Endoscopic Hyperspectral Imaging System to Discriminate Tissue Characteristics in Tissue Phantom and Orthotopic Mouse Pancreatic Tumor Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of the eHSI System
2.2. Tissue Phantom Imaging with the eHSI System
2.3. Image Analysis
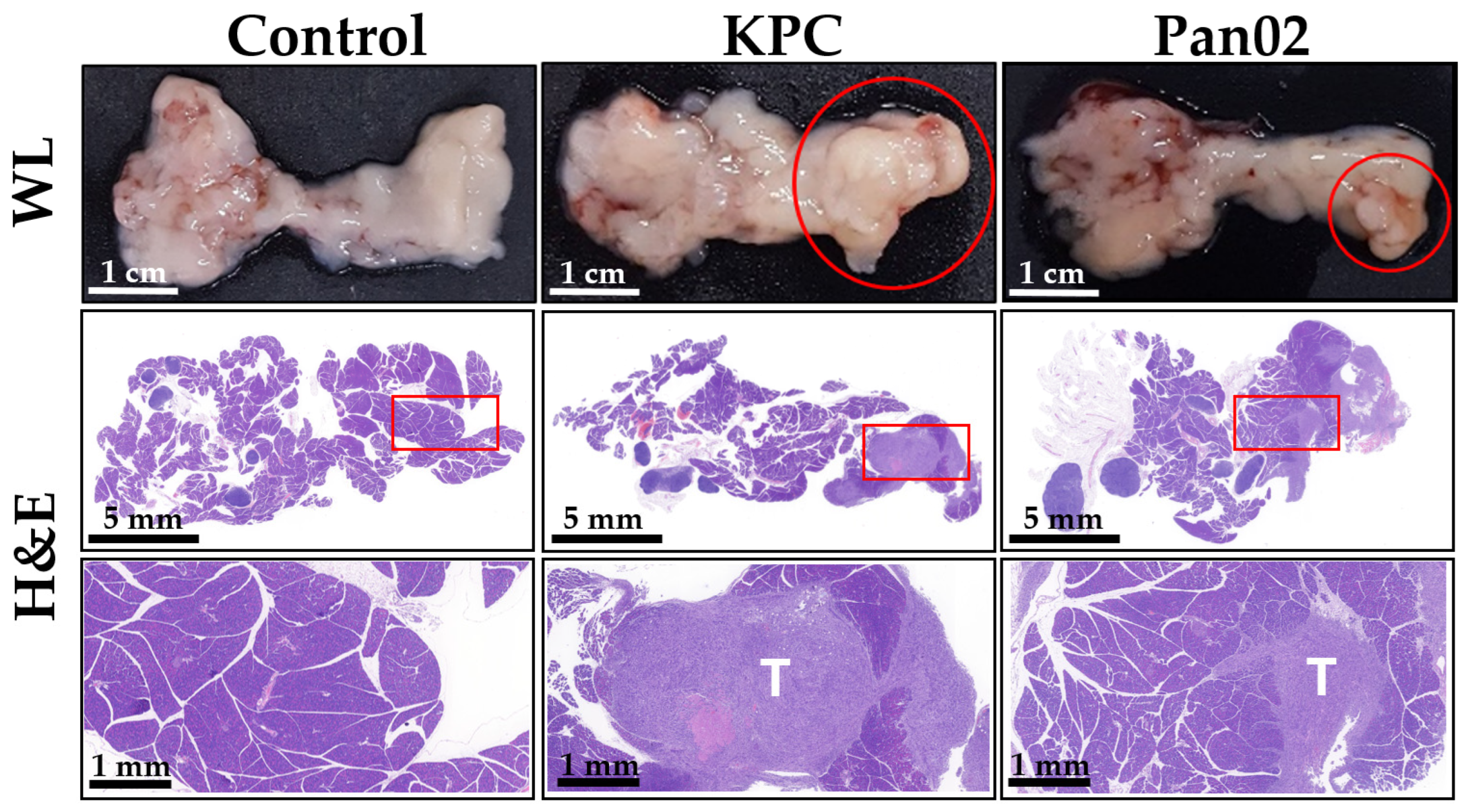
2.4. Generation of Orthotopic Mouse Pancreatic Tumor Model
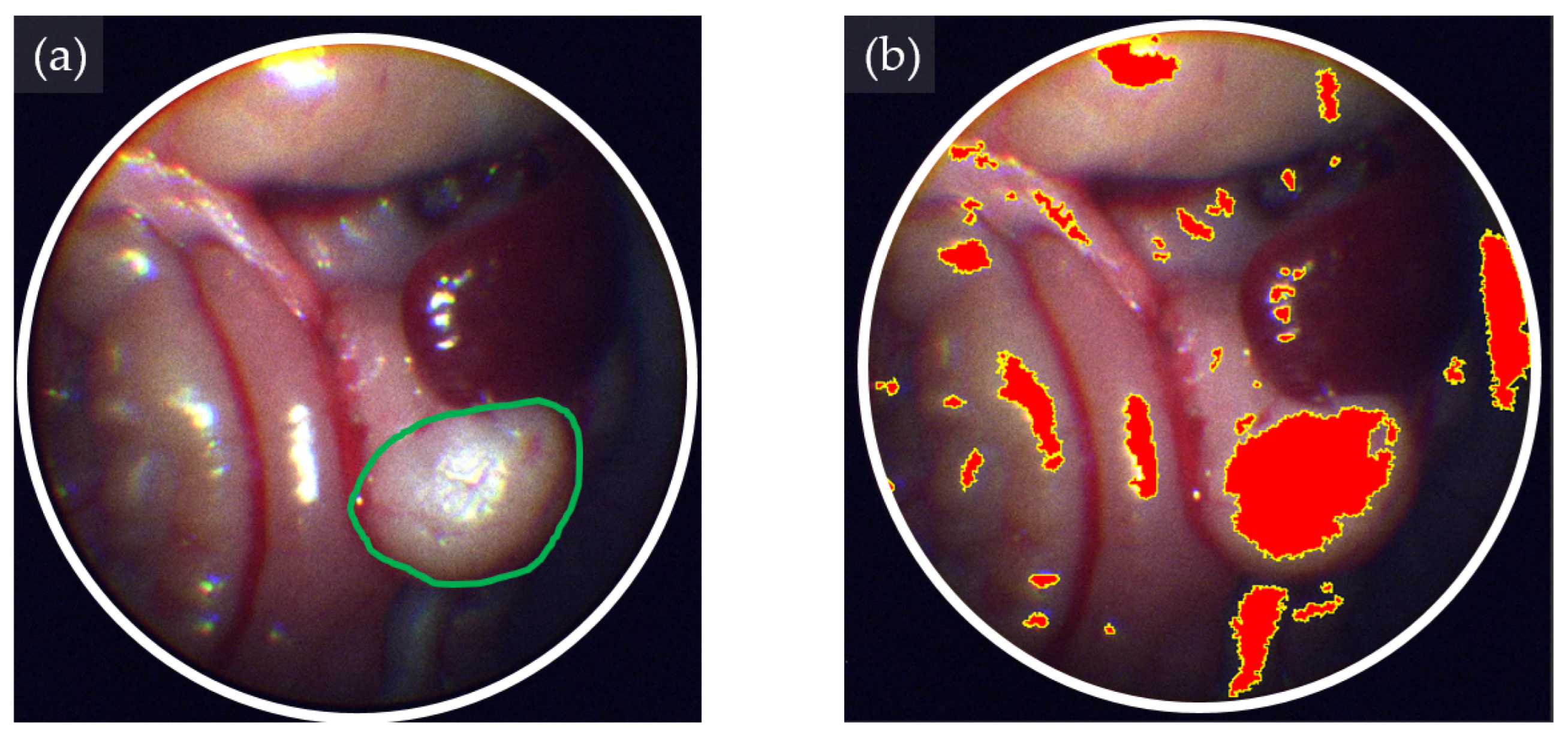
2.5. eHSI in Orthotopic Pancreatic Tumors
3. Results
3.1. Tissue Classification Performance of eHSI Images in Tissue Phantoms
3.2. Tissue Classification Performance of eHSI in Orthotopic Pancreatic Tumors
4. Discussions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cronin, K.A.; Scott, S.; Firth, A.U.; Sung, H.; Henley, S.J.; Sherman, R.L.; Siegel, R.L.; Anderson, R.N.; Kohler, B.A.; Benard, V.B.; et al. Annual report to the nation on the status of cancer, part 1: National cancer statistics. Cancer 2022, 128, 4251–4284. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute, Cancer Stat Facts: Pancreatic Cancer. 2024. Available online: https://seer.cancer.gov/ (accessed on 8 January 2024).
