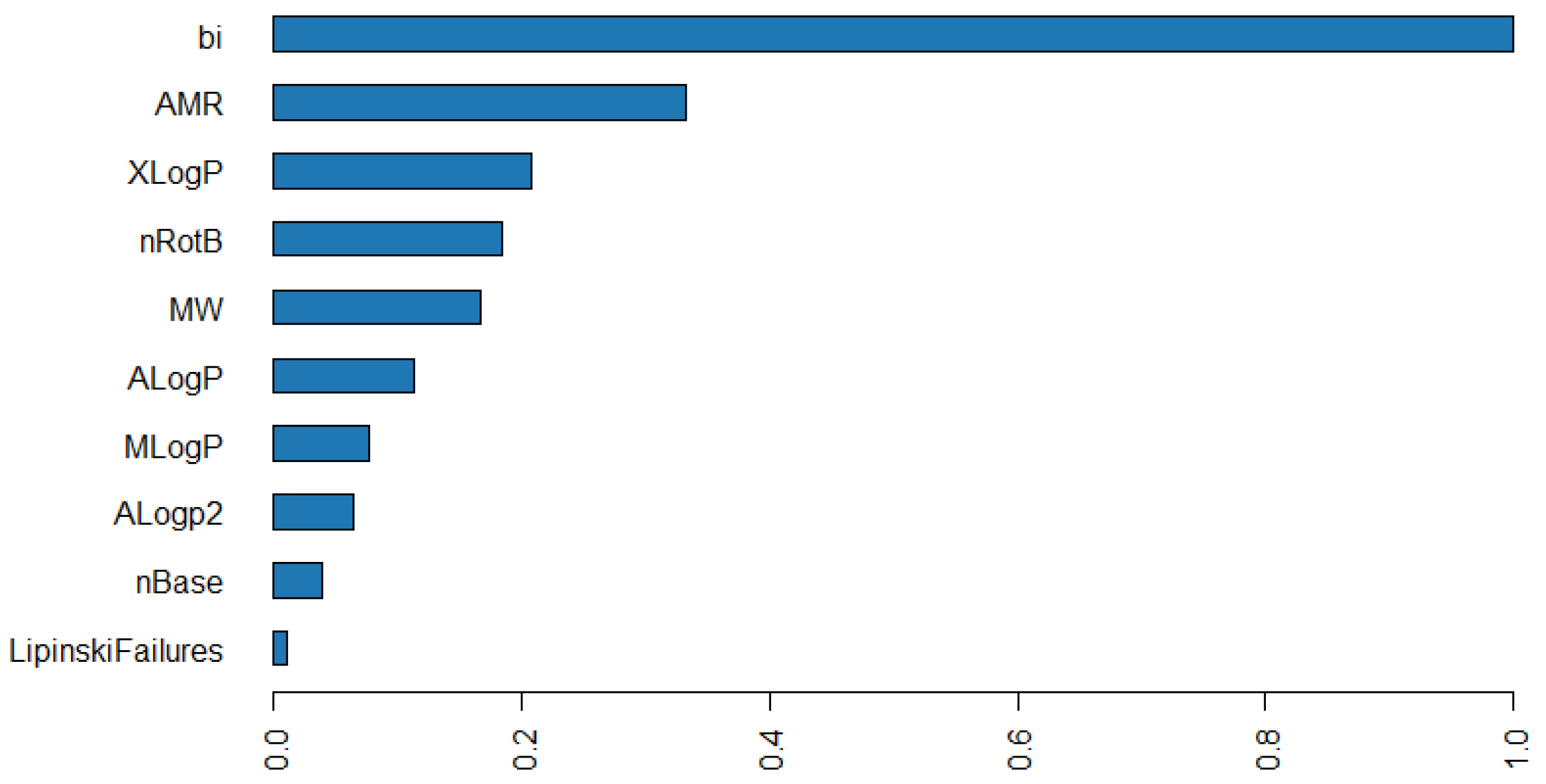
3.2. Descriptor Contributions to the Model Development
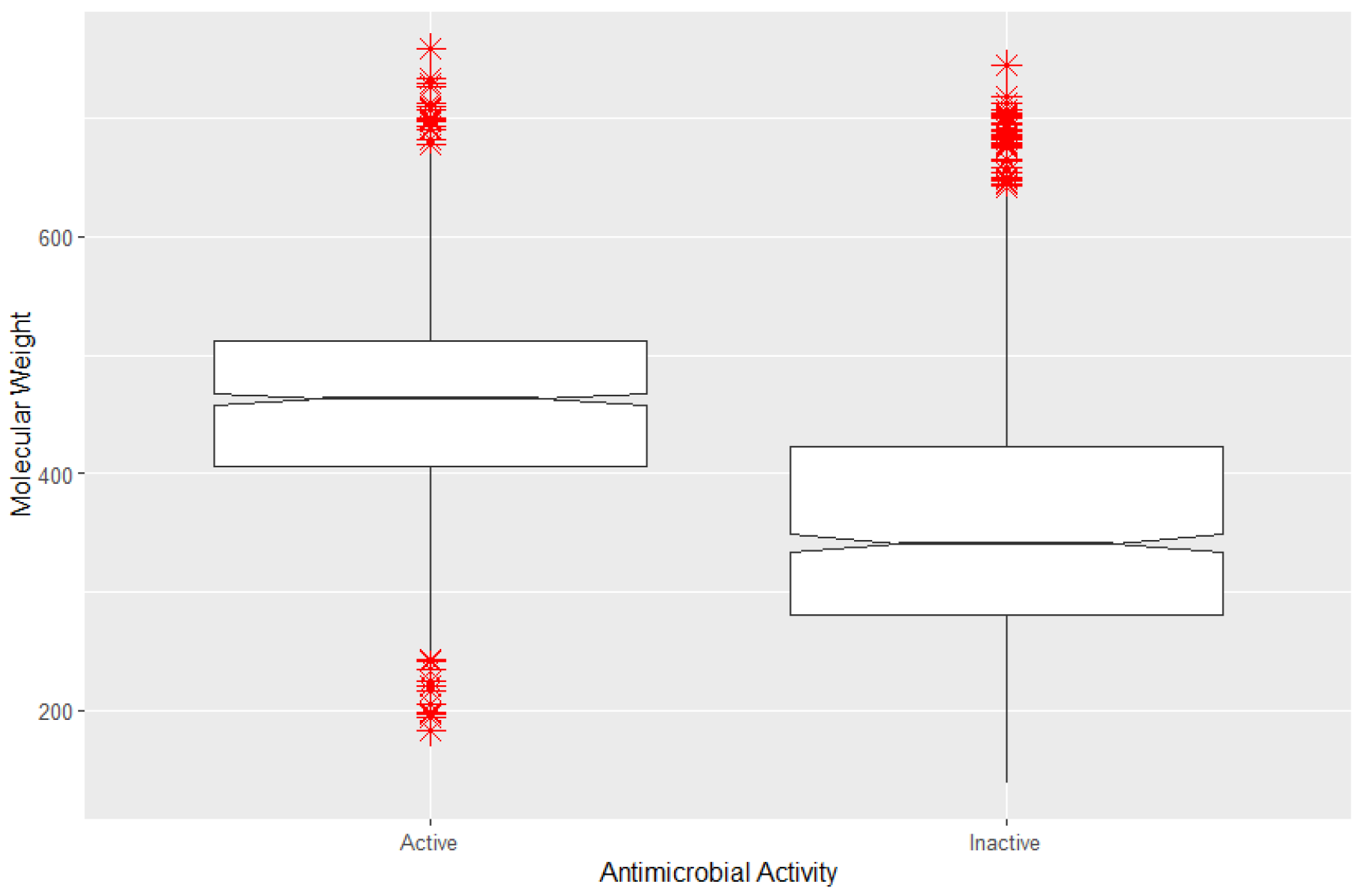
3.3. Analysis of Variable Importance Underscores the Value of the Bitterness Index
3.3.1. The Bitterness Index as a Watershed Differentiating Variable
3.3.2. Eliminating the Bitterness Index Impoverishes Model Performance
3.4. An Analysis of Model Development
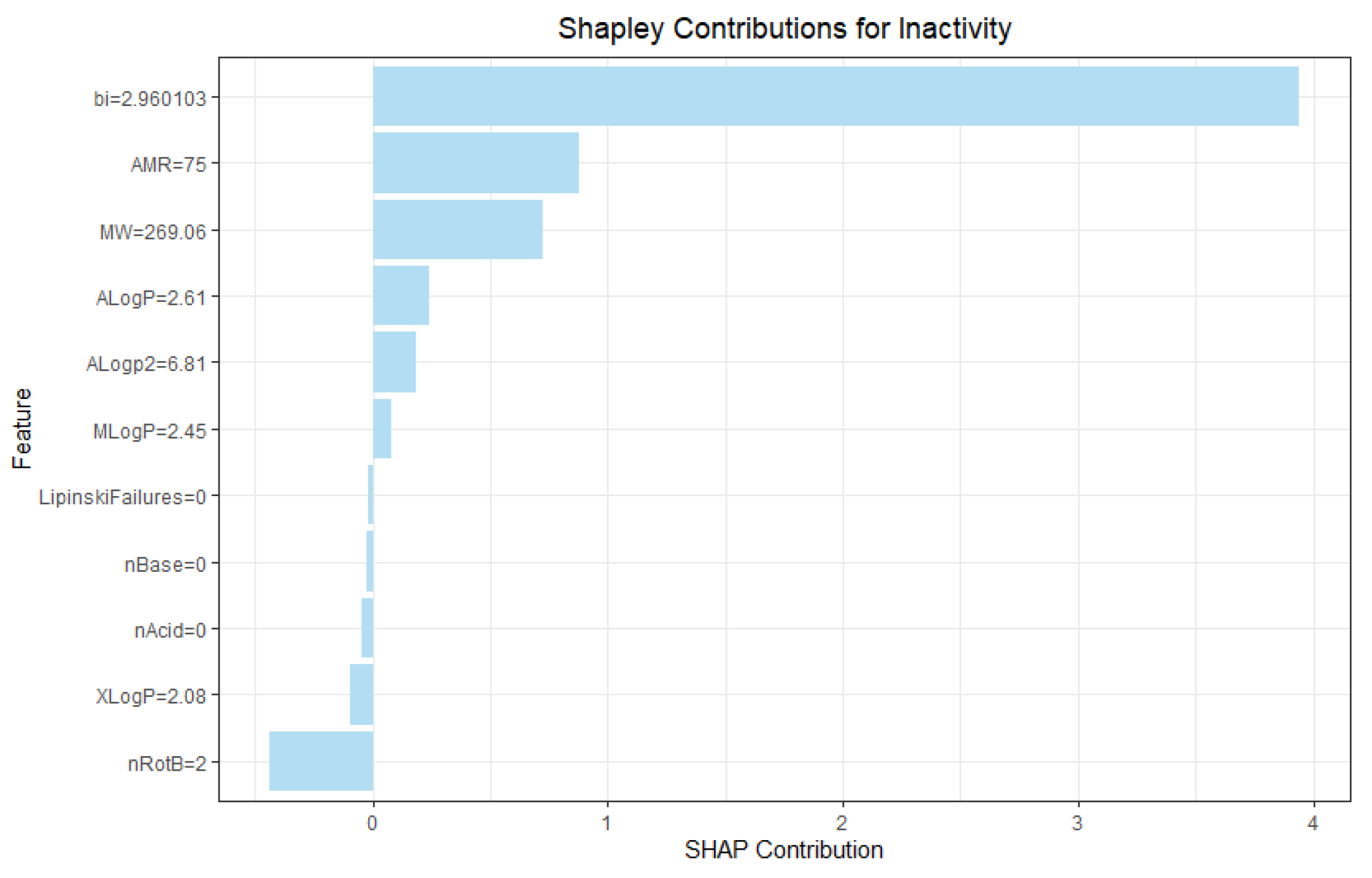
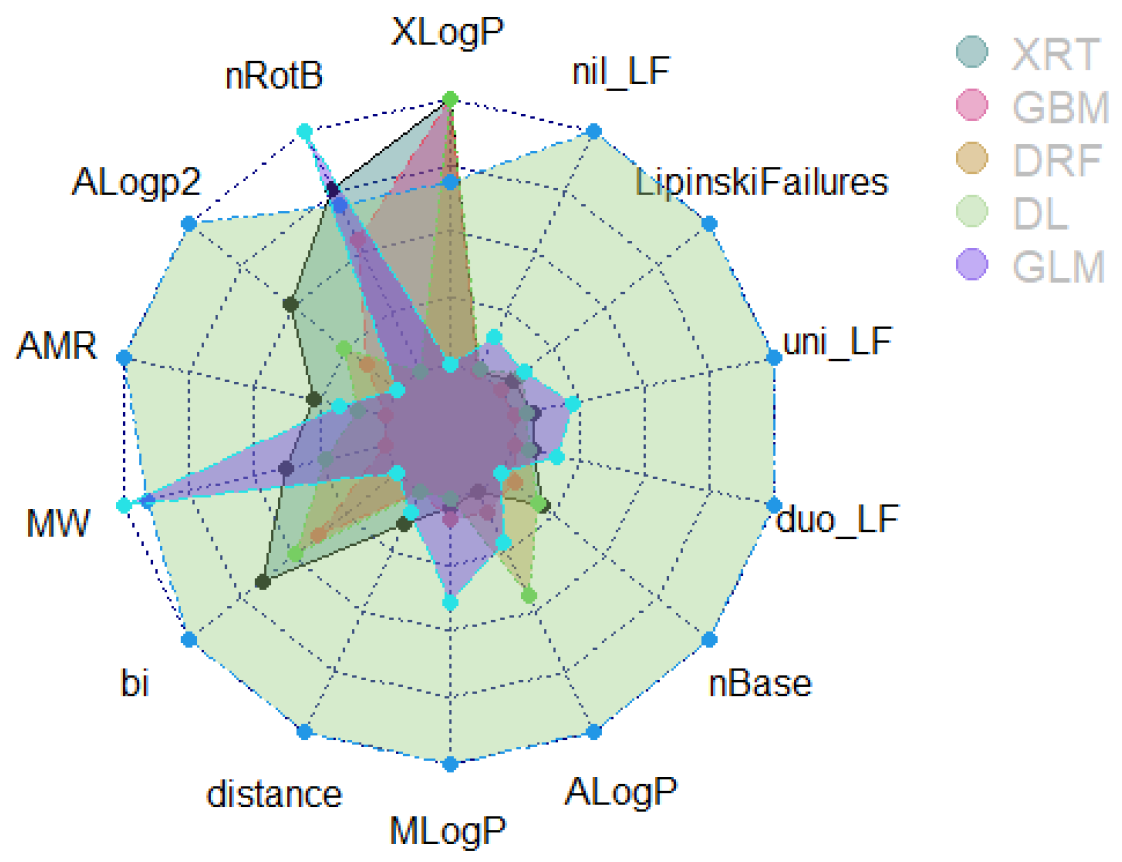
3.4.1. Contrastive Evaluation of Cooperative Contributions Recapitulates Feature Selection
3.4.2. Comparison of Modelling Methods Underscores High Signal in the Dataset Compilation
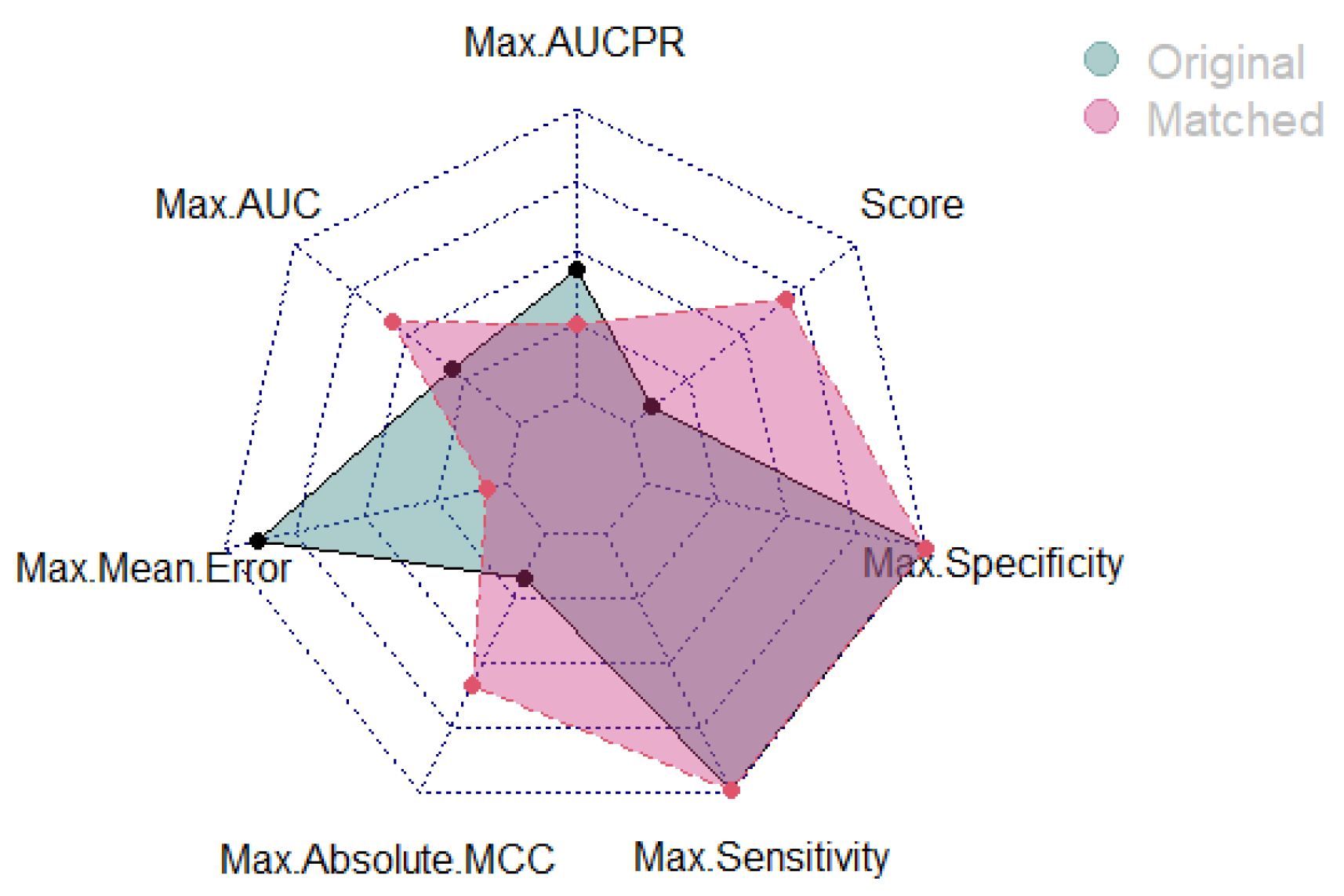
3.4.3. Statistical Matching with Lipinski’s Rules for De-Confounded Model Learning
4. Compound Synthesizability and Commercial Availability
5. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ganganwar, V. An overview of classification algorithms for imbalanced datasets. Int. J. Emerg. Technol. Adv. Eng. 2012, 2, 42–47. [Google Scholar]
- Clark, A.A.; Liggett, S.B.; Munger, S.D. Extraoral bitter taste receptors as mediators of off-target drug effects. FASEB J. 2012, 26, 4827–4831. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Sulli, C.; Davidson, E.; Berdougo, E.; Phillips, M.; Puffer, B.A.; Paes, C.; Doranz, B.J.; Rucker, J.B. The Bitter Taste Receptor TAS2R16 Achieves High Specificity and Accommodates Diverse Glycoside Ligands by using a Two-faced Binding Pocket. Sci. Rep. 2017, 7, 7753. [Google Scholar] [CrossRef] [PubMed]
- Sambu, S. The determinants of chemoreception as evidenced by gradient boosting machines in broad molecular fingerprint spaces. PeerJ Org. Chem. 2019, 1, e2. [Google Scholar] [CrossRef]
- Woolf, L.I. The heterozygote advantage in phenylketonuria. Am. J. Hum. Genet. 1986, 38, 773. [Google Scholar]
- Meyerhof, W.; Batram, C.; Kuhn, C.; Brockhoff, A.; Chudoba, E.; Bufe, B.; Appendino, G.; Behrens, M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 2010, 35, 157–170. [Google Scholar] [CrossRef]
- Behrens, M.; Meyerhof, W. Bitter taste receptor research comes of age: From characterization to modulation of TAS2Rs. Semin. Cell Dev. Biol. 2013, 24, 215–221. [Google Scholar] [CrossRef]
- Yang, M.Y.; Kim, S.-K.; Kim, D.; Liggett, S.B.; Goddard, W.A.I. Structures and Agonist Binding Sites of Bitter Taste Receptor TAS2R5 Complexed with Gi Protein and Validated against Experiment. J. Phys. Chem. Lett. 2021, 12, 9293–9300. [Google Scholar] [CrossRef]
- Jackson, F.L.C. Two evolutionary models for the interactions of dietary organic cyanogens, hemoglobins, and falciparum malaria. Am. J. Hum. Biol. 1990, 2, 521–532. [Google Scholar] [CrossRef]
