Monitoring the Milk Composition, Milk Microbiota, and Blood Metabolites of Jersey Cows throughout a Lactation Period
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Milk Composition and Blood Metabolites Analyses
2.3. Bacterial DNA Extraction
2.4. 16S rRNA Gene Amplicon Sequencing
2.5. Bioinformatics and Statistical Analyses
3. Results
3.1. Milk Yield, Milk Composition, and Blood Metabolites Concentration
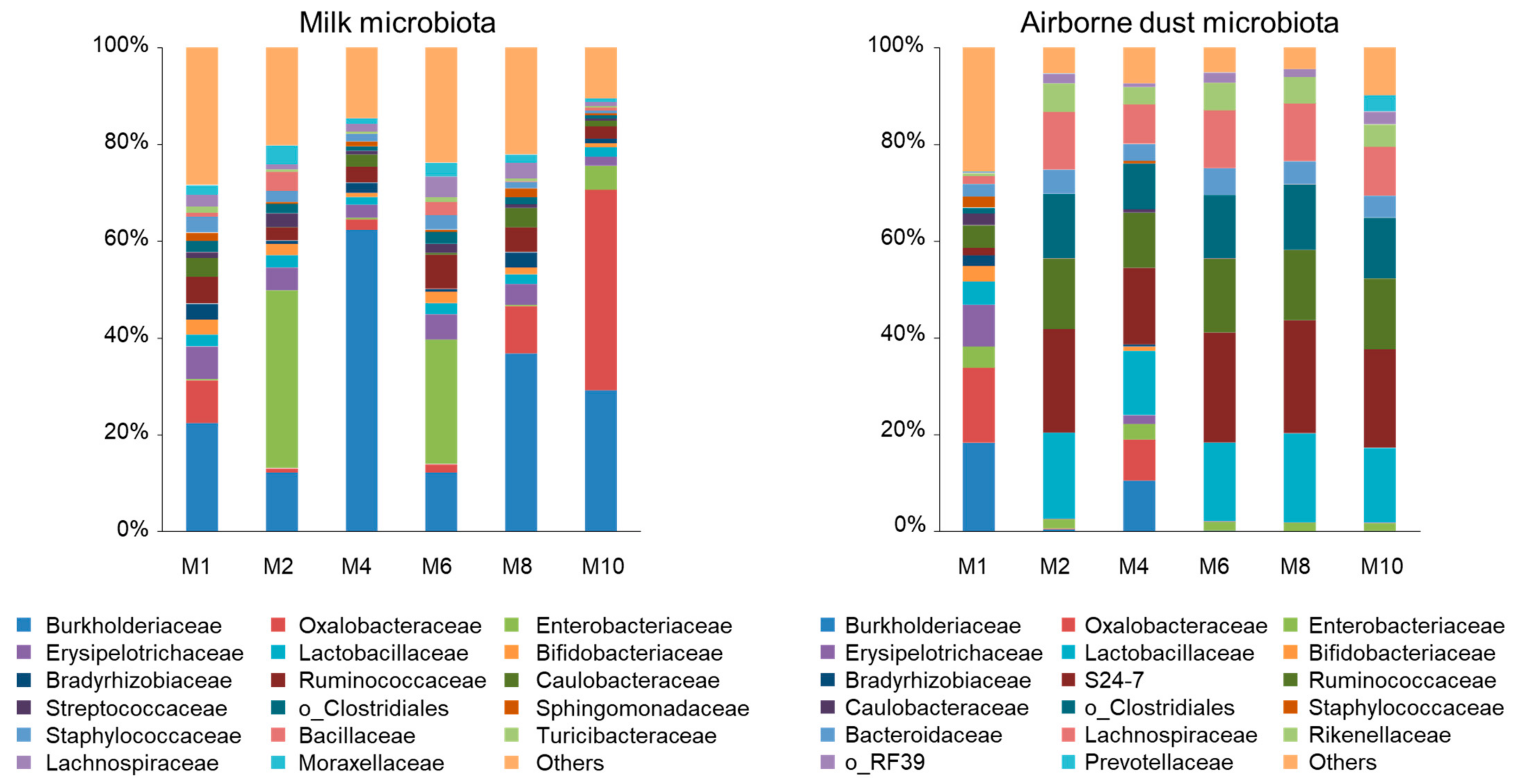
3.2. Milk Microbiota
3.3. Airborne Dust Microbiota
3.4. Relationship between Milk Composition, Blood Metabolite Concentrations, Milk Microbiota, and Airborne Dust Microbiota
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mackle, T.R.; Parr, C.R.; Stakelum, G.K.; Bryant, A.M.; MacMillan, K.L. Feed conversion efficiency, daily pasture intake, and milk production of primiparous Friesian and Jersey cows calved at two different live weights. N. Z. J. Agric. Res. 1996, 39, 357–370. [Google Scholar] [CrossRef]
- Prendiville, R.; Pierce, K.M.; Delaby, L.; Buckley, F. Animal performance and production efficiencies of Holstein-Friesian, Jersey and Jersey × Holstein-Friesian cows throughout lactation. Livest. Sci. 2011, 138, 25–33. [Google Scholar] [CrossRef]
- Washburn, S.P.; White, S.L.; Green, J.T.; Benson, G.A. Reproduction, mastitis, and body condition of seasonally calved Holstein and Jersey cows in confinement or pasture systems. J. Dairy Sci. 2002, 85, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Alstrup, L.; Nielsen, M.O.; Lund, P.; Sehested, J.; Larsen, M.K.; Weisbjerg, M.R. Milk yield, feed efficiency and metabolic profiles in Jersey and Holstein cows assigned to different fat supplementation strategies. Livest. Sci. 2015, 178, 165–176. [Google Scholar] [CrossRef]
- Huson, H.J.; Sonstegard, T.S.; Godfrey, J.; Hambrook, D.; Wolfe, C.; Wiggans, G.; Blackburn, H.; VanTassell, C.P.A. Genetic investigation of island Jersey cattle, the foundation of the Jersey breed: Comparing population structure and selection to Guernsey, Holstein, and United States Jersey cattle. Front. Genet. 2020, 11, 366. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Song, M.; Park, W.; Oh, S.; Ham, J.S.; Jeong, S.G.; Kim, Y. A comparison of quality characteristics in dairy products made from Jersey and Holstein milk. Food Sci. Anim. Res. 2019, 39, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Lim, D.H.; Mayakrishnan, V.; Lee, H.J.; Ki, K.S.; Kim, T.I.; Kim, Y. A comparative study on milk composition of Jersey and Holstein dairy cows during the early lactation. J. Anim. Sci. Technol. 2020, 62, 565–576. [Google Scholar] [CrossRef]