- Daamen, L.A.; Groot, V.P.; Besselink, M.G.; Bosscha, K.; Busch, O.R.; Cirkel, G.A.; van Dam, R.M.; Festen, S.; Groot Koerkamp, B.; Haj Mohammad, N.; et al. Detection, Treatment, and Survival of Pancreatic Cancer Recurrence in the Netherlands: A Nationwide Analysis. Ann. Surg. 2022, 275, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Mieog, J.S.D.; Achterberg, F.B.; Zlitni, A.; Hutteman, M.; Burggraaf, J.; Swijnenburg, R.J.; Gioux, S.; Vahrmeijer, A.L. Fundamentals and developments in fluorescence-guided cancer surgery. Nat. Rev. Clin. Oncol. 2022, 19, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; van den Berg, N.S.; Martin, B.A.; Nishio, N.; Hart, Z.P.; van Keulen, S.; Fakurnejad, S.; Chirita, S.U.; Raymundo, R.C.; Yi, G.; et al. Tumour-specific fluorescence-guided surgery for pancreatic cancer using panitumumab-IRDye800CW: A phase 1 single-centre, open-label, single-arm, dose-escalation study. Lancet Gastroenterol. Hepatol. 2020, 5, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J. Hyperspectral imaging for clinical applications. BioChip J. 2022, 16, 1–12. [Google Scholar] [CrossRef]
- Fei, B. Hyperspectral imaging in medical applications. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 32, pp. 523–565. [Google Scholar]
- Karim, S.; Qadir, A.; Farooq, U.; Shakir, M.; Laghari, A.A. Hyperspectral Imaging: A Review and Trends towards Medical Imaging. Curr. Med. Imaging 2022, 19, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Halig, L.; Wang, D.; Qin, X.; Chen, Z.G.; Fei, B. Spectral-spatial classification for noninvasive cancer detection using hyperspectral imaging. J. Biomed. Opt. 2014, 19, 106004. [Google Scholar] [CrossRef]
- Gerstner, A.O.; Laffers, W.; Bootz, F.; Farkas, D.L.; Martin, R.; Bendix, J.; Thies, B. Hyperspectral imaging of mucosal surfaces in patients. J. Biophotonics 2012, 5, 255–262. [Google Scholar] [CrossRef]
- Yoon, J.; Joseph, J.; Waterhouse, D.J.; Luthman, A.S.; Gordon, G.S.D.; di Pietro, M.; Januszewicz, W.; Fitzgerald, R.C.; Bohndiek, S.E. A clinically translatable hyperspectral endoscopy (HySE) system for imaging the gastrointestinal tract. Nat. Commun. 2019, 10, 1902. [Google Scholar] [CrossRef]
- Modir, N.; Shahedi, M.; Dormer, J.; Ma, L.; Ghaderi, M.; Sirsi, S.; Cheng, Y.L.; Fei, B. LED-based Hyperspectral Endoscopic Imaging. Proc. SPIE Int. Soc. Opt. Eng. 2022, 11954, 1195408. [Google Scholar]
- Waterhouse, D.J.; Bano, S.; Januszewicz, W.; Stoyanov, D.; Fitzgerald, R.C.; di Pietro, M.; Bohndiek, S.E. First-in-human pilot study of snapshot multispectral endoscopy for early detection of Barrett’s-related neoplasia. J. Biomed. Opt. 2021, 26, 106002. [Google Scholar] [CrossRef]
- Jang, M.; Kim, S.S.; Lee, J. Cancer cell metabolism: Implications for therapeutic targets. Exp. Mol. Med. 2013, 45, e45. [Google Scholar] [CrossRef]
- Krafft, C.; Popp, J. Opportunities of optical and spectral technologies in intraoperative histopathology. Optica 2023, 10, 214–231. [Google Scholar] [CrossRef]
- Wilson, R.H.; Chandra, M.; Scheiman, J.; Simeone, D.; McKenna, B.; Purdy, J.; Mycek, M.A. Optical spectroscopy detects histological hallmarks of pancreatic cancer. Opt. Express 2009, 17, 17502–17516. [Google Scholar] [CrossRef]
