Diagnostic Screening of Bovine Mastitis Using MALDI-TOF MS Direct-Spotting of Milk and Machine Learning
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Sample Preparation
2.1.1. Raw Milk Samples
2.1.2. Sample Preparation and Spotting
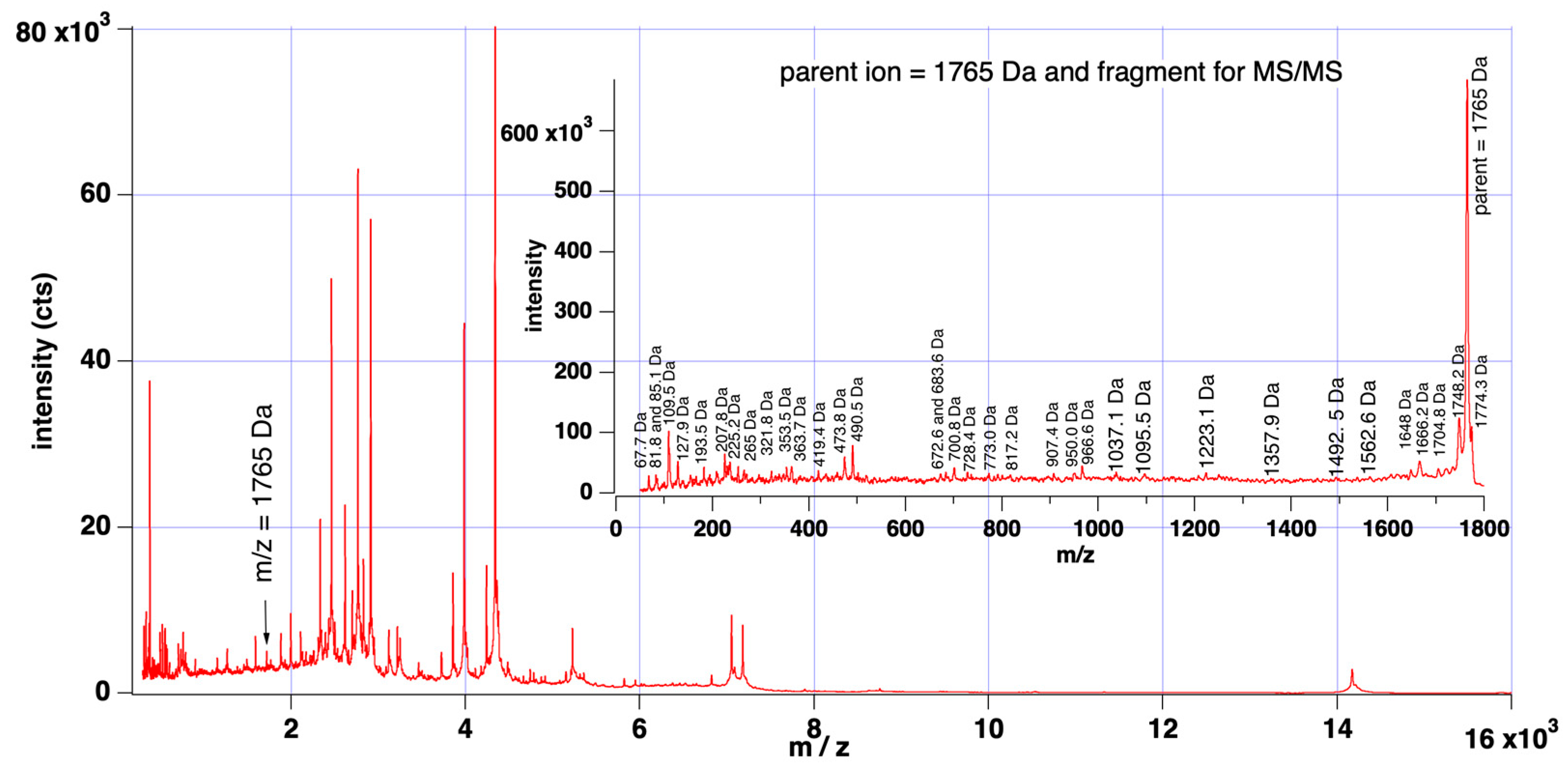
2.2. MALDI-TOF-MS Analysis
MALDI-TOF Mass Spectra Acquisition