- Tardon, D.C.; Hoffmann, C.; Santos, F.C.R.; Decaris, N.; Pinheiro, F.A.; Queiroz, L.L.; Hurley, D.J.; Gomes, V. Relationshhips among indicators of metabolism, mammnary health and the microbiomes of periparturient Holstein cows. Animals 2022, 12, 3. [Google Scholar] [CrossRef]
- Janovick Guretzky, N.A.; Carlson, D.B.; Garrett, J.E.; Drackley, J.K. Lipid metabolite profiles and milk production for Holstein and Jersey cows fed rumen-protected choline during the periparturient period. J. Dairy Sci. 2006, 89, 188–200. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.L.; Cassell, B.G.; McGilliard, M.L.; Hanigan, M.D.; Gwazdauskas, F.C. Hormones, metabolites, and reproduction in Holsteins, Jerseys, and their crosses. J. Dairy Sci. 2012, 95, 698–707. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Nguyen, Q.D.; Tran, T.T.M.; Tang, M.T.; Tsuruta, T.; Nishino, N. Rumen fluid, feces, milk, water, feed, airborne dust, and bedding microbiota in dairy farms managed by automatic milking systems. Anim. Sci. J. 2019, 90, 445–452. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Wu, H.; Nishino, N. An investigation of seasonal variations in the microbiota of milk, feces, bedding, and airborne dust. Asian-Australas. J. Anim. Sci. 2020, 33, 1858–1865. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, Q.D.; Tsuruta, T.; Nishino, N. Examination of milk microbiota, fecal microbiota, and blood metabolites of Jersey cows in cool and hot seasons. Anim. Sci. J. 2020, 91, e13441. [Google Scholar] [CrossRef]
- Bainbridge, M.L.; Cersosimo, L.M.; Wright, A.D.G.; Kraft, J. Content and composition of branched-chain fatty acids in bovine milk are affected by lactation stage and breed of dairy cow. PLoS ONE 2016, 11, e0150386. [Google Scholar] [CrossRef] [Green Version]
- Bytyqi, H.; Zaugg, U.; Sherififi, K.; Hamidi, A.; Gjonbalaj, M.; Muji, S.; Mehmeti, H. Influence of management and physiological factors on somatic cell count in raw cow milk in Kosova. Vet. Arh. 2010, 80, 173–183. [Google Scholar]
- Hammond, C. Use of BUN and MUN as guides for protein and energy in cattle. Rev. Corpoica 1998, 2, 44–48. [Google Scholar] [CrossRef] [Green Version]
- Rajala-Schultz, P.J.; Saville, W.J.A. Sources of variation in milk urea nitrogen in Ohio dairy herds. J. Dairy Sci. 2003, 86, 1653–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafsson, A.H.; Palmquist, D.L. Diurnal variation of rumen ammonia, serum urea, and milk urea in dairy cows at high and low yields. J. Dairy Sci. 1993, 76, 475–484. [Google Scholar] [CrossRef]
- Grum, D.E.; Drackley, J.K.; Clark, J.H. Fatty acid metabolism in liver of dairy cows fed supplemental fat and nicotinic acid during an entire lactation. J. Dairy Sci. 2002, 85, 3026–3034. [Google Scholar] [CrossRef] [Green Version]