- Rutkowski, R.; Schuster, M.; Unger, J.; Seebauer, C.; Metelmann, H.; Woedtke, T.V.; Weltmann, K.; Daeschlein, G. Hyperspectral imaging for in vivo monitoring of cold atmospheric plasma effects on microcirculation in treatment of head and neck cancer and wound healing. Clin. Plasma Med. 2017, 7, 52–57. [Google Scholar] [CrossRef]
- Aref, M.H.; Aboughaleb, I.H.; El-Sharkawy, Y.H. Tissue characterization utilizing hyperspectral imaging for liver thermal ablation. Photodiagn. Photodyn. Ther. 2020, 31, 101899. [Google Scholar] [CrossRef]
- McCormack, D.R.; Walsh, A.J.; Sit, W.; Arteaga, C.L.; Chen, J.; Cook, R.S.; Skala, M.C. In vivo hyperspectral imaging of microvessel response to trastuzumab treatment in breast cancer xenografts. Biomed. Opt. Express 2014, 5, 2247–2261. [Google Scholar] [CrossRef]
- Lindsley, E.H.; Wachman, E.S.; Farkas, D.L. The hyperspectral imaging endoscope: A new tool for in vivo cancer detection. In Proceedings of the Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues II, San Jose, CA, USA, 1 July 2004; pp. 75–82. [Google Scholar]
- Hohmann, M.; Kanawade, R.; Klampfl, F.; Douplik, A.; Mudter, J.; Neurath, M.F.; Albrecht, H. In-vivo multispectral video endoscopy towards in-vivo hyperspectral video endoscopy. J. Biophotonics 2017, 10, 553–564. [Google Scholar] [CrossRef] [PubMed]
- an, Z.; Zhang, A.; Wang, X.; Sun, Z.; Wang, M.D.; Xie, T. In vivo use of hyperspectral imaging to develop a noncontact endoscopic diagnosis support system for malignant colorectal tumors. J. Biomed. Opt. 2016, 21, 16001. [Google Scholar]
- Kumashiro, R.; Konishi, K.; Chiba, T.; Akahoshi, T.; Nakamura, S.; Murata, M.; Tomikawa, M.; Matsumoto, T.; Maehara, Y.; Hashizume, M. Integrated Endoscopic System Based on Optical Imaging and Hyperspectral Data Analysis for Colorectal Cancer Detection. Anticancer Res. 2016, 36, 3925–3932. [Google Scholar]
- Regeling, B.; Thies, B.; Gerstner, A.O.; Westermann, S.; Muller, N.A.; Bendix, J.; Laffers, W. Hyperspectral Imaging Using Flexible Endoscopy for Laryngeal Cancer Detection. Sensors 2016, 16, 1288. [Google Scholar] [CrossRef]
- Wu, I.C.; Syu, H.Y.; Jen, C.P.; Lu, M.Y.; Chen, Y.T.; Wu, M.T.; Kuo, C.T.; Tsai, Y.Y.; Wang, H.C. Early identification of esophageal squamous neoplasm by hyperspectral endoscopic imaging. Sci. Rep. 2018, 8, 13797. [Google Scholar] [CrossRef]
- Thomassen, M.T.; Kohler, H.; Pfahl, A.; Stelzner, S.; Mehdorn, M.; Thieme, R.; Jansen-Winkeln, B.; Gockel, I.; Chalopin, C.; Moulla, Y. In vivo evaluation of a hyperspectral imaging system for minimally invasive surgery (HSI-MIS). Surg. Endosc. 2023, 37, 3691–3700. [Google Scholar] [CrossRef]
- Barberio, M.; Collins, T.; Bencteux, V.; Nkusi, R.; Felli, E.; Viola, M.G.; Marescaux, J.; Hostettler, A.; Diana, M. Deep Learning Analysis of In Vivo Hyperspectral Images for Automated Intraoperative Nerve Detection. Diagnostics 2021, 11, 1508. [Google Scholar] [CrossRef] [PubMed]
- Wang, L. Support Vector Machines: Theory and Applications; Springer Science & Business Media: Auckland, New Zealand, 2005; Volume 177. [Google Scholar]
- Jozdani, S.E.; Johnson, B.A.; Chen, D. Comparing deep neural networks, ensemble classifiers, and support vector machine algorithms for object-based urban land use/land cover classification. Remote Sens. 2019, 11, 1713. [Google Scholar] [CrossRef]