2.3. Data Mining
2.3.1. Data Formatting
2.3.2. Data Mining Models and Runs
3. Results
3.1. Machine Learning Models and Model Evaluation
3.1.1. Decision Trees
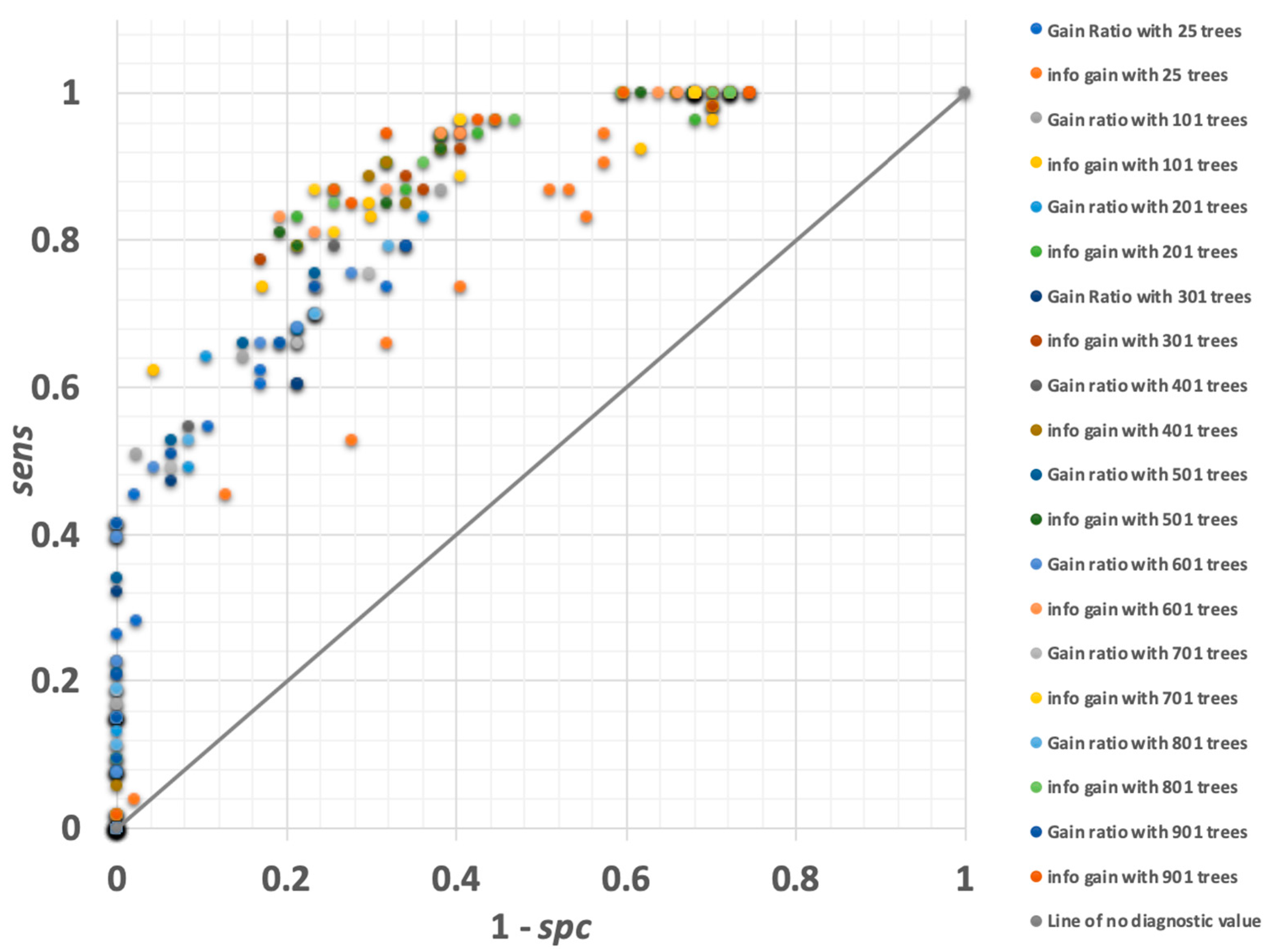
3.1.2. Random Forest
3.1.3. Naïve Bayes Model
3.1.4. Generalized Linear Model
3.1.5. Fast Large-Margin Model
3.1.6. Gradient-Boosted Trees