- Kaneene, J.B.; Miller, R.A.; Herdt, T.H.; Gardiner, J.C. The association of serum nonesterified fatty acids and cholesterol, management and feeding practices with peripartum disease in dairy cows. Prevent. Vet. Med. 1997, 31, 59–72. [Google Scholar] [CrossRef]
- Ospina, P.A.; Nydam, D.V.; Stokol, T.; Overton, T.R. Evaluation of nonesterified fatty acids and β-hydroxybutyrate in transition dairy cattle in the northeastern United States: Critical thresholds for prediction of clinical diseases. J. Dairy Sci. 2010, 93, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Stojević, Z.; Piršljin, J.; Milinković-Tur, S.; Zdelar-Tuk, M.; Ljubić, B.B. Activities of AST, ALT and GGT in clinically healthy dairy cows during lactation and in the dry period. Vet. Arh. 2005, 75, 67–73. [Google Scholar]
- Chan, J.P.W.; Chu, C.C.; Fung, H.P.; Chuang, S.T.; Lin, Y.C.; Chu, R.M.; Lee, S.L. Serum haptoglobin concentration in cattle. J. Vet. Med. Sci. 2004, 66, 43–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, L.; Wang, H.; Ding, Y.; Li, J.; Li, J. Changes in the blood routine, biochemical indexes and the pro-inflammatory cytokine expressions of peripheral leukocytes in postpartum dairy cows with metritis. BMC Vet. Res. 2019, 15, 157. [Google Scholar] [CrossRef] [Green Version]
- Oikonomou, G.; Addis, M.F.; Chassard, C.; Nader-Macias, M.E.F.; Grant, I.; Delbès, C.; Bogni, C.I.; Le Loir, Y.; Even, S. Milk Microbiota: What are we exactly talking about? Front. Microbiol. 2020, 11, 60. [Google Scholar] [CrossRef] [Green Version]
- Cremonesi, P.; Ceccarani, C.; Curone, G.; Severgnini, M.; Pollera, C.; Bronzo, V.; Riva, F.; Addis, M.F.; Filipe, J.; Amadori, M.; et al. Milk microbiome diversity and bacterial group prevalence in a comparison between healthy Holstein Friesian and Rendena cows. PLoS ONE 2018, 13, e0205054. [Google Scholar] [CrossRef] [Green Version]
- Metzger, S.A.; Hernandez, L.L.; Skarlupka, J.H.; Walker, T.M.; Suen, G.; Ruegg, P.L. A Cohort study of the milk microbiota of healthy and inflamed bovine mammary glands from dryoff through 150 days in milk. Front. Vet. Sci. 2018, 14, 247. [Google Scholar] [CrossRef] [Green Version]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The core and seasonal microbiota of raw bovine milk in tanker trucks and the impact of transfer to a milk processing facility. MBio 2016, 7, e00836-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahlberg, J.; Sun, L.; Waller, K.P.; Ostensson, K.; Mcguire, M.; Agenas, S.; Dicksved, J. Microbiota data from low bio-mass milk samples is markedly affected by laboratory and reagent contamination. PLoS ONE 2019, 14, e0218257. [Google Scholar] [CrossRef] [Green Version]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| TMR for High Milk Yield | TMR for Low Milk Yield | |