- Lee, R.J.; Cohen, N.A. Bitter and sweet taste receptors in the respiratory epithelium in health and disease. J. Mol. Med. 2014, 92, 1235–1244. [Google Scholar] [CrossRef]
- Lee, R.J.; Hariri, B.M.; McMahon, D.B.; Chen, B.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; Kennedy, D.W.; Jiang, P.; Margolskee, R.F.; et al. Bacterial d-amino acids suppress sinonasal innate immunity through sweet taste receptors in solitary chemosensory cells. Sci. Signal. 2017, 10, eaam7703. [Google Scholar] [CrossRef] [PubMed]
- Agüero-Chapin, G.; Galpert-Cañizares, D.; Domínguez-Pérez, D.; Marrero-Ponce, Y.; Pérez-Machado, G.; Teijeira, M.; Antunes, A. Emerging Computational Approaches for Antimicrobial Peptide Discovery. Antibiotics 2022, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Duarte-Mata, D.I.; Salinas-Carmona, M.C. Antimicrobial peptides’ immune modulation role in intracellular bacterial infection. Front. Immunol. 2023, 14, 1119574. [Google Scholar] [CrossRef] [PubMed]
- Barham, H.; Cooper, S.; Anderson, C.; Tizzano, M.; Kingdom, T.; Finger, T.; Kinnamon, S.; Ramakrishnan, V. Solitary chemosensory cells and bitter taste receptor signaling in human sinonasal mucosa. Int. Forum Allergy Rhinol. 2013, 3, 450–457. [Google Scholar] [CrossRef]
- Tizzano, M.; Gulbransen, B.D.; Vandenbeuch, A.; Clapp, T.R.; Herman, J.P.; Sibhatu, H.M.; Churchill, M.E.A.; Silver, W.L.; Kinnamon, S.C.; Finger, T.E. Nasal chemosensory cells use bitter taste signaling to detect irritants and bacterial signals. Proc. Natl. Acad. Sci. USA 2010, 107, 3210–3215. [Google Scholar] [CrossRef]
- Jugder, B.-E.; Batista, J.H.; Gibson, J.A.; Cunningham, P.M.; Asara, J.M.; Watnick, P.I. Vibrio cholerae high cell density quorum sensing activates the host intestinal innate immune response. Cell Rep. 2022, 40, 111368. [Google Scholar] [CrossRef]
- Zagidullin, B.; Wang, Z.; Guan, Y.; Pitkänen, E.; Tang, J. Comparative analysis of molecular fingerprints in prediction of drug combination effects. Brief. Bioinform. 2021, 22, bbab291. [Google Scholar] [CrossRef]
- Chakravarti, S.K.; Alla, S.R.M. Descriptor Free QSAR Modeling Using Deep Learning With Long Short-Term Memory Neural Networks. Front. Artif. Intell. 2019, 2, 17. [Google Scholar] [CrossRef]
- Torres-Barceló, C. Phage Therapy Faces Evolutionary Challenges. Viruses 2018, 10, 323. [Google Scholar] [CrossRef]
- AID 573—Primary Antimicrobial Assay for E. coli BW25113 ∆tolC::kan Protocol for 384-Well HTS—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/bioassay/573 (accessed on 17 October 2022).