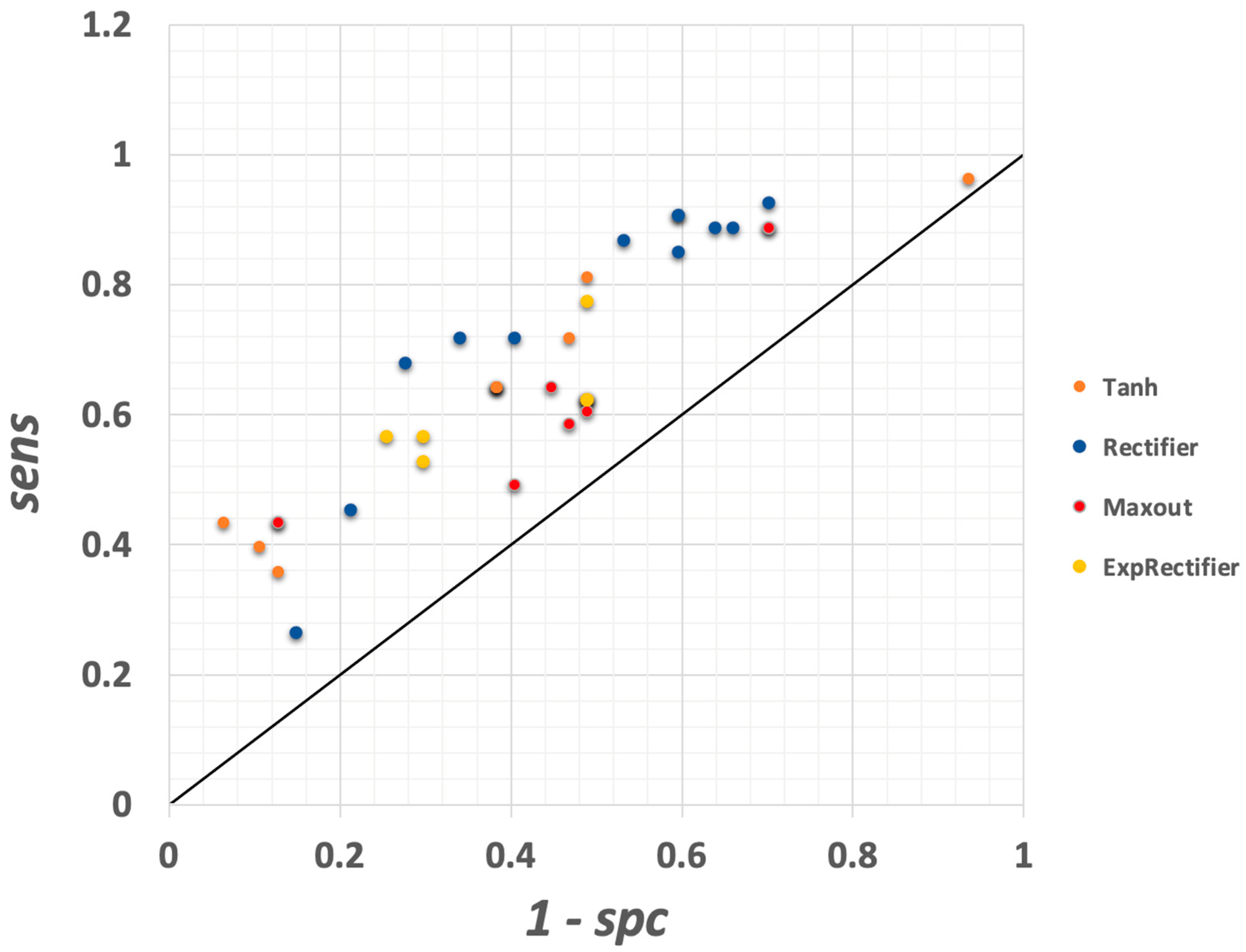
3.1.7. Deep Learning Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mastitis in Cattle—Reproductive System—Merck Veterinary Manual. Available online: https://www.merckvetmanual.com/reproductive-system/mastitis-in-large-animals/mastitis-in-cattle (accessed on 15 October 2022).