|---|---|---|
| Diet ingredients | ||
| Grass silage | 301 | 359 |
| Grass hay | 26.2 | 28.6 |
| Legume hay | 13.1 | 14.3 |
| Wet brewers’ grains | 22.9 | 25.0 |
| Soybean meal | 23.1 | 25.2 |
| Beet pulp | 52.6 | 57.4 |
| Compound feed | 561 | 490 |
| Chemical composition | ||
| Crude protein | 166 | 166 |
| Rumen degradable protein | 97.9 | 105 |
| Ether extract | 36.2 | 35.3 |
| aNDFom | 298 | 327 |
| Starch | 306 | 263 |
| NEL (Mcal/kgDM) | 1.67 | 1.64 |
| Lactation Period and Season at Sampling | M1 September | M2 October | M4 December | M6 February | M8 April | M10 June | SE |
|---|---|---|---|---|---|---|---|
| Milk yield (kg) | 27.0 a | 27.3 a | 25.9 ab | 20.4 bc | 20.3 bc | 14.8 c | 0.67 |
| Milk composition | |||||||
| Fat (%) | 4.49 c | 4.69 bc | 5.31 ab | 5.31 ab | 5.49 ab | 5.85 a | 0.08 |
| Protein (%) | 3.39 b | 3.47 b | 4.23 a | 4.54 a | 4.54 a | 4.46 a | 0.04 |
| Solids-not-fat (%) | 9.01 b | 9.20 b | 9.83 a | 10.0 a | 9.97 a | 9.83 a | 0.04 |
| SCC (× 103 cells/mL) | 103 | 70.1 | 87.6 | 122 | 202 | 169 | 5.94 |
| MUN (mg/dL) | 8.96 c | 3.86 d | 8.71 c | 12.3 a | 10.3 bc | 11.6 ab | 0.20 |
| Blood metabolites | |||||||
| Albumin (g/dL) | 3.85 c | 4.03 bc | 4.23 bc | 7.04 a | 4.33 b | 4.35 b | 0.04 |
| BUN (mg/dL) | 6.13 c | 5.57 c | 9.47 b | 14.2 a | 9.02 b | 10.6 b | 0.27 |
| Total cholesterol (mg/dL) | 160 b | 229 a | 214 ab | 209 ab | 196 ab | 180 ab | 5.65 |
| NEFA (mEq/L) | 0.39 a | 0.29 b | 0.11 c | 0.11 c | 0.09 c | 0.14 c | 13.59 |
| Calcium (mg/dL) | 8.28 bc | 9.25 ab | 8.15 c | 10.0 a | 8.94 abc | 8.61 bc | 0.10 |
| Phosphorus (mg/dl) | 3.43 b | 4.37 ab | 5.27 a | 4.52 ab | 5.45 a | 5.77 a | 0.15 |
| AST (U/L) | 112 a | 76.0 c | 116 a | 76.3 bc | 87.6 bc | 98.1 ab | 2.02 |
| ALT (U/L) | 16.5 c | 16.3 c | 33.6 a | 28.0 ab | 22.7 bc | 19.6 c | 0.63 |
| Haptoglobin (μg/L) | 62.6 a | 51.9 ab | 47.5 ab | 13.3 b | 15.3 b | 10.6 b | 3.68 |
| Lactation Period and Season at Sampling | M1 September | M2 October | M4 December | M6 February | M8 April | M10 June | SE |
|---|---|---|---|---|---|---|---|
| Diversity indices | |||||||
| Chao1 | 405 a | 313 ab | 270 ab | 388 a | 374 a | 217 b | 13.1 |
| Shannon | 6.01 a | 4.87 a | 3.56 b | 5.75 a | 5.17 a | 3.37 b | 0.12 |
| Phyla/Families | |||||||
| Proteobacteria | 48.1 c | 59.1 bc | 74.3 ab | 47.2 c | 61.4 bc | 80.5 a | 1.44 |
| Burkholderiaceae | 22.4 cd | 12.2 d | 62.3 a | 12.2 d | 36.7 b | 29.2 bc | 0.97 |