- Urbanos, G.; Martin, A.; Vazquez, G.; Villanueva, M.; Villa, M.; Jimenez-Roldan, L.; Chavarrias, M.; Lagares, A.; Juarez, E.; Sanz, C. Supervised Machine Learning Methods and Hyperspectral Imaging Techniques Jointly Applied for Brain Cancer Classification. Sensors 2021, 21, 3827. [Google Scholar] [CrossRef]
- Okere, E.E.; Ambaw, A.; Perold, W.J.; Opara, U.L. Early bruise detection on pomegranate (Punica granatum L.), using hyperspectral imaging coupled with artificial neutral network algorithm. Technol. Hortic. 2023, 3, 27. [Google Scholar] [CrossRef]
- Halicek, M.; Little, J.V.; Wang, X.; Patel, M.; Griffith, C.C.; Chen, A.Y.; Fei, B. Tumor Margin Classification of Head and Neck Cancer Using Hyperspectral Imaging and Convolutional Neural Networks. Proc. SPIE Int. Soc. Opt. Eng. 2018, 10576, 17–27. [Google Scholar]
- Ahamad, M.M.; Aktar, S.; Uddin, M.J.; Rahman, T.; Alyami, S.A.; Al-Ashhab, S.; Akhdar, H.F.; Azad, A.; Moni, M.A. Early-Stage Detection of Ovarian Cancer Based on Clinical Data Using Machine Learning Approaches. J. Pers. Med. 2022, 12, 1211. [Google Scholar] [CrossRef]
- Kim, H.; Jeon, J.; Han, Y.J.; Joo, Y.; Lee, J.; Lee, S.; Im, S. Convolutional Neural Network Classifies Pathological Voice Change in Laryngeal Cancer with High Accuracy. J. Clin. Med. 2020, 9, 3415. [Google Scholar] [CrossRef]





| SVM 1 | NN 2 | LGBM 3 | |
|---|---|---|---|
| Precision | 98.3% | 97.7% | 96.0% |
| Recall | 93.4% | 93.8% | 97.2% |
| F-score | 95.8% | 95.7% | 96.6% |
| KPC Tumor Bearing Model | Pan02 Tumor Bearing Model | |||||
|---|---|---|---|---|---|---|
| SVM 1 | NN 2 | LGBM 3 | SVM 1 | NN 2 | LGBM 3 | |
| Precision | 91.5% | 91.7% | 89.6% | 83.0% | 82.5% | 73.3% |
| Recall | 58.5% | 57.9% | 61.7% | 50.9% | 50.8% | 48.8% |
| F-score | 71.4% | 71.0% | 73.1% | 63.1% | 62.9% | 58.6% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mun, N.E.; Tran, T.K.C.; Park, D.H.; Im, J.H.; Park, J.I.; Le, T.D.; Moon, Y.J.; Kwon, S.-Y.; Yoo, S.W. Endoscopic Hyperspectral Imaging System to Discriminate Tissue Characteristics in Tissue Phantom and Orthotopic Mouse Pancreatic Tumor Model. Bioengineering 2024, 11, 208. https://doi.org/10.3390/bioengineering11030208
Mun NE, Tran TKC, Park DH, Im JH, Park JI, Le TD, Moon YJ, Kwon S-Y, Yoo SW. Endoscopic Hyperspectral Imaging System to Discriminate Tissue Characteristics in Tissue Phantom and Orthotopic Mouse Pancreatic Tumor Model. Bioengineering. 2024; 11(3):208. https://doi.org/10.3390/bioengineering11030208
Chicago/Turabian StyleMun, Na Eun, Thi Kim Chi Tran, Dong Hui Park, Jin Hee Im, Jae Il Park, Thanh Dat Le, Young Jin Moon, Seong-Young Kwon, and Su Woong Yoo. 2024. "Endoscopic Hyperspectral Imaging System to Discriminate Tissue Characteristics in Tissue Phantom and Orthotopic Mouse Pancreatic Tumor Model" Bioengineering 11, no. 3: 208. https://doi.org/10.3390/bioengineering11030208
APA StyleMun, N. E., Tran, T. K. C., Park, D. H., Im, J. H., Park, J. I., Le, T. D., Moon, Y. J., Kwon, S.-Y., & Yoo, S. W. (2024). Endoscopic Hyperspectral Imaging System to Discriminate Tissue Characteristics in Tissue Phantom and Orthotopic Mouse Pancreatic Tumor Model. Bioengineering, 11(3), 208. https://doi.org/10.3390/bioengineering11030208