- Ho, D.; Imai, K.; King, G.; Stuart, E.; Whitworth, A.; Greifer, N. MatchIt: Nonparametric Preprocessing for Parametric Causal Inference. J. Stat. Softw. 2022, 42, 8. [Google Scholar]
- Liszt, K.I.; Wang, Q.; Farhadipour, M.; Segers, A.; Thijs, T.; Nys, L.; Deleus, E.; Van der Schueren, B.; Gerner, C.; Neuditschko, B. Human intestinal bitter taste receptors regulate innate immune responses and metabolic regulators in obesity. J. Clin. Investig. 2022, 132, e144828. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, I.; Murugesan, K.; Balasubramanian, L.; Arumugam, M. Interpretation of Artificial Intelligence Algorithms in the Prediction of Sepsis; 2019 Computing in Cardiology (CinC); IEEE: Singapore, 2019; p. 1. [Google Scholar]
- Dinga, R.; Schmaal, L.; Penninx, B.W.J.H.; Veltman, D.; Marquand, A. Controlling for effects of confounding variables on machine learning prediction. Biorxiv 2020, 1, 2020-08. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead-and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Soares, K.M.; Blackmon, N.; Shun, T.Y.; Shinde, S.N.; Takyi, H.K.; Wipf, P.; Lazo, J.S.; Johnston, P.A. Profiling the NIH Small Molecule Repository for Compounds That Generate H2O2 by Redox Cycling in Reducing Environments. Assay Drug Dev. Technol. 2010, 8, 152–174. [Google Scholar] [CrossRef] [PubMed]




| ACTUAL | ||||
|---|---|---|---|---|
| PREDICTED | ACTIVE | INACTIVE | ERROR | |
| ACTIVE | 94% | 6% | 6% | |
| INACTIVE | 12% | 88% | 12% | |
| TOTALS | 58% | 42% | 9% | |
| Descriptor | Function | Contribution | Rationale |
|---|---|---|---|
| Mass | Molecular weight | 10% | Diffusion rate across biomembranes and cytosol |
| Hydrophobicity | Multiple | 40% | Partitioning across biomembranes |
| Electro-topological | Polarizability | 10% | Charge separation in response to an external electric field: may influence coulombic interactions with cellular environment |
| Binding degrees of freedom (D.F.) | Multiple features | 20% | Solvation and motion free energies determined by acid/base pairs and rotatable bonds |
| Combinatorial | Multiple features | 20% | Drug design rules and psychophysical response indices |
| Model without bi | Model with bi | |
| Max. AUCPR | 0.8910 | 0.9931 |
| Max. AUC | 0. 9006 | 0.9940 |
| Max. Mean Error | 0.1866 | 0.022 |
| Max. Absolute MCC | 0.6413 | 0.9966 |
| Max. Sensitivity (TPR/Precision) | 0.9999 | 0.9999 |
| Max. Specificity (TNR) | 0.9999 | 0.9999 |
| Overall Score | 2/6 | 6/6 |
| GBM | DRF | XRT | |
| Max. AUCPR | 0.9666 | 0.9597 (↓ 0.71%) | 0.9605 (↓ 0.63%) |
| Max. AUC | 0.9650 | 0.9534 (↓ 1.2%) | 0.9590 (↓ 0.62%) |
| Max. Mean Error | 0.093 (↑ 4.5%) | 0.089 | 0.09 (↑ 1.2%) |
| Max. Absolute MCC | 0.8324 (↓ 1.2%) | 0.8428 | 0.8324 (↓ 1.2%) |
| Max. Sensitivity (TPR/Precision) | 0.9999 | 0.9999 | 0.9999 |
| Max. Specificity (TNR) | 0.9999 | 0.9999 | 0.9966 (↓ 0.33%) |
| Overall Score | 4/6 | 4/6 | 1/6 (↓ 75%) |
| Traditional Model | Statistically Matched | |
| Max. AUCPR | 0.9666 | 0.9553 (↓ 1.17%) |
| Max. AUC | 0.9650 | 0.9786 (↑ 1.41%) |
| Max. Mean Error | 0.093 | 0.0440 (↓ 52.69 %) |
| Max. Absolute MCC | 0.8324 | 0.9111 (↑9.45%) |
| Max. Sensitivity (TPR/Precision) | 0.9999 | 0.9999 |
| Max. Specificity (TNR) | 0.9999 | 0.9999 |
| Overall Score | 3/6 (↓ 40%) | 5/6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sambu, S. TAS2R Receptor Response Helps Design New Antimicrobial Molecules for the 21st Century. ChemEngineering 2024, 8, 96. https://doi.org/10.3390/chemengineering8050096
Sambu S. TAS2R Receptor Response Helps Design New Antimicrobial Molecules for the 21st Century. ChemEngineering. 2024; 8(5):96. https://doi.org/10.3390/chemengineering8050096
Chicago/Turabian StyleSambu, Sammy. 2024. "TAS2R Receptor Response Helps Design New Antimicrobial Molecules for the 21st Century" ChemEngineering 8, no. 5: 96. https://doi.org/10.3390/chemengineering8050096
APA StyleSambu, S. (2024). TAS2R Receptor Response Helps Design New Antimicrobial Molecules for the 21st Century. ChemEngineering, 8(5), 96. https://doi.org/10.3390/chemengineering8050096