| Oxalobacteraceae | 8.75 b | 0.87 b | 2.21 b | 1.71 b | 9.81 b | 41.4 a | 1.00 |
| Bradyrhizobiaceae | 3.31 a | 0.64 c | 2.09 ab | 0.53 c | 3.20 a | 0.96 bc | 0.10 |
| Enterobacteriaceae | 0.32 c | 36.8 a | 0.29 c | 25.8 b | 0.26 c | 4.88 c | 0.99 |
| Moraxellaceae | 1.95 | 3.84 | 1.14 | 2.80 | 1.67 | 0.76 | 0.26 |
| Caulobacteraceae | 3.84 ab | 0.16 d | 2.54 bc | 0.41 d | 4.05 a | 1.13 cd | 0.11 |
| Firmicutes | 33.6 a | 26.6 ab | 15.8 bc | 36.4 a | 23.4 abc | 12.6 c | 1.10 |
| Erysipelotrichaceae | 6.70 a | 4.66 ab | 2.72 ab | 5.14 ab | 4.26 ab | 1.90 b | 0.38 |
| Ruminococcaceae | 5.51 | 2.65 | 3.27 | 7.09 | 5.05 | 2.55 | 0.42 |
| Lactobacillaceae | 2.40 | 2.59 | 1.53 | 2.30 | 2.00 | 1.96 | 0.18 |
| Streptococcaceae | 1.26 b | 2.81 a | 0.75 b | 1.85 ab | 0.80 b | 0.51 b | 0.12 |
| o_Clostridiales | 2.35 a | 2.04 ab | 0.96 b | 2.54 a | 1.45 ab | 0.67 b | 0.12 |
| Bacillaceae | 0.81 b | 3.94 a | 0.06 b | 2.70 a | 0.13 b | 0.65 b | 0.13 |
| Lachnospiraceae | 2.45 | 1.04 | 1.61 | 4.25 | 3.26 | 0.80 | 0.34 |
| Staphylococcaceae | 3.27 | 2.32 | 1.49 | 3.10 | 1.40 | 0.52 | 0.34 |
| Actinobacteria | 5.16 a | 3.77 ab | 1.68 b | 3.25 ab | 2.64 ab | 1.38 b | 0.23 |
| Bifidobacteriaceae | 3.14 | 2.35 | 0.88 | 2.38 | 1.39 | 0.75 | 0.22 |
| Bacteroidetes | 9.19 a | 7.80 ab | 4.45 ab | 8.39 a | 5.91 ab | 3.09 b | 0.41 |
| Bacteroidaceae | 1.81 ab | 0.92 b | 0.96 b | 2.29 a | 1.26 ab | 0.66 b | 0.13 |
| Others | 0.59 | 0.83 | 1.50 | 2.46 | 3.56 | 1.30 | 0.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gathinji, P.K.; Yousofi, Z.; Akada, K.; Wali, A.; Nishino, N. Monitoring the Milk Composition, Milk Microbiota, and Blood Metabolites of Jersey Cows throughout a Lactation Period. Vet. Sci. 2023, 10, 226. https://doi.org/10.3390/vetsci10030226
Gathinji PK, Yousofi Z, Akada K, Wali A, Nishino N. Monitoring the Milk Composition, Milk Microbiota, and Blood Metabolites of Jersey Cows throughout a Lactation Period. Veterinary Sciences. 2023; 10(3):226. https://doi.org/10.3390/vetsci10030226
Chicago/Turabian StyleGathinji, Peter Kiiru, Zabiallah Yousofi, Karin Akada, Ajmal Wali, and Naoki Nishino. 2023. "Monitoring the Milk Composition, Milk Microbiota, and Blood Metabolites of Jersey Cows throughout a Lactation Period" Veterinary Sciences 10, no. 3: 226. https://doi.org/10.3390/vetsci10030226
APA StyleGathinji, P. K., Yousofi, Z., Akada, K., Wali, A., & Nishino, N. (2023). Monitoring the Milk Composition, Milk Microbiota, and Blood Metabolites of Jersey Cows throughout a Lactation Period. Veterinary Sciences, 10(3), 226. https://doi.org/10.3390/vetsci10030226