- Janzen, J.J. Economic Losses Resulting from Mastitis. A Review. J. Dairy Sci. 1970, 53, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Arnold, L.M.; Stowe, C.J.; Harmon, R.J.; Bewley, J.M. Estimating US Dairy Clinical Disease Costs with a Stochastic Simulation Model. J. Dairy Sci. 2017, 100, 1472–1486. [Google Scholar] [CrossRef] [PubMed]
- Rollin, E.; Dhuyvetter, K.C.; Overton, M.W. The Cost of Clinical Mastitis in the First 30 Days of Lactation: An Economic Modeling Tool. Prev. Vet. Med. 2015, 122, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.A.M.; Martins, V.C.; Cardoso, F.A.; Germano, J.; Rodrigues, M.; Duarte, C.; Bexiga, R.; Cardoso, S.; Freitas, P.P. Biosensors for On-Farm Diagnosis of Mastitis. Front. Bioeng. Biotechnol. 2019, 7, 186. [Google Scholar] [CrossRef] [PubMed]
- Peek, S.F.; Divers, T.J. Rebhun’s Diseases of Dairy Cattle, 3rd ed.; Saunders: St. Louis, MO, USA, 2018. [Google Scholar]
- Schukken, Y.H.; Wilson, D.J.; Welcome, F.; Garrison-Tikofsky, L.; Gonzalez, R.N. Monitoring Udder Health and Milk Quality Using Somatic Cell Counts. Vet. Res. 2003, 34, 579–596. [Google Scholar] [CrossRef]
- dos Reis, C.B.M.; Barreiro, J.R.; Moreno, J.F.G.; Porcionato, M.A.F.; Santos, M.V. Evaluation of Somatic Cell Count Thresholds to Detect Subclinical Mastitis in Gyr Cows. J. Dairy Sci. 2011, 94, 4406–4412. [Google Scholar] [CrossRef]
- Sargeant, J.M.; Leslie, K.E.; Shirley, J.E.; Pulkrabek, B.J.; Lim, G.H. Sensitivity and Specificity of Somatic Cell Count and California Mastitis Test for Identifying Intramammary Infection in Early Lactation. J. Dairy Sci. 2001, 84, 2018–2024. [Google Scholar] [CrossRef]
- Kandeel, S.A.; Morin, D.E.; Calloway, C.D.; Constable, P.D. Association of California Mastitis Test Scores with Intramammary Infection Status in Lactating Dairy Cows Admitted to a Veterinary Teaching Hospital. J. Vet. Intern. Med. 2018, 32, 497–505. [Google Scholar] [CrossRef]
- Antanaitis, R.; Juozaitienė, V.; Jonike, V.; Baumgartner, W.; Paulauskas, A. Milk Lactose as a Biomarker of Subclinical Mastitis in Dairy Cows. Animals 2021, 11, 1736. [Google Scholar] [CrossRef]
- Zhang, L.; Boeren, S.; van Hooijdonk, A.C.M.; Vervoort, J.M.; Hettinga, K.A. A Proteomic Perspective on the Changes in Milk Proteins Due to High Somatic Cell Count. J. Dairy Sci. 2015, 98, 5339–5351. [Google Scholar] [CrossRef]
- Smolenski, G.; Haines, S.; Kwan, F.Y.S.; Bond, J.; Farr, V.; Davis, S.R.; Stelwagen, K.; Wheeler, T.T. Characterisation of Host Defence Proteins in Milk Using a Proteomic Approach. J. Proteome Res. 2007, 6, 207–215. [Google Scholar] [CrossRef]
- Chen, X.-F.; Hou, X.; Xiao, M.; Zhang, L.; Cheng, J.-W.; Zhou, M.-L.; Huang, J.-J.; Zhang, J.-J.; Xu, Y.-C.; Hsueh, P.-R.; et al. Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis for the Identification of Pathogenic Microorganisms: A Review Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) Analysis for the Identification of Pathogenic Microorganisms: A Review. Microorganisms 2021, 9, 1536. [Google Scholar] [CrossRef]
- Li, D.; Yi, J.; Han, G.; Qiao, L. MALDI-TOF Mass Spectrometry in Clinical Analysis and Research. ACS Meas. Sci. Au 2022, 2022, 385–404. [Google Scholar] [CrossRef]
- Roscher, R.; Bohn, B.; Duarte, M.F.; Garcke, J. Explainable Machine Learning for Scientific Insights and Discoveries. IEEE Access 2020, 8, 42200–42216. [Google Scholar] [CrossRef]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive Analysis of Machine Learning Models for Prediction of Sub-Clinical Mastitis: Deep Learning and Gradient-Boosted Trees Outperform Other Models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef]
- Vervier, K.; Mahé, P.; Veyrieras, J.-B.; Vert, J.-P. Benchmark of Structured Machine Learning Methods for Microbial Identification from Mass-Spectrometry Data; Cornell University Press: Ithaca, NY, USA, 2015. [Google Scholar]
- Weis, C.V.; Jutzeler, C.R.; Borgwardt, K. Machine Learning for Microbial Identification and Antimicrobial Susceptibility Testing on MALDI-TOF Mass Spectra: A Systematic Review. Clin. Microbiol. Infect. 2020, 26, 1310–1317. [Google Scholar] [CrossRef]
- Esener, N.; Guerra, A.M.; Giebel, K.; Lea, D.; Green, M.J.; Bradley, A.J.; Dottorini, T. Mass Spectrometry and Machine Learning for the Accurate Diagnosis of Benzylpenicillin and Multidrug Resistance of Staphylococcus Aureus in Bovine Mastitis. PLoS Comput. Biol. 2021, 17, e1009108. [Google Scholar] [CrossRef]
- Tran, N.K.; Howard, T.; Walsh, R.; Pepper, J.; Loegering, J.; Phinney, B.; Salemi, M.R.; Rashidi, H.H. Novel Application of Automated Machine Learning with MALDI-TOF-MS for Rapid High-Throughput Screening of COVID-19: A Proof of Concept. Sci. Rep. 2021, 11, 8219. [Google Scholar] [CrossRef]
- Piras, C.; Hale, O.J.; Reynolds, C.K.; Jones, A.K.; Taylor, N.; Morris, M.; Cramer, R. LAP-MALDI MS Coupled with Machine Learning: An Ambient Mass Spectrometry Approach for High-Throughput Diagnostics. Chem. Sci. 2022, 13, 1746. [Google Scholar] [CrossRef]
- Stevenson, M. An Introduction to Veterinary Epidemiology; Massey University: Palmerston North, New Zealand, 2008. [Google Scholar]
- Thomas, F.C.; Mullen, W.; Tassi, R.; Ramírez-Torres, A.; Mudaliar, M.; McNeilly, T.N.; Zadoks, R.N.; Burchmore, R.; David Eckersall, P. Mastitomics, the Integrated Omics of Bovine Milk in an Experimental Model of Streptococcus Uberis Mastitis: 1. High Abundance Proteins, Acute Phase Proteins and Peptidomics. Mol. Biosyst. 2016, 12, 2735–2747. [Google Scholar] [CrossRef]
- Youden, W.J. Index for Rating Diagnostic Tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Schepers, A.J.; Lam, T.J.G.M.; Schukken, Y.H.; Wilmink, J.B.M.; Hanekamp, W.J.A. Estimation of Variance Components for Somatic Cell Counts to Determine Thresholds for Uninfected Quarters. J. Dairy Sci. 1997, 80, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Mudaliar, M.; Tassi, R.; Thomas, F.C.; McNeilly, T.N.; Weidt, S.K.; McLaughlin, M.; Wilson, D.; Burchmore, R.; Herzyk, P.; Eckersall, P.D.; et al. Mastitomics, the Integrated Omics of Bovine Milk in an Experimental Model of Streptococcus Uberis Mastitis: 2. Label-Free Relative Quantitative Proteomics. Mol. Biosyst. 2016, 12, 2748–2761. [Google Scholar] [CrossRef]
- Yu, W.; Zhao, S.-L. LC/MS/MS Analysis of Melamine in Liquid Milk and Milk Powder with Bond ElutPlexa PCX. Available online: https://www.agilent.com/chem (accessed on 19 September 2022).



| Model | sens | spc | Jmax |
|---|---|---|---|
| Decision tree | 0.85 | 0.66 | 0.51 |
| Random forest | 0.83 | 0.81 | 0.64 |
| Naïve Bayes | 0.87 | 0.38 | 0.25 |
| Gen. linear | 0.87 | 0.49 | 0.37 |
| Fast large-margin | 0.58 | 0.85 | 0.43 |
| Grad. boosted trees | 0.89 | 0.81 | 0.70 |
| Deep learning | 0.68 | 0.72 | 0.40 |
| Comparison case SCC * [10] | 0.59 | 0.72 | 0.31 |
| Comparison case CMT ** [12] | 0.27 | 0.85 | 0.12 |
| Comparison case SCC [26] | 0.74 | 0.90 | 0.64 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, J.; Everhart Nunn, S.L.; Sarkar, S.; Clayton, B. Diagnostic Screening of Bovine Mastitis Using MALDI-TOF MS Direct-Spotting of Milk and Machine Learning. Vet. Sci. 2023, 10, 101. https://doi.org/10.3390/vetsci10020101
Thompson J, Everhart Nunn SL, Sarkar S, Clayton B. Diagnostic Screening of Bovine Mastitis Using MALDI-TOF MS Direct-Spotting of Milk and Machine Learning. Veterinary Sciences. 2023; 10(2):101. https://doi.org/10.3390/vetsci10020101
Chicago/Turabian StyleThompson, Jonathan, Savana L. Everhart Nunn, Sumon Sarkar, and Beth Clayton. 2023. "Diagnostic Screening of Bovine Mastitis Using MALDI-TOF MS Direct-Spotting of Milk and Machine Learning" Veterinary Sciences 10, no. 2: 101. https://doi.org/10.3390/vetsci10020101
APA StyleThompson, J., Everhart Nunn, S. L., Sarkar, S., & Clayton, B. (2023). Diagnostic Screening of Bovine Mastitis Using MALDI-TOF MS Direct-Spotting of Milk and Machine Learning. Veterinary Sciences, 10(2), 101. https://doi.org/10.3390/vetsci10020101








